Abstract
RNA transcript levels in the syphilis spirochete Treponema pallidum subsp. pallidum (Nichols) isolated from experimentally infected rabbits were determined by the use of DNA microarray technology. This characterization of the T. pallidum transcriptome during experimental infection provides further insight into the importance of gene expression levels for the survival and pathogenesis of this bacterium.
Treponema pallidum subsp. pallidum is the causative agent of syphilis, a sexually transmitted disease characterized by multiple stages, widespread dissemination, persistent infection for years to decades, and varied clinical manifestations (18). Closely related spirochetes cause yaws, pinta, and endemic syphilis. This group of bacteria has not been cultured continuously under in vitro conditions, although some multiplication can be obtained in a tissue culture system (8, 17). T. pallidum can be maintained by intratesticular or intradermal inoculation of rabbits, causing an experimental infection that in many ways resembles the primary stage of syphilis. The inability to culture T. pallidum in vitro prevents the use of mutational analysis and other common molecular genetic approaches to study this bacterium and has limited the information available on the physiology and pathogenesis of the organism.
The 1.14-Mbp genome of T. pallidum subsp. pallidum Nichols was sequenced in 1998, and 1,039 open reading frames (ORFs) were predicted (12, 30). The availability of the complete genome sequence permits the use of genomic approaches to study this organism. DNA microarray-based gene expression profiling has been used to study differences in gene expression in several pathogenic and nonpathogenic bacteria under various environmental conditions (19-21, 24, 27). Moreover, DNA microarray techniques have been employed to map the genomic differences among strains of the same bacterial species (3, 7, 13).
The intradermal or intratesticular inoculation of rabbits with T. pallidum results in lesions that in many ways resemble the primary lesions of human syphilis (8, 28). As in human chancres, T. pallidum can reach extremely high concentrations in experimentally infected rabbits, permitting the isolation of 1010 to 1011 organisms from infected testicular tissue at 10 to 12 days postinoculation. This source of host-adapted T. pallidum permits analyses of gene expression during infection. In this study, we report the construction of a T. pallidum DNA microarray and the profiling of T. pallidum subsp. pallidum Nichols gene transcript levels during experimental infections of rabbits.
T. pallidum subsp. pallidum (Nichols) was maintained by rabbit inoculation and purified by Hypaque gradient centrifugation as described previously (2, 12). Chromosomal DNA was prepared as described by Fraser et al. (12). For RNA isolation, T. pallidum (Nichols) was purified from rabbit testicular tissue at 10 or 11 days postinoculation by Hypaque centrifugation, and RNAs were purified by the RNAzol B method based on guanidine thiocyanate phenol-chloroform extraction (TelTest, Friendswood, Tex.). Care was taken to maintain the samples at 4°C at all times and to minimize the processing time (3 to 4 h). RNA processing is unlikely to occur during the guanidine and phenol RNA extraction procedure. Gel electrophoresis of the samples was performed to evaluate the quality of RNA isolation. A smear with two strong bands corresponding to 23S and 16S RNAs and with discrete bands representing precursor RNAs was observed, indicating the integrity of the RNA molecules. To further assess the quality of RNA samples, transcript levels of 93 genes in two independent RNA preparations were examined by reverse transcription and real-time PCR amplification. A quantitation of gene expression yielded a high degree of correlation (r = 0.94), indicating that there was no bias in the relative amounts of mRNA species in individual RNA isolates.
We developed a microarray chip for T. pallidum subsp. pallidum Nichols containing all 1,039 predicted ORF PCR products together with a set of control PCR products from Escherichia coli, Shigella sp., and Enterococcus sp. (see the supplemental material). The annotation of ORFs was taken from the genome sequence (12), and most of the PCR products were amplified from the pUNI-D-TOPO cloning vector containing individual cloned T. pallidum genes (15). Some of the genes were amplified either from treponemal cosmid clones (23) or from isolated T. pallidum chromosomal DNA. Most of the individual PCR products covered the complete gene length, whereas products for genes with predicted signal sequences lacked the gene segment corresponding to those sequences (15). Negative control spots with no DNA were also included. The PCR products were purified by the use of Multiscreen 96-well filter plates (Millipore, Bedford, Mass.) according to the manufacturer's recommendations. Alternatively, some of the PCR products were purified by use of a PCR purification kit (QIAGEN, Valencia, Calif.).
T. pallidum chromosomal DNA (0.25 to 0.75 μg) was labeled by use of the Klenow enzyme (New England Biolabs, Beverly, Mass.) and random nonamers with a CyScribe first-strand cDNA labeling kit (Amersham Pharmacia Biotech, Piscataway, N.J.) protocol, with minor modifications. Labeling was performed at room temperature for 2.5 h. The total RNA (10 to 20 μg) was labeled with Superscript II reverse transcriptase (Invitrogen, Carlsbad, Calif.) as described in the supplemental material. The pretreated slides were hybridized simultaneously with labeled DNA and cDNAs by use of a CyScribe first-strand cDNA labeling kit (Amersham Pharmacia Biotech). Slides were then scanned with a ScanArray 5000 scanner (Packard BioScience, Meriden, Conn.), and the fluorescence intensities of individual spots were measured with ImaGene software (BioDiscovery, Marina del Rey, Calif.). The data analysis method was adapted from the work of Revel et al. (20) (see the supplemental material). The microarray data represent the means of three separate hybridizations (of two different RNA preparations), i.e., nine possible hybridizations for each gene.
The T. pallidum gene microarray used for these studies was constructed on glass slides by the use of PCR products. The 1,039 annotated T. pallidum ORFs (12) were named TP0001 to TP1041 (the loci TP0495 and TP0635 are not valid; for more information, see www.tigr.org). The average gene length of T. pallidum genes is 1,023 bp. Of the 1,039 ORFs, 1,034 gave positive signals for both labeled chromosomal DNA and cDNAs labeled with the total RNA as a template. The microarray signals of five ORFs (TP0156, TP0161, TP0518, TP0777, and TP1032) were below the threshold limit of the DNA channel and were omitted. All of these ORFs are relatively short (402, 60, 690, 222, and 432 bp, respectively) and code for conserved hypothetical (TP0156 and TP0518) or hypothetical (TP0161, TP0777, and TP1032) proteins. The cohybridization of Cy3-labeled T. pallidum DNA and Cy5-labeled cDNAs (or the reverse) yielded highly reproducible results in three independent experiments performed for this study. Moreover, comparable data were obtained with different RNA preparations, indicating that minor variations in the RNA preparations did not affect the overall results. Data were calculated as average expression ratios representing average, normalized ratios of cDNA to DNA fluorescent signals for replicate spots on each microarray and for replicate experiments. A value of 1 corresponds to the mean transcript level for all genes on the array. When expressed as a log value, the mean value is 0; values of >0 represent higher than average transcript levels, whereas those of <0 are below the average level. The cohybridization of differentially labeled cDNA and DNA preparations with each microarray chip permitted the normalization of effects resulting from different gene lengths, varied quantities of target DNA in each spot and of total RNA per hybridization, and other factors (29). This approach allowed us to compare gene expression levels among treponemal genes under given conditions. Gene expression profiling experiments performed with many organisms that can be grown under different conditions (10, 19-21, 24, 27) cannot be performed with T. pallidum because this spirochete cannot be continuously grown under in vitro conditions. Thus far, we have been unable to isolate sufficient RNAs after intradermal inoculations of rabbits and time-limited cultivation of T. pallidum under in vitro conditions to perform DNA microarray experiments. Although the cDNA signal depends on the efficiency of reverse transcription, which is different for different mRNA species, the random priming used in this study to label mRNA may minimize these differences. cDNA signals were detected for all treponemal genes, and observed differences in their expression varied by >2 orders of magnitude. This was consistent with the microarray experimental design (1). The fact that positive cDNA signals were detected for nearly all genes suggests that all genes of T. pallidum are expressed during experimental infection. This might imply that all of the genes that are not necessary for interacting with the mammalian host were lost during adaptation.
The expression of treponemal genes along the chromosome varied to a wide extent; the average log ratios in windows containing 10 genes are shown in Fig. 1. The region with the highest average transcription rate was located between genes TP0187 and TP0250, which encompass a large ribosomal protein operon. Interestingly, this region was previously described to be underrepresented in a bacterial artificial chromosome library of T. pallidum DNA in E. coli (23).
FIG. 1.
Gene expression of treponemal genes along the chromosome computed for windows containing 10 genes. The highest average transcription rate was located between genes TP0187 and TP0250, comprising a large ribosomal gene operon.
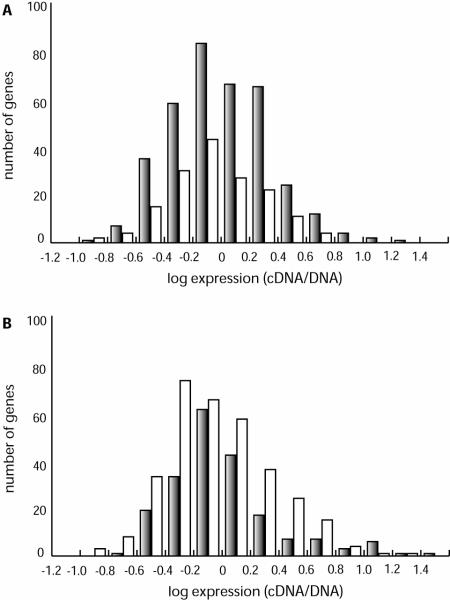
The coordinates and gene numbering of the T. pallidum genome sequence begin at the predicted origin of replication; thus, roughly the first 520 genes are replicated in the forward (+) direction, whereas the other half of the genome is replicated in the reverse (−) direction. In most bacterial genomes, the majority of genes are transcribed in the same direction as that in which DNA replication proceeds (22), and this replication transcription codirectionality appears to apply especially for highly transcribed genes (31). The distribution of treponemal genes in the expression level intervals for each chromosomal half is shown in Fig. 2. The tendency toward replication transcription codirectionality was also found for T. pallidum, but the higher transcription of genes oriented in the direction of replication was not observed. Given the long doubling time of T. pallidum (>30 h) in vivo and in vitro (9, 11), it is possible that exceptionally high transcription rates are not needed to keep up with cellular multiplication.
FIG. 2.
Histograms of treponemal genes sorted according to their gene expression levels. (A) Distribution of treponemal genes in gene expression intervals for the first chromosomal half (TP0001 to TP0521). Shaded columns represent numbers of genes with the same direction of transcription and replication in log expression intervals. Open columns represent genes transcribed in the opposite direction. (B) Distribution of treponemal genes in gene expression intervals for the second chromosomal half (TP0522 to TP1041). Shaded columns represent numbers of genes transcribed in the opposite direction relative to the DNA replication direction. Open columns represent genes oriented in the direction of replication.
T. pallidum genes were sorted according to their relative degrees of expression standardized to the amount of T. pallidum chromosomal DNA. The 100 genes with the highest transcript levels are shown in Table 1. For brevity and clarity, ORFs encoding hypothetical proteins (41 ORFs) were omitted from the table, but they are included in the supplemental material (Tables S1 and S2). Forty-one ORFs encoding hypothetical proteins (with an average ORF length of 624 bp) were randomly dispersed along the chromosome. Some of these formed clusters of two to three ORFs, indicating an operon organization. Besides the expected genes for ribosomal and polypeptide processing proteins (17 genes), the most highly transcribed genes were the flagellar filament and cytoplasmic filament protein genes (5 genes, including flaB-1-3, flaA, and cfpA), genes for lipoproteins or other prominent membrane proteins (11 genes, namely, tp47, TP0663, tpD, tmpC, tmpA, tpp15, tmpB, tpp17, p83/100h, tpn38b, and tap1), genes encoding chaperonins (3 genes, namely, groEL, groES, and dnaK), genes for proteins involved in redox balance (5 genes encoding alkyl hydroperoxidase C, flavodoxin, thioredoxin, pyruvate oxidoreductase, and desulfoferrodoxin), 4 genes for chemotaxis proteins (cheX, cheY, mcp2-1, and cheA), and genes encoding metabolic and other functions (14 genes). High gene expression activities were also detected for genes encoding glycolytic pathway enzymes (e.g., TP0844, encoding glyceraldehyde-3-phosphate dehydrogenase) and for one V-type ATPase operon (TP0424 to TP0430), whereas the other ATPase operon (TP0527 to TP0533) was transcribed at low levels. These findings may reflect the dependence of T. pallidum on the glycolytic pathway for energy production, on the maintenance of optimal redox conditions related to its microaerophilism, and on chemotaxis to localize to optimal tissue environments (17).
TABLE 1.
T. pallidum genes expressed most highly during infection
| T. pallidum ORF no.a | cDNA/DNA signal ratio | Coding strand | Putative gene function | cDNA/DNA signal ratio normalized to that of TP0426b
|
|
|---|---|---|---|---|---|
| DNA microarray | Real-time PCR | ||||
| TP0870 | 25.9 | + | Flagellar filament 31-kDa core protein (flaB3) | 20.0 | 19.4 |
| TP0574 | 17.7 | − | Carboxypeptidase, 47 kDa | ||
| TP0684 | 16.2 | + | Methylgalactoside ABC transporter, periplasmic galactose-binding protein (mglB-2) | 12.5 | 9.6 |
| TP0249 | 16.0 | + | Flagellar filament outer layer protein (flaA-1) | 12.4 | 18.3 |
| TP0509 | 15.5 | + | Alkyl hydroperoxide reductase (ahpC) | 12.0 | 41.7 |
| TP0663 | 13.9 | + | Putative outer membrane protein | ||
| TP0792 | 13.8 | + | Flagellar filament 33-kDa core protein (flaB2) | 10.6 | 54.2 |
| TP0971 | 12.6 | − | Membrane antigen, pathogen specific (tpd) | 9.7 | 12.4 |
| TP0356 | 10.7 | + | Putative RNA-binding protein | ||
| TP0868 | 10.4 | + | Flagellar filament 34.5-kDa core protein (flaB1) | 8.0 | 11.2 |
| TP0319 | 9.7 | + | Membrane lipoprotein (tmpC) | 7.5 | 12.9 |
| TP0925 | 9.4 | + | Flavodoxin | 7.2 | 13.2 |
| TP0844 | 9.0 | + | Glyceraldehyde-3-phosphate dehydrogenase (gap) | 7.0 | 19.0 |
| TP0030 | 9.0 | + | Heat shock protein (groEL) | 7.0 | 8.5 |
| TP0919 | 8.1 | − | Thioredoxin (trx) | 6.3 | 20.9 |
| TP0849 | 7.1 | − | Ribosomal protein L35 (rpmI) | ||
| TP0243 | 6.9 | + | Ribosomal protein S12 (rpsL) | 5.3 | 8.3 |
| TP0748 | 6.8 | − | Cytoplasmic filament protein A (cfpA) | 5.2 | 14.8 |
| TP0768 | 6.7 | + | Membrane protein (tmpA) | 5.2 | 8.3 |
| TP0171 | 5.9 | + | Lipoprotein, 15 kDa (tpp15) | ||
| TP0758 | 5.8 | − | Ribosomal protein S21 (rpsU) | ||
| TP1024 | 5.4 | − | Ribosomal protein S9 (rpsI) | ||
| TP0362 | 5.4 | + | Ribosomal protein L28 (rpmB) | ||
| TP0862 | 5.3 | − | Peptidyl-prolyl cis-trans isomerase, FKBP-type, 22 kDa (fklB) | ||
| TP0769 | 5.2 | + | Outer membrane protein (tmpB) | 4.0 | 2.2 |
| TP0435 | 5.1 | − | Lipoprotein, 17 kDa (tpp17) | 3.9 | 10.5 |
| TP0228 | 5.1 | − | Putative biotin synthase | ||
| TP0122 | 5.0 | − | Phosphoenolpyruvate carboxykinase (pckA) | 3.9 | 3.6 |
| TP1025 | 4.8 | − | Ribosomal protein L13 (rplM) | ||
| TP0424 | 4.7 | + | Putative V-type ATPase, subunit E | 3.6 | 4.4 |
| TP0662 | 4.6 | − | Fructose-bisphosphate aldolase (cbbA) | ||
| TP1013 | 4.6 | + | Chaperonin (groES) | 3.5 | 4.6 |
| TP0216 | 4.5 | + | Heat shock protein 70 (dnaK) | 3.5 | 10.3 |
| TP0365 | 4.5 | + | Chemotaxis protein (cheX) | ||
| TP0905 | 4.4 | + | Ribosomal protein S16 (rpsP) | ||
| TP0887 | 4.3 | − | Ribosomal protein S15 (rpsO) | ||
| TP0366 | 4.3 | + | Chemotaxis response regulator (cheY) | 3.3 | 3.4 |
| TP0234 | 4.2 | + | Ribosomal protein L33 (rpmG) | ||
| TP0807 | 4.1 | + | Ribosomal protein L32 (rpmF) | ||
| TP0606 | 4.0 | − | Ribosomal protein S2 (rpsB) | ||
| TP0609 | 4.0 | + | Asparaginyl-tRNA synthetase (asnS) | ||
| TP0888 | 3.9 | − | Riboflavin kinase/FMN adenylyltransferase (ribF) | ||
| TP0848 | 3.9 | − | Ribosomal protein L20 (rplT) | ||
| TP0115 | 3.7 | − | Phosphomethylpyrimidine kinase (thiD) | ||
| TP0488 | 3.3 | + | Methyl-accepting chemotaxis protein (mcp2-1) | ||
| TP0486 | 3.3 | − | Antigen, p83/100 | ||
| TP0298 | 3.3 | + | Exported protein (tpn38b) | ||
| TP0939 | 3.3 | − | Pyruvate oxidoreductase | 2.5 | 3.0 |
| TP0363 | 3.3 | + | Chemotaxis histidine kinase (cheA) | 2.5 | 2.2 |
| TP0823 | 3.2 | − | Desulfoferrodoxin-related protein | ||
| TP0209 | 3.2 | + | Ribosomal protein L36 (rpmJ-1) | ||
| TP0237 | 3.2 | + | Ribosomal protein L11 (rplK) | ||
| TP0244 | 3.1 | + | Ribosomal protein S7 | ||
| TP0850 | 3.0 | − | Translation initiation factor 3 (infC) | ||
| TP0361 | 3.0 | − | Putative lysophosphatidic acid acyltransferase | 2.3 | 3.1 |
| TP1041 | 3.0 | − | ATP-dependent Clp protease proteolytic component (clpP-2) | ||
| TP0817 | 3.0 | − | Enolase (eno) | 2.3 | 1.6 |
| TP0729 | 2.9 | − | Treponemal aqueous protein (tap1) | ||
| TP0746 | 2.9 | − | Pyruvate, phosphate dikinase | ||
Forty-one ORFs coding for hypothetical proteins (including conserved hypothetical proteins) were omitted from this table. For a complete listing, see Tables S1 and S2 in the supplemental material.
The TP0426 ORF was used as a reference because of its consistent cDNA/DNA ratio of close to 1.0 obtained by either a DNA microarray or real-time PCR approach.
Most of the prominent proteins recognized by two-dimensional gel electrophoresis (16) were shown to be encoded by genes with high degrees of gene expression (Fig. 3). Two relatively minor spots corresponded to an average (1.01) gene transcription level (TP1016 [tpn39b], coding for a basic membrane protein) and to a below-average level (0.41) (TP0545 [mglB-1]), coding for a periplasmic galactose binding protein). Overall, there appears to be a strong correlation between the level of gene transcripts and protein abundance in T. pallidum.
FIG. 3.
Two-dimensional sodium dodecyl sulfate-polyacrylamide gel showing identified treponemal proteins. The corresponding treponemal genes are shown, with cDNA/DNA signal ratios in parentheses. Prominent protein spots correspond to high transcription levels of the relevant genes. (Reprinted from reference 16.)
T. pallidum genes with the lowest cDNA/DNA ratios, coding for 62 proteins with assigned functions, are shown in Table 2. Genes encoding flagellar biosynthesis and other components of the cell envelope (12 genes), components of DNA metabolism (11 genes, e.g., genes encoding subunits of DNA polymerases I and III), and metabolic and other functions (11 genes) constituted the major groups. Eight genes for transporters (with specificities for oligopeptides, K+, thiamine, carnitine, and sugars), eight genes encoding various cellular processes (putative hemolysins and cell division proteins), four genes involved in transcription and translation, and four regulators were found within this group. In addition, four genes with internal authentic frame shifts were detected. Possible reasons for these low transcription levels range from constitutively weak promoters to tightly regulated genes under in vivo conditions. The latter explanation might apply to at least some of the genes encoding transporters, components of the cell envelope, and proteins for DNA metabolism. In contrast to genes encoding components of periplasmic flagella (14) (flaB-1, flaB-2, flaB-3, and flaA), which were the most highly expressed genes during infection, low transcription levels were observed for some biosynthetic flagellar genes (fliQ, fliR, flhA, and flhB). These last genes encode proteins with significant homologies to bacterial proteins associated with the flagellar export apparatus (25). The remaining 38 ORFs encoding hypothetical proteins were localized along the whole chromosome, with few clusters. The average ORF length within this group (1,276 bp) was considerably longer than the corresponding average length of ORFs encoding hypothetical proteins with high transcript levels, suggesting that these ORFs represent weakly transcribed genes. Investigations of changes in gene expression by more sensitive methods for these genes in T. pallidum grown under different conditions (e.g., in time-limited in vitro cultures) will be needed to detect changes induced by environmental changes and to unravel the regulatory mechanisms.
TABLE 2.
T. pallidum genes that are weakly expressed during infection
| T. pallidum ORF no.a | cDNA/DNA signal ratio | Coding strand | Putative gene function | cDNA/DNA signal ratio normalized to that of TP0426
|
|
|---|---|---|---|---|---|
| DNA microarray | Real-time PCR | ||||
| TP0585 | 0.36 | + | Oligopeptide ABC transporter, periplasmic binding protein (oppA) | ||
| TP0717 | 0.36 | − | Flagellar biosynthetic protein (fliQ) | ||
| TP0627 | 0.36 | + | Exonuclease (sbcC) | ||
| TP0999 | 0.36 | + | Putative cell division protein | ||
| TP0045 | 0.36 | − | Putative adenosine deaminase | ||
| TP0140 | 0.35 | − | K+ transport protein (ntpJ) | ||
| TP0141 | 0.35 | + | Methylated-DNA-protein-cysteine S-methyltransferase (dat) | ||
| TP0647 | 0.35 | − | Seryl-tRNA synthetase (serS) | ||
| TP0626 | 0.34 | + | Putative exonuclease | ||
| TP0016 | 0.34 | + | ATP-dependent protease LA (lon-1) | ||
| TP0446 | 0.34 | + | GcpE protein (gcpE) | ||
| TP0812 | 0.34 | + | Long-chain-fatty-acid-CoA ligase, authentic frame shift | ||
| TP0694 | 0.33 | + | Putative 5,10-methenyltetrahydrofolate synthetase | ||
| TP0393 | 0.33 | + | Smf protein (smf) | ||
| TP0805 | 0.33 | − | Exoribonuclease II (rnb) | ||
| TP0078 | 0.33 | + | Spore coat polysaccharide biosynthesis protein (spsC) | ||
| TP0003 | 0.33 | + | RecF protein (recF) | ||
| TP0902 | 0.32 | − | Carboxylesterase (est) | 0.25 | 0.16 |
| TP0035 | 0.32 | + | ABC transporter, ATP-binding protein | ||
| TP0142 | 0.32 | − | Putative thiamine ABC transporter, ATP-binding protein | ||
| TP0270 | 0.32 | + | Polynucleotide adenylyltransferase (pcnA) | ||
| TP0009 | 0.31 | − | Tpr protein A, authentic frame shift (tprA) | 0.24 | 0.13 |
| TP0077 | 0.31 | + | Capsular polysaccharide biosynthesis protein (cap5D) | ||
| TP0218 | 0.31 | + | Putative sigma factor SigB regulation protein | ||
| TP0146 | 0.31 | + | Chromate resistance protein A, authentic frame shift | ||
| TP1005 | 0.31 | − | DNA polymerase III, subunits gamma and tau (dnaH) | ||
| TP0500 | 0.30 | + | Penicillin-binding protein (pbp-1) | ||
| TP0027 | 0.30 | + | Putative hemolysin | ||
| TP0578 | 0.29 | + | Signal recognition particle-docking protein FtsY (ftsY) | ||
| TP0687 | 0.29 | − | DNA recombinase (recG) | 0.23 | 0.28 |
| TP0501 | 0.29 | + | Rod shape-determining protein (rodA) | 0.22 | 0.10 |
| TP0080 | 0.29 | + | Quinoline 2-oxidoreductase | ||
| TP0581 | 0.29 | + | ABC transporter, ATP-binding protein | ||
| TP0105 | 0.28 | + | DNA polymerase I (polA) | ||
| TP0344 | 0.27 | − | Transcription repair coupling factor (trcF) | ||
| TP0387 | 0.27 | + | Cell division protein (ftsW) | 0.21 | 0.16 |
| TP0881 | 0.27 | − | ABC transporter, ATP-binding protein (natA) | ||
| TP0448 | 0.27 | + | Putative uracil phosphoribosyltransferase | ||
| TP0521 | 0.27 | − | Putative DNA polymerase III, gamma and tau subunits | ||
| TP0450 | 0.27 | + | Translation elongation factor G (fusA-1) | ||
| TP0716 | 0.27 | − | Flagellar biosynthetic protein (fliR) | ||
| TP0670 | 0.26 | − | d-Alanine-d-alanine ligase (ddlA) | ||
| TP0933 | 0.26 | − | UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase (murE) | 0.20 | 0.26 |
| TP0106 | 0.26 | − | Putative carnitine transporter | ||
| TP0804 | 0.26 | + | Sugar ABC transporter, ATP-binding protein (ugpC) | ||
| TP0340 | 0.26 | − | Folylpolyglutamate synthetase (folC) | ||
| TP0288 | 0.25 | + | Spore coat polysaccharide biosynthesis protein (spsF) | ||
| TP0337 | 0.24 | + | Dimethyladenosine transferase (ksgA) | ||
| TP0028 | 0.23 | + | Putative hemolysin | 0.18 | 0.09 |
| TP0514 | 0.23 | + | Excinuclease ABC, subunit A (uvrA) | ||
| TP0689 | 0.22 | − | GTP-binding protein | ||
| TP0147 | 0.21 | − | Putative alpha-amylase 1 | ||
| TP0336 | 0.20 | + | Putative ComE protein | ||
| TP0671 | 0.20 | − | Putative sn-1,2-diacylglycerol cholinephosphotransferase | ||
| TP0566 | 0.20 | − | Alginate O-acetylation protein (algI) | ||
| TP0516 | 0.19 | − | Virulence factor (mviN) | 0.15 | 0.15 |
| TP0688 | 0.18 | − | Immunity protein (mccF) | ||
| TP0715 | 0.17 | − | Flagellar biosynthetic protein (flhB) | ||
| TP0714 | 0.17 | − | Flagellar biosynthesis protein (flhA) | ||
| TP0520 | 0.16 | − | Sensory transduction histidine kinase, authentic frame shift | ||
| TP0519 | 0.15 | − | Response regulatory protein (atoC) | 0.11 | 0.07 |
| TP0517 | 0.13 | − | Holliday junction nuclease (ruvC) | ||
Thirty-eight ORFs coding for hypothetical proteins (including conserved hypothetical proteins) were omitted from this table. For a complete listing, see Tables S1 and S2 in the supplemental material.
A set of genes encoding possible virulence factors of T. pallidum was predicted and identified by Weinstock et al. (30). The corresponding expression levels of these genes are shown in Table 3. Since the treponemes investigated for this study were isolated from rabbit testes, expression profiling showed gene expression during infection of an animal host. The most highly transcribed genes code for membrane or surface-exposed proteins, and a majority of them are known antigens. Genes encoding putative hemolysins and regulators and genes involved in polysaccharide biosynthesis and other functions were found to be expressed at moderate or relatively low levels. Similarly, the T. pallidum repeat (tpr) genes were found to be expressed at relatively low levels. The tpr genes encode paralogous proteins with sequence similarity to the major surface protein (Msp) of Treponema denticola (12). This multigene family is found only in the genus Treponema, induces an antibody response during infection, and exhibits heterogeneity both within and between the T. pallidum subspecies and strains examined (4-6); therefore, it is thought that the Tpr proteins may be involved in pathogenesis and/or immune evasion. Our results indicate that the tpr genes are expressed at relatively low levels, consistent with the difficulties encountered in detecting Tpr protein expression in T. pallidum (J. M. Hardham and S. J. Norris, unpublished data).
TABLE 3.
Possible virulence factors of T. pallidum (30) and corresponding gene expression levels in treponemes isolated from rabbit testes
| ORF or protein category | cDNA/DNA signal ratio | Coding strand | Putative gene function | cDNA/DNA signal ratio normalized to that of TP0426
|
|
|---|---|---|---|---|---|
| DNA microarray | Real-time PCR | ||||
| Tpr proteins | |||||
| TP0897 | 1.1 | 0 | Tpr protein K (tprK) | ||
| TP0610 | 0.92 | 1 | Tpr protein H (tprH) | 0.71 | 0.74 |
| TP1031 | 0.76 | 1 | Tpr protein L (tprL) | 0.58 | 3.44 |
| TP0621 | 0.75 | 0 | Tpr protein J (tprJ) | 0.58 | 0.15 |
| TP0317 | 0.74 | 0 | Tpr protein G (tprG) | 0.57 | 0.88 |
| TP0117 | 0.74 | 0 | Tpr protein C (tprC) | ||
| TP0131 | 0.70 | 0 | Tpr protein D (tprD) | ||
| TP0313 | 0.70 | 1 | Tpr protein E (tprE) | 0.54 | 0.20 |
| TP0316 | 0.68 | 0 | Tpr protein F, authentic frame shift (tprF) | 0.52 | 0.69 |
| TP0620 | 0.65 | 0 | Tpr protein I (tprI) | 0.50 | 0.21 |
| TP0011 | 0.39 | 1 | Tpr protein B (tprB) | ||
| TP0009 | 0.32 | 0 | Tpr protein A, authentic frame shift (tprA) | 0.24 | 0.13 |
| Hemolysins | |||||
| TP1037 | 1.1 | 0 | Hemolysin III (hlyIII) | 0.83 | 0.49 |
| TP0936 | 0.78 | 0 | Putative hemolysin | 0.60 | 0.74 |
| TP0649 | 0.57 | 0 | Hemolysin (tlyC) | ||
| TP0027 | 0.30 | 1 | Putative hemolysin | ||
| TP0028 | 0.23 | 1 | Putative hemolysin | 0.18 | 0.09 |
| Regulators | |||||
| TP0981 | 0.98 | 1 | Putative sensory transduction histidine kinase | ||
| TP0454 | 0.94 | 0 | Hypothetical protein (regA) | ||
| TP0038 | 0.79 | 0 | Regulatory protein (pfoS/R) | ||
| TP0980 | 0.73 | 1 | Histidine phosphokinase/phosphatase (ntrB) | ||
| TP0877 | 0.41 | 1 | Conserved hypothetical protein (regC) | ||
| TP0516 | 0.19 | 0 | Virulence factor (mviN) | 0.15 | 0.15 |
| TP0520 | 0.16 | 0 | Sensory transduction histidine kinase, authentic frame shift | ||
| TP0519 | 0.15 | 0 | Response regulatory protein (atoC) | 0.11 | 0.08 |
| Polysaccharide biosynthesis | |||||
| TP0562 | 1.5 | 1 | Spore coat polysaccharide biosynthesis protein (spsE) | ||
| TP0440 | 0.89 | 1 | Putative spore coat polysaccharide biosynthesis protein | ||
| TP0283 | 0.79 | 1 | Lipopolysaccharide core biosynthesis protein (kdtB) | ||
| TP0107 | 0.44 | 0 | LicC protein (licC) | ||
| TP0078 | 0.33 | 1 | Spore coat polysaccharide biosynthesis protein (spsC) | ||
| TP0077 | 0.31 | 1 | Capsular polysaccharide biosynthesis protein (cap5D) | ||
| TP0288 | 0.25 | 1 | Spore coat polysaccharide biosynthesis protein (spsF) | ||
| Potential membrane or surface-exposed proteins | |||||
| TP0574 | 17.7 | 0 | Carboxypeptidase, 47 kDa | ||
| TP0971 | 12.6 | 0 | Membrane antigen, pathogen specific (tpd) | 9.7 | 12.4 |
| TP0319 | 9.7 | 1 | Membrane lipoprotein (tmpC) | 7.5 | 12.8 |
| TP0768 | 6.7 | 1 | Membrane protein (tmpA) | 5.2 | 8.3 |
| TP0171 | 5.9 | 1 | Lipoprotein, 15 kDa (tpp15) | ||
| TP0769 | 5.2 | 1 | Outer membrane protein (tmpB) | 4.0 | 2.2 |
| TP0435 | 5.1 | 0 | Lipoprotein, 17 kDa (tpp17) | 3.9 | 10.5 |
| TP0486 | 3.3 | 0 | Antigen, p83/100 | ||
| TP0298 | 3.3 | 1 | Exported protein (tpn38b) | ||
| TP0470 | 3.1 | 0 | Conserved hypothetical protein | ||
| TP0729 | 2.9 | 0 | Treponemal aqueous protein (tap1) | ||
| TP0006 | 2.5 | 1 | Tp75 protein | ||
| TP0993 | 2.5 | 0 | Putative rare lipoprotein A | ||
| TP0225 | 2.3 | 1 | Leucine-rich repeat protein TpLRR | ||
| TP0702 | 2.2 | 0 | Conserved hypothetical protein | ||
| TP0020 | 2.2 | 1 | 76K protein | ||
| TP1038 | 2.1 | 0 | Bacterioferrin (tpF1) | 1.7 | 3.4 |
| TP0821 | 1.5 | 0 | Lipoprotein (tpn32) | ||
| TP0571 | 1.3 | 0 | Tp70 protein | ||
| TP0163 | 1.1 | 1 | ABC transporter, periplasmic binding protein (troA) | ||
| TP1016 | 1.0 | 0 | Basic membrane protein (tpn39b) | ||
| TP0957 | 0.78 | 1 | Tp33 protein | ||
| TP0624 | 0.66 | 1 | Conserved hypothetical protein | ||
| TP0819 | 0.65 | 0 | Conserved hypothetical protein | ||
| TP0326 | 0.62 | 1 | Outer membrane protein | ||
| TP0989 | 0.58 | 0 | P26 | ||
| TP0567 | 0.54 | 0 | Conserved hypothetical protein | ||
| TP0327 | 0.52 | 1 | Cationic outer membrane protein (ompH) | ||
| TP0796 | 0.46 | 0 | Conserved hypothetical protein | ||
| TP0292 | 0.42 | 1 | Outer membrane protein (tpn50) | ||
| TP0034 | 0.36 | 0 | ABC transporter, periplasmic binding protein | ||
| Miscellaneous functions | |||||
| TP0835 | 0.51 | 0 | Putative ankyrin | ||
| TP0680 | 0.50 | 0 | o-Sialoglycoprotein endopeptidase (gcp) | ||
| TP0502 | 0.42 | 1 | Hypothetical protein | ||
| TP0580 | 0.19 | 1 | Conserved hypothetical integral membrane protein | ||
To determine the accuracy of the microarray data and to test their consistency, we performed real-time PCR amplifications of selected genes. Eighty-four genes with varied levels of expression were tested by real-time PCRs using the same RNA preparations as those used for the microarray experiments. Primers for real-time PCR amplifications were designed with the Primer3 program (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi/), and the lengths of PCR products were set to be between 150 and 250 bp. cDNA samples were prepared as described for microarray experiments, without the addition of Cy-labeled nucleotides. cDNAs were used at a 10-ng/μl concentration with primers (10 pmol of each/reaction) and SYBR green master mix (Applied Biosystems). The ABI 7900 sequence detection system (Applied Biosystems) was used according to the manufacturer's instructions. The relative quantitation method (ΔΔCT) was used to evaluate the real-time PCR data. All amplifications were performed at least three times. For each amplification, the resulting ΔΔCT values were normalized to the corresponding value obtained with the control gene TP0426 (encoding the V-type ATPase subunit A-1) and were log transformed. Average log ratios from three independent experiments were computed and correlated to the log ratios obtained by the DNA microarray method. For microarray experiments, TP0426 had a cDNA/DNA signal ratio of close to 1. A similar normalization was also applied to relevant ratios obtained for microarray experiments. A high degree of correlation (r = 0.94) was achieved, with a relatively narrow 95% confidence interval (± 0.46). The real-time reverse transcription-PCR (RT-PCR) data for the genes shown in Tables 1 to 3 can be compared to the ratios obtained by the use of DNA microarrays. A correlation between DNA microarray and real-time RT-PCR data for tpr genes (Table 3) suggests that expression ratios of individual paralogous genes sharing regions of high nucleotide homology can be altered in microarray experiments because of the cross-hybridization of cDNAs of other paralogous genes. It thus appears that real-time PCR amplifications can detect wider differences in gene expression and also render more accurate results for genes belonging to the same paralogous family. These two differences might be explained by the principal differences of both methods, i.e., their sequence specificities and effective concentration ranges. Microarray expression analysis as performed in this study detects transcripts along their entire length and thus may be subject to the confounding effects of cross-hybridization. In contrast, the quantitative RT-PCR method only detects gene transcripts recognized by primers (and the distances between primer sites) and thus is likely to be more specific for paralogous genes. Microarrays also have a more limited working range restricted to ∼2 to 3 orders of magnitude due to the relatively narrow range between the signal detection threshold and signal saturation (1).
This study represents the first global analysis of gene transcription in T. pallidum by utilizing organisms extracted from rabbit testicular tissue at the height of infection. As an obligate pathogen of humans, T. pallidum has evolved to occupy a relatively homeostatic environment and hence may have jettisoned most of the genes that are not necessary for interaction with the host and also its capacity to alter gene expression. The apparent lack of a heat shock response (16, 24) and the dramatic effect that temperature has on T. pallidum replication both in vivo and in vitro (8, 26) may exemplify this reduced adaptability. However, given the occurrence of latent infections (in both humans and experimentally infected rabbits) and late gummatous, neurologic, and cardiovascular forms of the disease in humans, it is likely that gene expression is altered in these different tissue environments. This question could be approached to some extent by examining the transcription levels of T. pallidum genes during intradermal rather than intratesticular infections of rabbits, although fewer organisms can be recovered from the resulting dermal lesions. Further analyses of the genes expressed at high levels during infection may provide insight into the potential roles of their products in the pathogenesis of syphilis.
Supplementary Material
Acknowledgments
This work was supported by grants from the U.S. Public Health Service to G.M.W. (R01 DE12488 and R01 DE13759), S.J.N. (R01 AI49252), and T.P. (AI45842) and by grants of the Grant Agency of the Czech Republic (310/04/0021) and the Ministry of Health of the Czech Republic (NI7351-3/2003) to D.S.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baldi, P., and G. W. Hatfield. 2002. DNA microarrays and gene expression. Cambridge University Press, Cambridge, United Kingdom.
- 2.Baseman, J. B., J. C. Nichols, O. Rumpp, and N. S. Hayes. 1974. Purification of Treponema pallidum from infected rabbit tissue: resolution into two treponemal populations. Infect. Immun. 10:1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Centurion-Lara, A., C. Castro, L. Barrett, C. Cameron, M. Mostowfi, W. C. Van Voorhis, and S. A. Lukehart. 1999. Treponema pallidum major sheath protein homologue TprK is a target of opsonic antibody and the protective immune response. J. Exp. Med. 189:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centurion-Lara, A., C. Godornes, C. Castro, W. C. Van Voorhis, and S. A. Lukehart. 2000. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect. Immun. 68:824-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centurion-Lara, A., E. S. Sun, L. K. Barrett, C. Castro, S. A. Lukehart, and Van W. C. Voorhis. 2000. Multiple alleles of Treponema pallidum repeat gene D in T. pallidum isolates. J. Bacteriol. 182:2332-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa de Oliveira, R., G. M. Yanai, N. H. Muto, D. B. Leite, A. A. de Souza, H. D. Coletta Filho, et al. 2002. Competitive hybridization on spotted microarrays as a tool to conduct comparative genomic analyses of Xylella fastidiosa strains. FEMS Microbiol. Lett. 216:15-21. [DOI] [PubMed] [Google Scholar]
- 8.Cox, D. L. 1994. Culture of Treponema pallidum. Methods Enzymol. 236:390-405. [DOI] [PubMed] [Google Scholar]
- 9.Cumberland, M. C., and T. B. Turner. 1945. The rate of multiplication of Treponema pallidum in normal and immune rabbits. Am. J. Syph. 33:201-211. [PubMed] [Google Scholar]
- 10.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 11.Fieldsteel, A. H., D. L. Cox, and R. A. Moeckli. 1981. Cultivation of virulent Treponema pallidum in tissue culture. Infect. Immun. 32:908-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 13.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holodniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 14.Li, C., L. Corum, D. Morgan, E. L. Rosey, T. B. Stanton, and N. W. Charon. 2000. The spirochete FlaA periplasmic flagellar sheath protein impacts flagellar helicity. J. Bacteriol. 182:6698-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKevitt, M., K. Patel, D. Šmajs, M. Marsh, M. McLoughlin, S. J. Norris, G. M. Weinstock, and T. Palzkill. 2003. Systematic cloning of Treponema pallidum open reading frames for protein expression and antigen discovery. Genome Res. 13:1665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norris, S. J. 1993. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Treponema pallidum Polypeptide Research Group. Microbiol. Rev. 57:750-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norris, S. J., D. L. Cox, and G. M. Weinstock. 2001. Biology of Treponema pallidum: correlation of functional activities with genome sequence data. J. Mol. Microbiol. Biotechnol. 3:37-62. [PubMed] [Google Scholar]
- 18.Norris, S. J., V. Pope, R. E. Johnson, and S. A. Larsen. 2003. Treponema and other human host-associated spirochetes, p. 955-971. In P. R. Murray (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 19.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid, M. B., and J. R. Roth. 1987. Gene location affects expression level in Salmonella typhimurium. J. Bacteriol. 169:2872-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Šmajs, D., M. McKevitt, L. Wang, J. K. Howell, S. J. Norris, T. Palzkill, and G. M. Weinstock. 2002. BAC library of T. pallidum DNA in E. coli. Genome Res. 12:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, et al. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamm, L. V., and H. L. Bergen. 2001. Molecular characterization of the Treponema denticola fliQ region. DNA Seq. 12:463-467. [DOI] [PubMed] [Google Scholar]
- 26.Stamm, L. V., F. C. Gherardini, E. A. Parrish, and C. R. Moomaw. 1991. Heat shock response of spirochetes. Infect. Immun. 59:1572-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner, T. B., and D. H. Hollander. 1957. Biology of the treponematoses. World Health Organization, Geneva, Switzerland.
- 29.Wei, Y., J. M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinstock, G. M., J. M. Hardham, M. P. McLeod, E. J. Sodergren, and S. J. Norris. 1998. The genome of Treponema pallidum: new light on the agent of syphilis. FEMS Microbiol. Rev. 22:323-332. [DOI] [PubMed] [Google Scholar]
- 31.Zivanovic, Y., P. Lopez, H. Philippe, and P. Forterre. 2002. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 30:1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.