Abstract
Root colonization by Agrobacterium tumefaciens was measured by using tomato and Arabidopsis thaliana roots dipped in a bacterial suspension and planted in soil. Wild-type bacteria showed extensive growth on tomato roots; the number of bacteria increased from 103 bacteria/cm of root length at the time of inoculation to more than 107 bacteria/cm after 10 days. The numbers of cellulose-minus and nonattaching attB, attD, and attR mutant bacteria were less than 1/10,000th the number of wild-type bacteria recovered from tomato roots. On roots of A. thaliana ecotype Landsberg erecta, the numbers of wild-type bacteria increased from about 30 to 8,000 bacteria/cm of root length after 8 days. The numbers of cellulose-minus and nonattaching mutant bacteria were 1/100th to 1/10th the number of wild-type bacteria recovered after 8 days. The attachment of A. tumefaciens to cut A. thaliana roots incubated in 0.4% sucrose and observed with a light microscope was also reduced with cel and att mutants. These results suggest that cellulose synthesis and attachment genes play a role in the ability of the bacteria to colonize roots, as well as in bacterial pathogenesis.
Infections of wound sites on dicotyledonous plants by the soil bacterium Agrobacterium tumefaciens result in the formation of crown gall tumors. An early step in tumor formation is the attachment of the bacteria to the plant cell surface. This attachment is required for bacterial virulence. All nonattaching mutants currently described are reduced in virulence (6, 17, 30).
In previous studies of the surface interactions between bacteria and plant cells leading to tumor formation, we identified two regions of the bacterial chromosome containing genes involved in tight binding of the bacteria to host cells (17, 20). The bacterium binds to host cells in a two-step process (19). The first step results in loose binding to the plant cell surface. At this stage the bacteria can be removed by shear forces, such as water washing or vortexing of tissue culture cells. Mutants in this step are avirulent. A series of genes required for this step have been located on the bacterial chromosome and are referred to as att genes (17). The genes attA1 to attH encode proteins with homology to ABC transporter systems in several bacteria, including the pot genes of Escherichia coli (21). These genes appear to be required for signaling between the bacteria and the plant host (18). Other att genes (including attR) appear to be required for the synthesis of surface molecules which may play a role in the actual attachment of the bacteria to the plant cell surface (25).
The second step in bacterial attachment is the synthesis of cellulose fibrils by the bacteria. This step results in tight binding of the bacteria; they can no longer be removed from the plant cells by shear forces (16). The genes required for bacterial cellulose synthesis (cel genes) are located on the bacterial chromosome near, but not contiguous with, the att genes (26). Cellulose-minus bacterial mutants show reduced virulence (between 10 and 1,000 times more bacteria are required to induce a tumor [22]).
The study described here was undertaken to explore the possible role of these genes in the interaction of the bacteria with intact roots.
MATERIALS AND METHODS
Bacterial strains and growth.
Wild-type virulent A. tumefaciens C58 and mutants derived from it were grown in Luria broth at 25°C. The cellulose-minus mutants C58::1 and C58::A60 have been described previously (20), as have the attachment-minus mutants C58::N004, C58::B123, and C58::A205 (21). The locations of the transposon insertions in these mutants are shown in Fig. 1. Spontaneous rifampin-resistant mutants of each of these strains were selected and used for root colonization experiments. The growth characteristics of the parent and mutant strains in liquid media (Luria broth or minimal medium) were not significantly different.
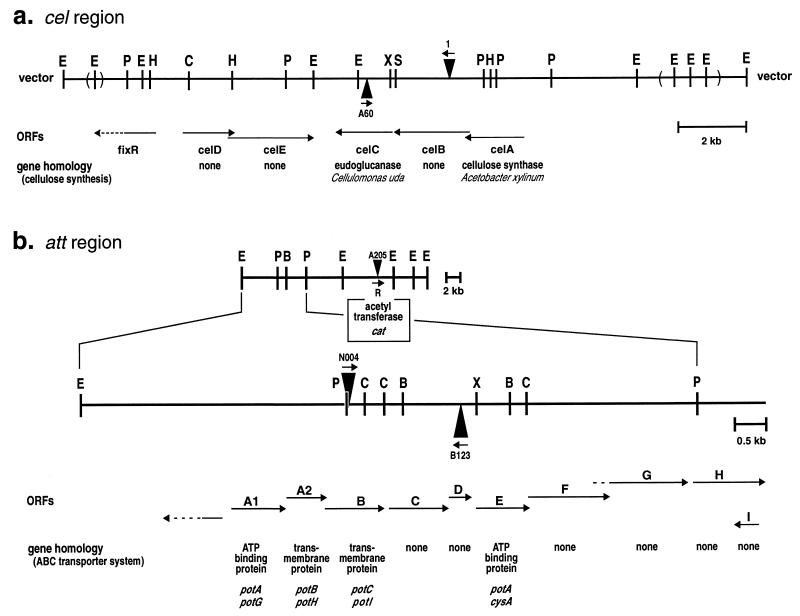
FIG. 1.
Positions of transposon insertions in mutants used in this study. The cel and att regions of the bacterial chromosome are shown. Both of these regions have been sequenced (20, 21). An enlargement of the left end of the att region is shown below the diagram of the larger clone. Triangles indicate the sites of transposon insertion in mutants used in this study. The small arrows with the triangles indicate the orientation of the promoterless β-galactosidase gene contained in Tn3HoHo1. The locations and directions of open reading frames (ORFs) are indicated by the larger arrows. The genes which have the highest homology to the open reading frames are indicated below the arrows (20, 21). Restriction sites are indicated as follows: E, EcoRI; C, ClaI; P, PstI; B, BamHI; X, XbaI; H, HindIII; S, SacI.
Growth and inoculation of plants.
Seeds of tomato (Lycopersicon esculentum cv. Marglobe) were surface sterilized in 0.16% sodium hypochlorite and germinated on 1% water agar in petri dishes. Seeds of Arabidopsis thaliana ecotype Landsberg erecta were surface sterilized in 1.6% sodium hypochlorite, stored at 4°C for 3 days, and planted in petri dishes containing MS salts, Gamborg’s B5 vitamin mixture, and 2% sucrose in 0.8% Phytagar (all obtained from GIBCO Laboratories, Grand Island, N.Y.).
When the tomato roots were 1 to 2 cm long and the A. thaliana roots were 0.25 to 0.5 cm long, the roots were dipped for 1 min into a suspension containing 105 bacteria per ml of 0.1 M sodium phosphate buffer (pH 7.0)–0.1% peptone (washing buffer [14]). The intact plants were then placed in soil in Conetainers (Stuewe and Sons, Inc., Corvallis, Oreg.) (tomato) or in 1.5-ml Eppendorf tubes which had a hole pierced in the tip (A. thaliana). The soil was a Gilead loamy sand which had been pasteurized by heating it in a microwave oven in sealed bags and stored for more than 4 weeks before use according to the procedure of Ferriss (7, 13). The bottom of each container was sealed with Parafilm, and the plant and the top of the container were covered with Parafilm. The plants were grown without additional water at 22 to 24°C with 14 h of light per day for up to 10 days, at which time they were too large to remain covered in their containers. After 10 days the tomato roots were between 8 and 13 cm long. The A. thaliana roots were between 2 and 3 cm long when they were harvested after 8 days.
Sampling of roots.
At different times after inoculation the plants and soil were removed from the containers, the soil was carefully separated from the roots, and the roots were gently shaken. Each whole plant was placed in 5 ml (tomato) or 2 ml (A. thaliana) of washing buffer in a vial. The vial was sealed and inverted 10 times. The washing buffer was removed, and the procedure was repeated. The number of viable bacteria per milliliter of washing buffer was determined by viable cell counts using Luria agar containing 50 μg of rifampin per ml. The viable cell counts in the first two washes were combined to give the number of bacteria loosely bound to the roots. Each plant was then placed in 5 or 2 ml of washing buffer and sonicated with a Branson ultrasonic cleaner (model B220) for 60 s (tomato) or 30 s (A. thaliana). The viable cell counts for the bacteria released into the washing buffer were determined, and the resulting data are reported below as the number of tightly bound bacteria. The plants were then removed from the buffer, placed on Luria agar plates containing rifampin, and covered with soft (0.7%) agar. The root lengths were measured. The plates were incubated until colonies were apparent. The number of colonies per centimeter of root length was determined with a dissecting microscope, and the resulting data are reported below as the number of irreversibly bound bacteria. The results are expressed as CFU per centimeter of root length rather than CFU per gram of root weight since the drying necessary to obtain reproducible root weights interfered with the recovery of viable bacteria which were bound to the roots. The number of CFU per centimeter of root length was calculated and was transformed to a log10 CFU/centimeter value (14) before the means and standard deviations were calculated. Experiments were repeated four times with a minimum of four plants per time interval.
Microscopic observation of bacterial attachment to cut A. thaliana roots.
Microscopic observation of bacterial attachment to cut roots of A. thaliana was carried out as described previously (23). Briefly, cut roots floating in 2 ml of 0.4% sucrose–1 mM CaCl2 were inoculated with approximately 108 bacteria. The roots were incubated at room temperature, and bacterial attachment was scored after 24, 48, and 72 h with a microscope (Zeiss photoscope 2) equipped with Nomarski optics.
RESULTS
Bacterial binding to cut A. thaliana roots.
Wild-type strain C58 bacteria bound to cut roots of A. thaliana within 24 h. Large numbers of bacteria were visibly bound to the epidermis, root hairs, and cut end of each root segment (Fig. 2). These surfaces appeared to be uniformly covered with bacteria; no preferential sites of attachment were observed. Fewer bacteria were seen on the root cap, although numerous bacteria were observed attached to the border cells behind the root cap. Bacterial aggregates were observed on the root hairs and epidermis (data not shown). Very few cellulose-minus mutant bacteria were observed at any location on the surfaces of roots after 24 h of incubation. No bacterial aggregates were seen. Similarly, very few attB, attD, or attR mutant bacteria were seen at any location on the root surface. These results suggest that the cel, attB, attD, and attR genes may be involved in bacterial attachment to intact surfaces of roots, as well as to cut plant surfaces and to tissue culture cells.
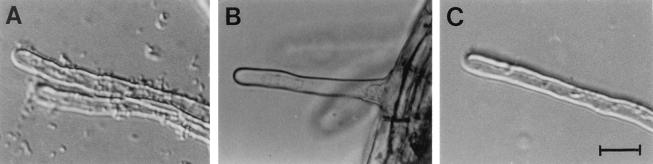
FIG. 2.
Attachment of bacteria to A. thaliana root hairs. Wild-type and mutant A. tumefaciens strains were incubated with roots of A. thaliana in 0.4% sucrose for 2 days. (A) Wild-type strain C58. Note the numerous bacteria attached to the root hairs. (B) Cellulose-minus strain C58::1. (C) Attachment-minus mutant C58::A205. The attachment of the cel and att mutants was much reduced compared with the attachment of the wild type. Similar results were observed for bacteria attached to the root epidermis. Bar = 5 μm.
Bacterial colonization of tomato roots.
Wild-type strain C58 bacteria were able to colonize the roots of tomato plants grown in soil. After an initial lag during the first 2 days following inoculation, there was an increase in the number of bacteria recovered in all three categories (loosely bound, tightly bound, and irreversibly bound). After 9 days there had been a 10,000-fold increase in the number of loosely and tightly bound bacteria per centimeter of root length and a 1,000-fold increase in the number of irreversibly bound bacteria (Table 1). After roots were placed in petri dishes and covered with soft agar, bacterial colonies were uniformly distributed throughout the length of the root, except that few colonies were observed at the root tip.
TABLE 1.
Numbers of wild-type strain C58 bacteria associated with tomato roots after incubation in soil for up to 10 days
| Day | No. of bacteria (mean log10 ± SD)a
|
||
|---|---|---|---|
| Loosely bound | Tightly bound | Irreversibly bound | |
| 0 | 2.7 ± 0.7 | 2.6 ± 0.7 | 0.7 ± 0.6 |
| 1 | 2.3 ± 0.5 | 2.4 ± 0.6 | 0.2 ± 0.3 |
| 2 | 2.7 ± 0.5 | 2.7 ± 0.7 | 0.8 ± 0.5 |
| 3 | 3.7 ± 0.8 | 3.6 ± 0.8 | 1.5 ± 0.7 |
| 4 | 4.4 ± 0.4 | 4.3 ± 0.3 | 1.5 ± 0.6 |
| 5 | 4.8 ± 0.7 | 4.9 ± 0.5 | 1.4 ± 0.3 |
| 6 | 5.0 ± 0.5 | 5.0 ± 0.6 | 1.6 ± 0.3 |
| 7 | 4.8 ± 0.7 | 5.3 ± 0.8 | 3.0 ± 0.3 |
| 8 | 6.9 ± 0.9 | 6.6 ± 0.6 | 3.1 ± 0.3 |
| 9 | 7.7 ± 0.7 | 7.7 ± 0.7 | 3.7 ± 0.2 |
| 10 | 6.7 ± 0.5 | 6.9 ± 0.8 | 3.4 ± 0.3 |
The method used to determine the numbers of loosely bound, tightly bound, and irreversibly bound bacteria is described in the text.
When cel and att mutant bacteria were inoculated onto roots, they failed to show much growth compared to the wild-type parent (Fig. 3). The numbers of loosely bound cellulose-minus mutant cells, as well as the numbers of tightly and irreversibly bound cellulose-minus mutant cells were reduced compared with the wild type (Table 2, C58::1 data; similar data were obtained for C58::A60). All three nonattaching mutants examined showed reduced attachment to tomato roots (Fig. 3B). Again, decreases were observed in all three categories of attachment, loosely bound, tightly bound, and irreversibly bound (data not shown). Thus, wild-type A. tumefaciens is capable of colonizing tomato roots, and colonization was reduced in cel and att mutants.
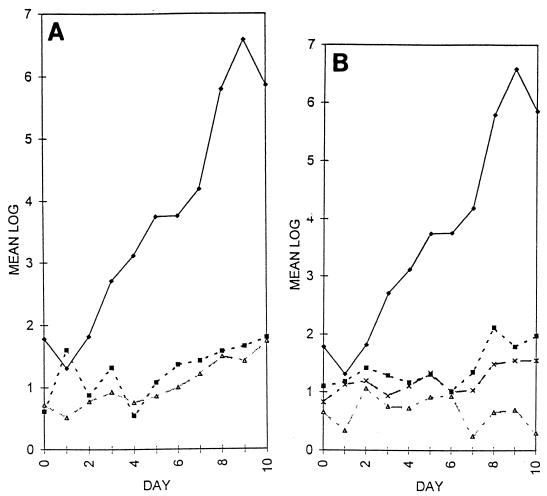
FIG. 3.
Numbers of bacteria recovered from tomato roots. (A) Log10 total number of bacteria per centimeter of root length recovered from tomato roots inoculated with wild-type strain C58 (⧫) and cellulose-minus mutants C58::1 (▪) and C58::A60 (▵). (B) Log10 total number of bacteria per centimeter of root length recovered from tomato roots inoculated with wild-type strain C58 (⧫) and attachment-minus mutants C58::A205 (▪), C58::N004 (▵), and C58::B123 (×). Note that cellulose-minus and attachment-minus bacteria had reduced abilities to colonize roots. These reductions were due to reductions in loosely bound, tightly bound, and irreversibly bound bacteria. The numbers shown are the means from a minimum of four separate experiments. The standard deviation of all points was less than 0.9.
TABLE 2.
Numbers of cellulose-minus C58::1 bacteria associated with tomato roots after incubation in soil for up to 10 days
| Day | No. of bacteria (mean log10 ± SD)a
|
||
|---|---|---|---|
| Loosely bound | Tightly bound | Irreversibly bound | |
| 0 | 2.1 ± 0.3 | 2.0 ± 0.4 | 0.2 ± 0.2 |
| 1 | 2.4 ± 0.8 | 2.1 ± 1.1 | 0.2 ± 0.2 |
| 2 | 2.0 ± 0.4 | 2.1 ± 0.4 | 0.4 ± 0.4 |
| 3 | 2.3 ± 0.5 | 2.3 ± 0.7 | 0.5 ± 0.5 |
| 4 | 2.4 ± 0.4 | 2.0 ± 1.0 | 1.1 ± 0.9 |
| 5 | 2.2 ± 0.5 | 2.0 ± 0.4 | 0.5 ± 0.5 |
| 6 | 2.2 ± 0.5 | 2.4 ± 0.6 | 0.7 ± 0.5 |
| 7 | 2.5 ± 0.6 | 2.5 ± 0.6 | 0.7 ± 0.5 |
| 8 | 2.4 ± 0.6 | 2.5 ± 0.6 | 0.8 ± 0.6 |
| 9 | 2.6 ± 0.6 | 2.6 ± 0.6 | 1.3 ± 0.2 |
| 10 | 3.2 ± 0.1 | 2.7 ± 0.3 | 1.1 ± 0.5 |
The method used to determine the numbers of loosely bound, tightly bound, and irreversibly bound bacteria is described in the text.
Bacterial colonization of A. thaliana roots.
The colonization of A. thaliana roots by wild-type bacteria, expressed as the number of bacteria per centimeter of root length, was less than the colonization of tomato roots. This may reflect the smaller diameter of the A. thaliana roots. The numbers of bacteria per centimeter of root length increased about 100-fold for loosely bound, tightly bound, and irreversibly bound bacteria (Fig. 4). In contrast to the data obtained for tomato roots, all of the mutants except C58::N004 showed some initial growth on A. thaliana roots. However, after 8 days the growth of the mutants lagged behind the growth of the wild-type strain. Colonization of cellulose-minus and attachment-minus mutants was reduced for all three binding categories (loosely, tightly, and irreversibly bound) (data not shown). Thus, colonization of A. thaliana roots by A. tumefaciens was reduced in att and cel mutants compared with the wild type.
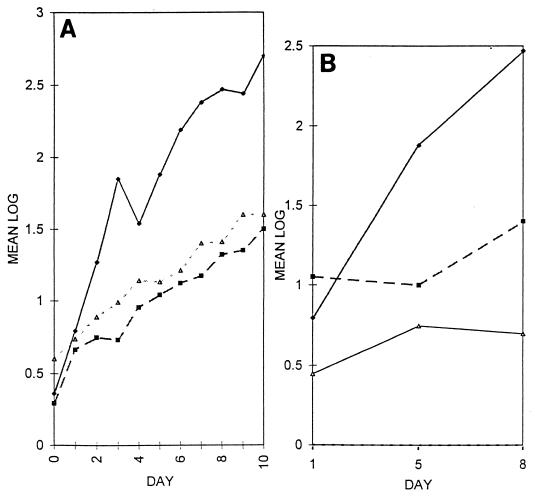
FIG. 4.
Numbers of bacteria recovered from A. thaliana roots. (A) Log10 total number of bacteria per centimeter of root length recovered from A. thaliana roots inoculated with wild-type strain C58 (⧫) and cellulose-minus mutants C58::1 (▪) and C58::A60 (▵). (B) Log10 total number of bacteria per centimeter of root length recovered from A. thaliana roots inoculated with wild-type strain C58 (⧫) and attachment-minus mutants C58::A205 (▪) and C58::N004 (▵). Note that cellulose-minus and attachment-minus bacteria had reduced abilities to colonize the roots after the first day. The reductions in the total numbers of bacteria were due to reductions in loosely bound, tightly bound, and irreversibly bound bacteria. The numbers shown are the means from a minimum of four separate experiments. The standard deviation of all points was less than 0.5.
DISCUSSION
Wild-type A. tumefaciens C58 colonized tomato roots in numbers comparable to the numbers observed for colonization of tomato roots by the biocontrol strains Agrobacterium rhizogenes K84 and J73 (15), for colonization of pea roots by A. tumefaciens A723 (8), and for colonization of many root systems by pseudomonads (3, 4, 11, 29). All regions of the root except the root tip were found to be colonized. Under the microscope bacteria were observed adhering to all regions of the root, including the border cells of the root cap. Adherence of agrobacteria to root cap cells has been described previously by Hawes and Pueppke (9). In the present colonization studies root cap cells were removed from the root by the washing steps, and the bacteria adhering to them were included in the data for the loosely bound bacteria.
Both tomato and A. thaliana are susceptible to crown gall tumor formation by A. tumefaciens. However, the bacterial colonization of A. thaliana roots was less than the bacterial colonization of tomato roots when data were expressed as number of bacteria per centimeter of root length. A. thaliana roots are much smaller than tomato roots and weigh between 40 and 100 times less per centimeter of root length than tomato roots. If this weight ratio is used to correct the numbers of bacteria found on A. thaliana roots, then the levels of bacterial colonization were roughly comparable for the two plants.
Mutations in genes involved in the synthesis of surface polysaccharides (cel and attR) and in genes involved in signaling between the plant and bacteria (attB and attD) reduced the ability of the bacteria to attach to and colonize intact roots. These genes are known to play a role in the pathogenic interaction of the bacteria with wound sites. Thus, it appears that the initial stages of pathogenesis and root colonization may depend on many of the same genes and processes.
The results reported here add to the number and type of genes known to be involved in root colonization. Previously identified genes include genes required for the synthesis of bacterial agglutinins of pseudomonads (2), flagellar, motility, and chemotaxis genes of rhizobia, agrobacteria, and pseudomonads (1, 5, 10, 24, 28), chvA and chvB of A. tumefaciens (8), the pilus genes of pseudomonads (31), genes required for biosynthesis of amino acids and thiamine (27, 28), genes required for synthesis of the lipopolysaccharide O-antigens of pseudomonads (28), and genes affecting siderophore synthesis in pseudomonads (12).
ACKNOWLEDGMENTS
This research was supported by grant 94-37303 from the U.S. Department of Agriculture Competitive Grants Program and by grant MCB-9405844 from NSF.
REFERENCES
- 1.Bowers J H, Parke J L. Colonization of pea (Pisum sativum L.) taproots by Pseudomonas fluorescens: effect of soil temperature and bacterial motility. Soil Biol Biochem. 1993;25:1693–1701. [Google Scholar]
- 2.Buell C R, Anderson A J. Genetic analysis of the aggA locus involved in agglutination and adherence of Pseudomonas putida, a beneficial fluorescent pseudomonad. Mol Plant Microbe Interact. 1992;5:154–162. doi: 10.1094/mpmi-5-154. [DOI] [PubMed] [Google Scholar]
- 3.Bull C T, Weller D M, Thomashow L S. Relationship between root colonization and suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens strain 2-79. Phytopathology. 1991;81:954–959. [Google Scholar]
- 4.de Freitas J R, Germida J J. A root tissue culture system to study winter wheat-rhizobacteria interactions. Appl Microbiol Biotechnol. 1990;33:589–595. [Google Scholar]
- 5.de Wegner L A, Van der Vlugt C I M, Wijfjes A H M, Bakker P A H M, Schippers B, Lugtenberg B. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J Bacteriol. 1987;169:2769–2773. doi: 10.1128/jb.169.6.2769-2773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas C J, Halperin W, Nester E W. Agrobacterium tumefaciens mutants affected in attachment to plants. J Bacteriol. 1982;152:1265–1275. doi: 10.1128/jb.152.3.1265-1275.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferriss R S. Effects of microwave oven treatment on microorganisms in soil. Phytopathology. 1984;74:121–126. [Google Scholar]
- 8.Hawes M C, Pueppke S G. Reduced rhizosphere colonization ability of Agrobacterium tumefaciens chromosomal virulence (chv) mutants. Plant Soil. 1989;113:129–132. doi: 10.1104/pp.91.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawes M C, Pueppke S G. Variation in binding and virulence of Agrobacterium tumefaciens chromosomal virulence (chv) mutant bacteria on different plant species. Plant Physiol. 1989;91:113–118. doi: 10.1104/pp.91.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawes M C, Smith L Y. Requirement for chemotaxis in pathogenicity of Agrobacterium tumefaciens on roots of soil-grown pea plants. J Bacteriol. 1989;171:5668–5671. doi: 10.1128/jb.171.10.5668-5671.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebbar K P, Davey A G, Merrin J, McLoughlin T J, Dart P J. Pseudomonas cepacia, a potential suppressor of maize soil-borne diseases—seed inoculation and maize root colonization. Soil Biol Biochem. 1997;24:999–1007. [Google Scholar]
- 12.Höfte M, Boelens J, Verstraete W. Survival and root colonization of mutants of plant growth-promoting pseudomonads affected in siderophore biosynthesis or regulation of siderophore production. J Plant Nutr. 1992;15:2253–2262. [Google Scholar]
- 13.Hord M J, Ristaino J B. Effect of the matrix component of soil water potential on infection of pepper seedlings in soil infested with oospores of Phytophthora capsici. Phytopathology. 1992;82:792–798. [Google Scholar]
- 14.Loper J E, Suslow T V, Schroth M N. Lognormal distribution of bacterial populations in the rhizosphere. Phytopathology. 1984;74:1454–1460. [Google Scholar]
- 15.Macrae S, Thomson J A, Van Staden J. Colonization of tomato plants by two agrocin-producing strains of Agrobacterium tumefaciens. Appl Environ Microbiol. 1988;54:3133–3137. doi: 10.1128/aem.54.12.3133-3137.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthysse A G. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J Bacteriol. 1983;154:906–915. doi: 10.1128/jb.154.2.906-915.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthysse A G. Characterization of nonattaching mutants of Agrobacterium tumefaciens. J Bacteriol. 1987;169:313–323. doi: 10.1128/jb.169.1.313-323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthysse A G. Conditioned medium promotes the attachment of Agrobacterium tumefaciens strain NT1 to carrot cells. Protoplasma. 1994;183:131–136. [Google Scholar]
- 19.Matthysse A G. Adhesion in the rhizosphere. In: Fletcher M, Savage D, editors. Molecular and ecological diversity of bacterial adhesion. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 129–153. [Google Scholar]
- 20.Matthysse A G, Lightfoot R, White S. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthysse A G, Yarnall H A, Young N. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J Bacteriol. 1996;178:5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minnemeyer S L, Lightfoot R, Matthysse A G. A semi-quantitative bioassay for relative virulence of Agrobacterium tumefaciens strains on Bryophyllum daigremontiana. J Bacteriol. 1991;173:7723–7724. doi: 10.1128/jb.173.23.7723-7724.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam J, Matthysse A G, Gelvin S B. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell. 1997;9:317–333. doi: 10.1105/tpc.9.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parco S Z, Dilworth M J, Glenn A R. Motility and the distribution of introduced root nodule bacteria on the root system of legumes. Soil Biol Biochem. 1994;26:297–300. [Google Scholar]
- 25.Reuhs B L, Kim J S, Matthysse A G. The attachment of Agrobacterium tumefaciens to carrot cells and Arabidopsis wound sites is correlated with the production of a cell-associated, acidic polysaccharide. J Bacteriol. 1997;179:5372–5379. doi: 10.1128/jb.179.17.5372-5379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson J L, Holliday T, Matthysse A G. Mapping of Agrobacterium tumefaciens chromosomal genes affecting cellulose synthesis and bacterial attachment to host cells. J Bacteriol. 1988;170:1408–1411. doi: 10.1128/jb.170.3.1408-1411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simons M, Permentier H P, de Wegner L A, Wijffelman C A, Lugtenberg B J J. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol Plant-Microbe Interact. 1997;10:102–106. [Google Scholar]
- 28.Simons M, van der Bij A J, Brand I, de Wegner L A, Wijffelman C A, Lugtenberg B J J. Gnotobiotic system for studying rhizosphere colonization by growth-promoting Pseudomonas bacteria. Mol Plant-Microbe Interact. 1996;9:600–607. doi: 10.1094/mpmi-9-0600. [DOI] [PubMed] [Google Scholar]
- 29.Stockwell V O, Moore L W, Loper J E. Fate of Agrobacterium radiobacter K84 in the environment. Appl Environ Microbiol. 1993;59:2112–2120. doi: 10.1128/aem.59.7.2112-2120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomashow M F, Karlinsky J E, Marks J R, Hurlburt R E. Identification of a new virulence locus in Agrobacterium tumefaciens that affects polysaccharide composition and plant cell attachment. J Bacteriol. 1987;169:3209–3216. doi: 10.1128/jb.169.7.3209-3216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vesper S J. Production of pili (fimbriae) by Pseudomonas fluorescens and correlation with attachment to corn roots. Appl Environ Microbiol. 1987;53:1397–1405. doi: 10.1128/aem.53.7.1397-1405.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]