Abstract
With the widespread appearance of antibiotic-resistant bacteria, there is an increasing demand for novel strategies to control infectious diseases. Furthermore, it has become apparent that the bacterial life style also contributes significantly to this problem. Bacteria living in the biofilm mode of growth tolerate conventional antimicrobial treatments. The discovery that many bacteria use quorum-sensing (QS) systems to coordinate virulence and biofilm development has pointed out a new, promising target for antimicrobial drugs. We constructed a collection of screening systems, QS inhibitor (QSI) selectors, which enabled us to identify a number of novel QSIs among natural and synthetic compound libraries. The two most active were garlic extract and 4-nitro-pyridine-N-oxide (4-NPO). GeneChip-based transcriptome analysis revealed that garlic extract and 4-NPO had specificity for QS-controlled virulence genes in Pseudomonas aeruginosa. These two QSIs also significantly reduced P. aeruginosa biofilm tolerance to tobramycin treatment as well as virulence in a Caenorhabditis elegans pathogenesis model.
Several bacteria show organized behavior when they establish themselves in the eukaryotic host (22). The invading bacteria express a battery of tissue-damaging virulence factors in accordance with their numbers in a process termed quorum sensing (QS) (16). This is accomplished by sensing the concentration of small, diffusible signal molecules produced by the bacteria themselves. In gram-negative bacteria, the signals are N-acyl homoserine lactones (AHLs), which are produced by the LuxI family of AHL synthases. The signal molecules differ with respect to the length of their side chains (C4 to C16) and with various degrees of substitution and saturation (34). Short-chain AHLs are freely diffusible over the cell membranes, whereas long-chain AHLs are the substrate of efflux pumps, such as mexAB-oprM (36). The AHLs are sensed by proteins belonging to the LuxR family of response regulators. LuxR homologues contain two domains, an AHL binding domain and a DNA binding domain. When AHL is bound, it alters the configuration of the LuxR homologue protein, enabling it to interact with DNA and act as a transcriptional activator (16). It should be noted that some LuxR homologues acts as repressors, blocking transcription in the absence of AHL and, when sufficient AHL is present, derepressing the target gene(s) (6). The two key components of the QS system, the luxI and luxR homologues, are often linked genes, whereas the QS target genes are localized elsewhere on the genome. In case of Vibrio fischeri, the AHL synthase gene itself is a target gene of the QS mechanism, creating an autoinduction loop, which at the triggering (or threshold) AHL concentration gives rise to a burst in AHL production and QS-controlled gene expression.
It has recently become evident that QS target genes are not generally activated at a certain threshold concentration but merely become activated as a continuum at different AHL-cell concentrations (23, 40). Pseudomonas aeruginosa utilizes at least two luxI-luxR homologous QS systems, las and rhl, to control expression of virulence factors, including elastase, (alkaline) proteases, rhamnolipids, pyocyanin, and cyanide (37). P. aeruginosa is involved in a number of acute and chronic infections of which the fatal pulmonary infection of cystic fibrosis (CF) patients is the best known (46). The significance of QS in relation to infection has become evident in several studies. Erickson et al. (15) have shown that QS signal molecules produced by P. aeruginosa can be detected in biologically significant concentrations in sputum from CF patients. Furthermore, another signal molecule, the quinolone signal, has been found in sputum, bronchoalveolar fluid, and mucopurulent fluid from CF patients (8). Using a burned mouse model of infection, Rumbaugh et al. (39) demonstrated that QS-deficient mutants were less lethal then their wild-type counterparts. Apart from taking part in control of expression of virulence factors, AHL signal molecules also interact directly with the host organism. Smith et al. (42) found that 3-oxo-C12-HSL induces COX-2, a membrane-associated prostaglandin E synthase, as well as prostaglandin E2, resulting in inflammation of the CF lung. Other studies revealed that the 3-oxo-C12-HSL molecules can interfere with components of the immune system such as interleukin-8 and interleukin-12, modulating the response to the presence of P. aeruginosa (11, 44).
Due to an increasing number of untreatable, persistent infections, there is a rising need to develop novel strategies which deal with this phenomenon. Such infections often involve the biofilm mode of growth, which adds to the bacterium's tolerance to conventional antimicrobial treatment (3). Since QS seems to be a key player in regulation of virulence and the formation of tolerant biofilms (9, 25), it is an intriguing target for future antimicrobial chemotherapy.
Several authors have suggested enzymatic degradation of AHLs as a strategy employed by several organisms, including several Bacillus species, in minimizing the detrimental effects caused by QS-regulated products (12, 14, 28). N-Acyl homoserine lactonase, encoded by aiiA, attacks the lactone bond, causing ring opening of AHLs (13). Transgenic plants harboring the aiiA gene from Bacillus thuringiensis were much less prone to maceration by Erwinia carotovora. Two genes, attM and aiiB, both encoding AHL hydrolases, are found on the Ti plasmid in Agrobacterium tumefaciens. As with AiiA, these two enzymes, expressed in E. carotovora, reduce the virulence of this bacterium (5). Other bacteria, including Arthrobacter sp., Klebsiella pneumoniae, Pseudomonas sp., Variovorax sp., Comamonas sp., and Rhodococcus erythropolis harbor enzymes capable of AHL destruction (27, 33, 45).
Probably the best-studied example of QS inhibition stems from a marine alga, Delisea pulchra. This organism produces several halogenated furanone compounds capable of interfering with AHL-mediated signaling in bacteria (18, 38). The furanones prevent the AHLs from binding to the luxR homologues and eventually cause a rapid turnover of these proteins (29, 31). Manefield et al. (30) demonstrated the potential of the halogenated furanones in preventing the pathogen Vibrio harveyi from infecting the commercially important black tiger prawn Paneus monodon. Furthermore, synthetic furanone derivatives are able to inhibit QS in vitro (23) and attenuate infection by P. aeruginosa in vivo (25). The proof of concept delivered by Hentzer et al. (25) has encouraged us to search for nontoxic compounds that might serve as scaffolds for the development of future QS inhibitor (QSI) drugs. To achieve this, we have developed a novel rapid screen for QSIs, which at the same time identifies growth inhibitory substances. The systems are termed QSI selectors (QSIS). When growth is not affected, there is no selection pressure for the development of resistant bacteria. Furthermore, QSI drugs are not expected to eliminate communities of beneficial bacteria present in the host.
MATERIALS AND METHODS
Bacterial strains.
P. aeruginosa PAO1 was obtained from the Pseudomonas genetic stock center (http://www.ecu.edu/pseudomonas; strain PAO0001). The lasI rhlI AHL− PAO-JP strain is deficient in the production of signal molecules (35).
Construction of plasmid pTBR2iB for QSIS1.
Plasmid pTBR2iB was constructed as follows. PCR amplification with the primer set PluxR_end (5′-TTAATTTTTAAAGTATGGGCAATCAATTG-3′) and Plux_start (5′-GCATACTCATGCTCATAGTTAATTTCTCCTCTTTAA-3′) and with pJBA89 (1) as a template produced a 1,070-bp fragment encoding the V. fischeri luxR gene, including the regulatory part of the lux operon. The 3′ end of the fragment contains the first 10 bp of the phlA gene from Serratia liquefaciens (19). A second PCR with the primer set PphlA(F) (5-AACTATGAGCATGAGTATGCCTTTAAGTTTTACCTCTGC-3′) and Pphla(R) (5′-GATGCCGAAACCGAGCAGCG-3′) and with pMG330 (19) as a template was performed. This reaction generated a 357-bp fragment encoding a promoterless phlA gene. The 5′ end contains the 10 bp immediately upstream of the luxI start codon in pJBA89. A third PCR was performed with PluxR_end and PphlA(R) as primers and the fragments from the first two PCRs as a template. The reaction generated a 1,427-bp fragment encoding luxR and phlA; the latter gene was fused to the promoter region of luxI from pJBA89. The fragment was filled in with the Klenow fragment of DNA polymerase I and subsequently cloned into the SmaI site of pUC18not (10) by standard methods, generating pTBR2iB. QSIS1 was created by electrophoration of pTBR2iB into Escherichia coli 1100 (endA thi) (21).
Construction of plasmid pLasB-SacB1 for QSIS2.
A 1.4-kb fragment containing the structural gene sacB encoding Bacillus subtilis levansucrase (43) flanked by SphI and HindIII sites was PCR amplified with the primers sacB fwd (5′-GCACATGCATGCACATCAAAAAGTTTGC-3′) and sacB rev (5′-GCAAGCTTGCGTTTTTATTTGTTAACTG-3′) and pEX100T as a template (41). The sacB fragment was ligated into the corresponding sites of pMHLB2 (23). This produced a fusion, harbored on the Pseudomonas-E. coli shuttle vector pUCP22Not (26), between the P. aeruginosa lasB promoter and the B. subtilis sacB gene, followed by two strong transcriptional terminators from phage λ and the E. coli rrnB1 operon. Along with the plasB-sacB1 plasmid, this selector strain harbors the pSM1990 plasmid carrying constitutive expressed lux genes (non-QS dependent), giving rise to bioluminescence (pSU2007 carrying PMTluxCDABE) (32).
Construction of plasmid pPK22 for QSIS3.
A promoterless npt gene was PCR amplified from pRL1063a (48) with the sense primer 5′-TTGTAAGCTTAAGAGACAGGATGAGGATCG-3′ and antisense primer 5′-TTGTAAGCTTTCATTTCGAACCCCAGAGTC-3′, both introducing HindIII restrictions sites (underlined). The HindIII-digested PCR product was ligated into the unique HindIII site of the high-copy-number plasmid pJBA25, creating a promoterless GFP-npt operon in the plasmid pPK10. pJBA25 is a derivative of pJBA28 (2), with the only difference that pJBA25 contains the gfpmut3 instead of gfpmut3* in pJBA28. An EcoRI/KpnI fragment from pJBA89 (1), containing the luxR gene with its own promoter and upstream of luxR with the luxI promoter orientated in the opposite direction, was ligated into EcoRI/KpnI digested pPK10, creating pPK11. The coding region of the E. coli lambda phage cI857 repressor was PCR amplified from pMG300 (21) with the sense primers 5′-CAGGTACCGATTTAACGTATGAGCAC-3′ and antisense primer 5′-AGGGATCCATTTACTATGTTATGTTCTG-3′ introducing KpnI and BamHI restriction sites, respectively (underlined). The KpnI/BamHI-digested PCR product was ligated into KpnI/BamHI-digested pPK11, creating plasmid pPK12. Due to read-through from the luxI promoter into the GFP-npt operon in pPK12, the orientation of luxR-PluxI-cI857 was reversed. First, a derivative of pPK10 was constructed by inserting an EcoRI-BamHI fragment from the pUC18 polylinker into pPK10, creating plasmid pPK20. The luxR-PluxI-cI857 fragment was isolated from pPK12 as an EcoRI/BamHI fragment, and the 5′ end overhangs were filled in with the Klenow fragment from DNA polymerase I. This blunt-end fragment was ligated into SmaI-digested pPK20, creating the plasmid pPK21. The orientation of the luxR-PluxI-cI857 fragment in pPK21 was confirmed by restriction digestion analysis.
The Pr promoter from the E. coli lambda phage cI was PCR amplified from pMG300 with the primers 5′-AGGGTACCTCTATCACCGCAAGGG-3′ and 5′-CGGGTACCAGATCTTTAGCTGTCTTGG-3′, both introducing a BamHI site (underlined). The BamHI-digested PCR product was finally ligated into BamHI-digested pPK21, creating the QSIS plasmid pPK22. The correct orientation of the Pr promoter in pPK22 was confirmed by monitoring green fluorescent protein (GFP) expression.
Growth medium and conditions.
The medium used in this study was either Luria-Bertani (LB) medium (4) or ABT minimal medium supplemented with 0.5% glucose and 0.5% Casamino Acids (AB medium, containing 2.5 mg of thiamine/liter) (7). The medium was supplemented with antibiotics where appropriate. Unless otherwise stated, all strains were incubated at 37°C.
QSIS assays. Preparation of QSIS1 plates was performed in the following way. A total of 250 ml of ABT medium containing 2% agar was melted and cooled to 45°C. 3-Oxo-C6-HSL (Sigma-Aldrich, Seelze, Germany), ampicillin, 5-bromo-4-chloro-3-indoxyl-β-d-galactopyranoside (X-Gal), and isopropyl-β-d-thiogalactoside (IPTG) were added in final concentrations of 100 nM, 100 μg/ml, 40 μg/ml, and 100 μM, respectively. After mixing, 1 ml of overnight culture of QSIS1 in ABT supplemented with 0.5% (wt/vol) glucose and 0.5% (wt/vol) Casamino Acids was added. Agar plates were subsequently made by pouring 25 ml of the mixture in petri dishes (each, 7 cm in diameter). Preparation of QSIS2 plates was performed the following way. A total of 250 ml of LB agar (2% [wt/vol]) was melted and cooled to 50°C, after which 25 ml of A10 (7) and 14 g of sucrose were added. Gentamicin, 3-oxo-C12-HSL, and C4-HSL (Sigma) were added to final concentrations of 80 μg/ml, 200 nM, and 200 nM, respectively. After being cooled to 43°C, 5 ml of QSIS2 overnight culture in ABT supplemented with 0.5% (wt/vol) glucose and 0.5% (wt/vol) Casamino Acids was added. Plates were poured as described above. Preparation of QSIS3 plates was performed the following way. The selector strain QSIS3 was grown overnight in LB with kanamycin (50 μg/ml) at 30°C. LB with agar was melted and cooled to 45°C. Next, 3-oxo-C6-HSL was added to a final concentration of 100 nM, kanamycin was added to a final concentration of 65 μg/ml, and an overnight culture of the selector strain was carried out at a 1,000-fold dilution.
The medium was allowed to solidify, after which wells (4 mm in diameter) were made. A total of 50 μl of test substance was added to each well, and the plates were left for 1 h at room temperature, after which they were incubated overnight at 30°C (QSIS1 and QSIS3) or 37°C (QSIS2). The appearance of a circular growth zone around the reservoir indicated the presence of QSI active compounds in the sample. In case of QSIS1, a blue zone emerged with growth due to the presence of hydrolyzed X-Gal in the plate. During the work with the selector strains, it was noted that a newly outgrown (16 h postinoculation) culture gave the best result with respect to low background growth. As there is a very strong selection for mutations in the genes comprising the selector systems, it is important that incubation times are kept as short as possible.
The pure compounds were tested in concentrations of 10 μM unless otherwise stated. Herbal medicine and food sources were loaded directly if in liquid form or were extracted with 2 volumes (vol/wt) of methanol for 24 h, unless otherwise stated.
Extraction of garlic.
Prior to extraction, the stem and dead leaves were removed from the garlic bulbs. A total of 150 g of garlic cloves was shredded with a standard kitchen blender along with 300 ml of toluene. After extraction overnight, the suspension was filtered through Whatman no. 1 filter paper. A total of 150 ml of sterile water was added, and the mixture was stirred for 24 h at room temperature, after which the two phases were allowed to form. The water phase was separated from the organic phase, sterile filtered, and used as raw extract in the present work.
Dose response.
To establish a dose-response relationship of a putative QSI compound or a QSI-containing extract, a twofold serial dilution was made with growth medium (ABT with 0.5% [wt/vol] Casamino Acids) in a microtiter dish. Each well contained 100 μl of QSI solution (diluted). Next, 200 μl of an overnight culture (diluted 1:100) of P. aeruginosa PAO1 lasB-gfp(ASV) (23) was added. Growth was monitored as the optical density at 450 nm (OD450) over a time course of 16 h, and GFP expression was measured at 515 nm.
TLC assay.
Two-dimensional thin-layer chromatography (TLC) was performed with RP-18 F254S sheets (Merck). A 50-μl spot of garlic extract was applied to the TLC plate, and the plate was developed. For the first dimension, a methanol:water (60:40) mixture was used. For the second dimension, an ethanol:water (70:30) mixture was used. After elution, the plates were overlaid with 250 ml of ABT agar (2% [wt/vol]) containing 0.5% (wt/vol) glucose, 0.5% (wt/vol) Casamino Acids, 100 μg of ampicillin/ml, 100 nM 3-oxo-C6-HSL, 40 μg of X-Gal/ml, 100 μM IPTG, and 1 ml of QSIS1 overnight culture. The TLC overlay was incubated overnight at 30°C.
DNA array analysis.
A total of 200 ml of ABT minimal medium supplemented with 0.5% (wt/vol) Casamino Acids was inoculated with exponentially growing P. aeruginosa PAO1 cells (OD600 < 0.5) to an optical density of 0.05. At a density of 0.7, the culture was split in two 100-ml cultures. The cultures were grown in 500-ml conical flasks in an orbital air shaker operated at 200 rpm at 37°C. Either 2% (vol/vol) garlic extract or 100 μM 4-nitro-pyridine-N-oxide (4-NPO) was added to one culture, whereas the second culture served as an untreated control. Treated and untreated cultures showed similar growth. Samples were retrieved at OD600 = 2.0, mixed with 2 volumes of RNAlater (Ambion), and stored at −80°C until RNA extraction. RNA extraction was performed with the QIAGEN RNeasy purification kit with the bacterial protocol. To remove all DNA, the purified RNA was treated for 1h with 11 U of DNase I, and RNA was retrieved with the QIAGEN RNeasy purification kit. Synthesis of cDNA was done by mixing 10 μg of RNA with 250 ng of random primers (Invitrogen Life Technologies) in a total volume of 30 μl. The rest of the assay was performed according to the protocol supplied by Affymetrix.
Biofilm.
Biofilms were cultivated in continuous-culture once-through flow chambers perfused with sterile ABtrace minimal medium containing 0.3 mM glucose as described previously (23). Garlic extract (1% [vol/vol]) was added to the ABtrace medium where appropriate. Biofilm growth and development were examined by scanning confocal laser microscopy with an LSM 510 system (Carl Zeiss GmbH, Jena, Germany) equipped with an argon laser and a helium-neon laser for excitation of fluorophores. Tolerance of biofilms to tobramycin was assessed by introducing tobramycin (340 μg/ml) to the influent medium to 3-day-old biofilms. After 24 h, biofilms were examined by scanning confocal laser microscopy. Bacterial viability in biofilm cultures was assessed using the LIVE/DEAD BacLight bacterial viability staining kit (Molecular Probes, Inc., Eugene, Oreg.) as described previously (24).
Caenorhabditis elegans nematode model.
P. aeruginosa strains, PAO1 wt. and PAO1 lasIrhlI (23), were grown in 5 ml of LB liquid culture in the presence or absence of 2% (vol/vol) garlic extract overnight with shaking. Similar cultures were made with 100 μM 4-NPO. Culture samples (each, 15 μl) were then spread on BHI agar plates (diameter of each, 55 mm) containing either no or 2% (vol/vol) garlic extract. Likewise, plates containing 100 μM 4-NPO were made. Following overnight incubation at 30°C, five to seven L4 or adult wild-type Bristol N2 worms were transferred to the plates, which then were sealed with parafilm and incubated at 20°C. The number of living worms per plate was determined at various time points with a Stemi SV 6 microscope (Zeiss) at a magnification of ×50. Nematodes were considered dead when they failed to respond to tapping of the plate against the microscope stage. All experiments were carried out five times.
RESULTS AND DISCUSSION
Construction of QSIS.
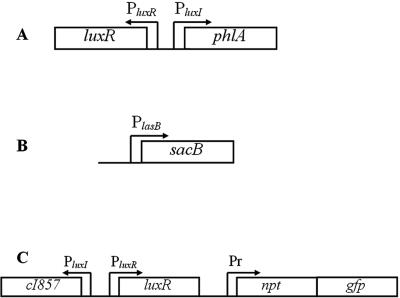
We devised two general types of QSIS systems, A and B (Fig. 1). The basic design of type A comprised a gene encoding a lethal protein (leading to growth arrest and cell death) fused to a QS-controlled promoter. Consequently, type A was unable to grow in the presence of AHL signal molecules unless a functional nontoxic QSI compound was present at a sufficiently high concentration. The basic design of type B employed an antibiotic resistance gene, which was controlled by a repressor. Expression of the repressor was in turn controlled by a QS-regulated promoter. In the presence of AHL, production of the repressor quenched expression of the antibiotic resistance gene, leading to growth inhibition in the presence of the appropriate antibiotic. However, if a QSI compound was present, downregulation of the repressor enabled growth of the bacteria.
FIG. 1.
The three QSIS systems. (A) QSIS1 phlA encodes the toxic gene product, expression of which is controlled by LuxR, the system established in E. coli. (B) In QSIS2, the LasR- and RhlR-regulated lasB promoter control expression of the sacB gene, expression of which leads to cell death in the presence of sucrose. The system was established in a lasI rhlI mutant of P. aeruginosa harboring a plasmid containing a constitutively expressed lux operon, which gives rise to expression of bioluminescence in growing cells. (C) The QSIS3 system is also based on LuxR regulation. The npt and gfp genes, conferring kanamycin resistance and green fluorescence, respectively, are controlled by the cI repressor, which in turn is regulated by QS through the luxI promoter. The system was established in E. coli.
QSIS bacteria supplemented with AHL signals were cast in agar plates according to the description in Materials and Methods. Test samples were applied to wells made in the agar. The samples diffused into the agar, creating a gradient with the highest concentration close to the well. The nature of the gradient depended on the molecules in the sample, i.e., large molecules diffused more slowly, less-soluble molecules diffused differently from easily solubilized ones, etc. Since the effect of the test compounds on growth of the bacterium is unknown, establishment of this concentration gradient is ideal, since all concentrations are tested at once.
The QSIS1 system was based on the luxI-gfp reporter made by Andersen et al. (1). The luxR gene and the promoter region of luxI were fused to phlA from S. liquefaciens MG1 (Fig. 1) (21). When expressed without its cognate antidote, PhlB, PhlA caused rapid lysis of the host cell (20). In the presence of a QSI compound and 3-oxo-C6 HSL, the majority of cells survived and grew at the position where the action of the AHL and the QSI counterbalanced each other. The active components of the QSIS1 system were present on an SmaI fragment in pUC18not, which in a lac+ E. coli background gave rise to a blue circle of growth on X-Gal-supplemented medium.
QSIS2 was based on the P. aeruginosa QS systems. The lasB promoter was fused to the levansucrase-encoding gene, sacB, leading to cell death in the presence of 3-oxo-C12-HSL, C4-HSL, and sucrose. This fusion is located on a pUCP22not plasmid harbored by a lasI rhlI mutant of PAO1 (Fig. 1B). The presence of a QSI compound rescued the cells, and a circle of growth arose at the position where the action of the AHL signals and the QSI counterbalanced each other. As an additional feature, we equipped the cells with bioluminescence for detection sensitivity.
In the presence of 3-oxo-C6-HSL, the QSIS3 system achieved repression of the npt gene by the luxR-PluxI quorum sensor (Fig. 1C). In the presence of a QSI, derepression of antibiotic resistance led to growth of the selector strain. We used the cI857 repressor and the corresponding Pr promoter to control the npt gene. As an additional feature of the selector, we inserted a promoterless gfp gene immediately downstream of the npt gene.
Calibration of the selector systems.
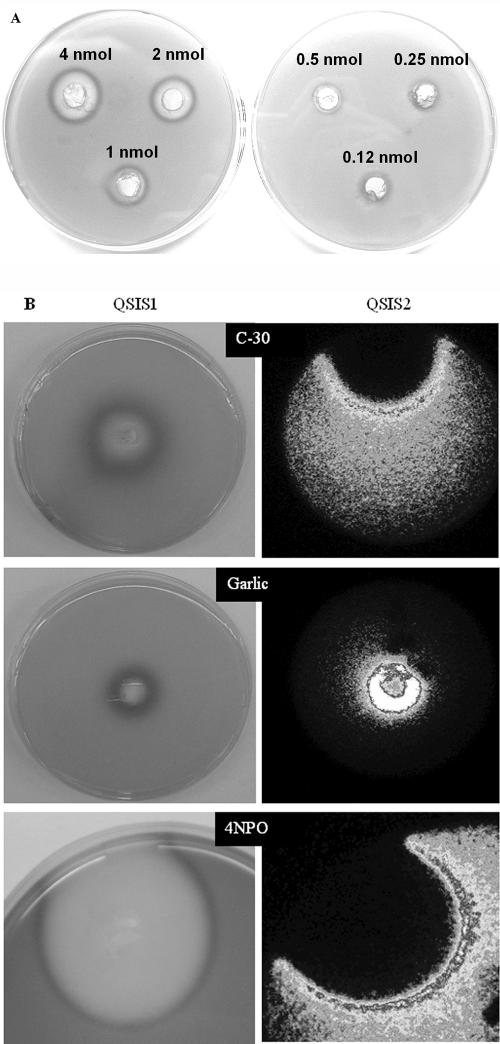
The halogenated furanones were indeed active with all three selector systems described. We used furanone 30 (25) and furanone 56 (23) to calibrate each system (Fig. 2A). The QSIS1 assay was optimized with respect to the concentration of 3-oxo-C6-HSL and inoculum size, as described in Material and Methods. The detection limit for furanone compound 30 with QSIS1 was 500 pmol.
FIG. 2.
(A) Calibration of QSIS 1 with different amounts (as indicated) of furanone compound 30. (B) QSIS1 (left) and QSIS2 (right) in action. The QSIS bacteria are cast into the agar along with appropriate AHLs, which activate the killing genes. Test samples as indicated were added to wells in the agar, and the compounds diffused from the well, creating a concentration gradient. Growth of the selector strains was visualized as blue (shown as a dark color, left) for QSIS1 (Lac+ phenotype on X-Gal agar) or as light emission (shown as a light color, right) for QSIS2 (constitutive bioluminescence expression) with a sensitive charge-coupled device camera.
QSIS2 was established in the lasI rhlI mutant of P. aeruginosa, and the assay was optimized with respect to C4-HSL and 3-oxo-C12-HSL, as well as to sucrose content and inoculum size (see Materials and Methods). We noted some false-positive results with this selector system. For example, high concentrations of glucose resulted in a strong positive result, probably due to interference with the sucrase killing system. Therefore, if QSIS2 is used to screen extracts that contain carbohydrates (such as extracts from fruit or berries) the results should be verified by QSIS1 or QSIS3.
The assay conditions for QSIS3 were optimized for kanamycin and 3-oxo-C6-HSL (see Materials and Methods). In the absence of AHL, the selector strain was able to grow in the presence of kanamycin in the range of 50 to 100 μg/ml. In the presence of both kanamycin and 3-oxo-C6-HSL (50 to 100 nM), the selector strain was unable to grow in LB agar at 30°C, but if the furanone compounds were added to the reservoir, growth was restored (data not shown).
Screening of extracts and/or compounds.
Since the recombinant lux QS system responds to a broad spectrum of signal molecules (2), we speculated that it would also respond to a broad spectrum of QSIs. Therefore, QSIS1 was used as the first screen. The las system responded almost exclusively to 3-oxo-C12-HSL (23), making it a more narrow-range screening system. Both QSIS1 and QSIS3 are based on the lux system; hence, they essentially screen for similar events—inhibition of LuxR activation. Since QSIS3 relies on the temperature-sensitive cI repressor, it is more difficult to handle in routine screenings and was therefore not used further in the present study.
We screened two different compound libraries; the first was based on extracts of several plants and herbs, and the second consisted of various pure chemical compounds (Table 1). Plants and herbs were extracted overnight with methanol, which does not in itself exhibit QSI activity in the screens (data not shown). The most active samples from the two groups, garlic and 4-NPO, were chosen for further studies (Fig. 2B). Extraction of garlic with different solvents revealed that toluene gave the most active extract (data not shown). Toluene did not exhibit QSI activity in our screens (data not shown). The crude toluene extract of garlic used in the initial screen exerted a strong growth inhibitory effect in addition to QSI activity. This is probably due to the high amount of allicin produced when garlic is crushed. We devised an extraction protocol to avoid most of the growth inhibitory compounds (see Materials and Methods). Extraction into an aqueous phase significantly reduced the growth inhibitory effect. We also tested pure, synthetic allicin and found that it did not give rise to a positive result with our systems (data not shown).
TABLE 1.
Compounds and extracts screened for QSI activitya
| Sample | QSI |
|---|---|
| Bean sprout | + |
| Blackberry | − |
| Brown onion | − |
| Chamomile | + |
| Carrot | + |
| Coffee | − |
| Cranberry | − |
| Poison ivy | − |
| Garlic | + |
| Gele Royal | − |
| Ginseng | − |
| Habanero | + |
| Honey | − |
| Clove | − |
| Leek | − |
| Mint tea | − |
| Propolis | + |
| Raspberry | − |
| Red chili | − |
| Spring onion | − |
| Tea tree oil | − |
| Water lily | + |
| Yellow pepper | + |
| Blood (plasma) | − |
| Stinging nettle | − |
| Anemone | − |
| Snowberry | − |
| 2-Methyl-imidazole | − |
| 2-Pyrrolidone | − |
| 4-Nitro-pyridine-N-oxide | + |
| 3-Hydroxy-benzaldehyde | − |
| P-Chlorobenzaldehyde | − |
| 2-Phenylcyclohexanone | − |
| P-Benzoquinone | + |
| Camphor | − |
| trans-1,2-Cyclohexandiole | − |
| Cycloheptane | − |
| Carbamic acid | − |
| 1,2,3-Benzo-triazole | − |
| 4-Hydroxymethyl-pyridine | − |
| trans-1,2-Diamino-cyclohexanesulfate | − |
| 2-Phenyl-imidazole | − |
| 2,4,5-Tri-bromo-imidazole | + |
| 2-Mercapto-benzimidazole | − |
| 3,6-Dipyridine | − |
| 1,4-Phenyl-1,2,3-triazole | − |
| Indole | + |
| 3-Amino-benzen-sulfonamide | − |
| Benzen-sulfon-amide | − |
| 3-Nitro-benzen-sulfonamide | + |
| 2-Oxazolidione | − |
| 2-Amino-4-methyl-pyridine | − |
| 1,4-Cyclohexan-dione | − |
| 4-Dimethyl-amino-pyridine | − |
Various food sources, herbal medicines, and a selection of pure chemical compounds are shown. QSIS1 was employed to screen the samples. +, sample has QSI activity; −, no activity.
Dose response of lasB expression.
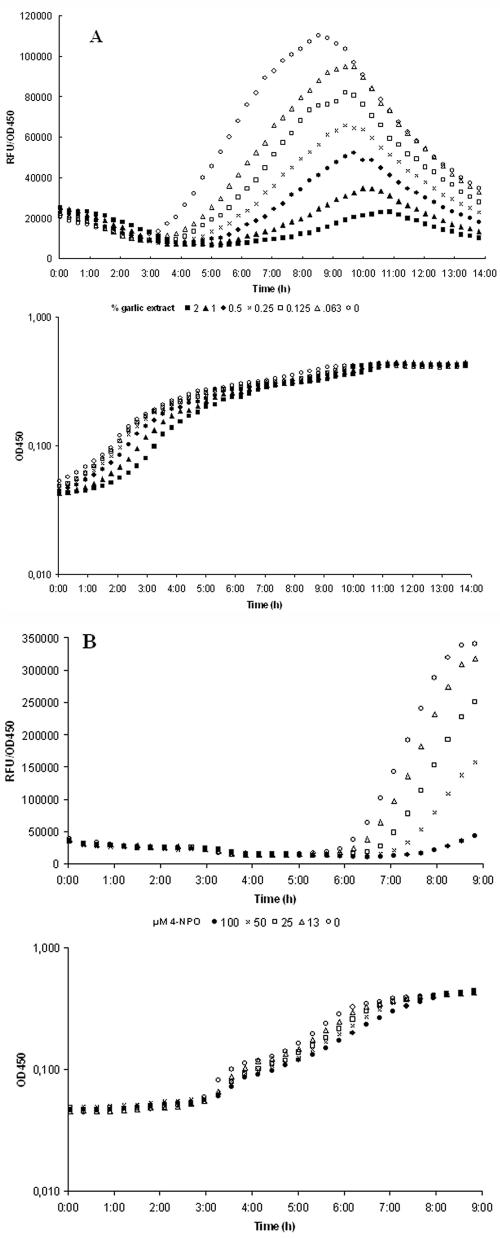
To further investigate the effect of the chosen substances on QS, we employed the lasB-gfp(ASV) QS monitor (23) harbored by PAO1. lasB is a type IV QS-controlled gene (48), which is maximally induced in the transition between exponential and stationary phases. This induction is reflected in a burst of green fluorescence. The presence of a QSI compound reduces this induction (23). As we only had a crude estimate of the QSI and subgrowth inhibitory concentrations from the selector assays, several different concentrations were tested. To determine the maximal concentration at which growth is not affected but maximum inhibition of the QS-regulated gene fusion is achieved, a 2-fold dilution series of each extract was incubated with a 100-fold dilution of an overnight culture of the QS monitor strain. Growth and fluorescence were monitored over time (Fig. 3A). During growth of this strain, lasB-gfp was induced more than 10 fold; however, when garlic extract was added to the growth medium, the onset, synthesis rate, and maximum induction were reduced in a concentration-dependent manner. At a concentration of 2% (vol/vol), the garlic extract significantly reduced the synthesis rate of GFP and lowered induction by about twofold. Usually our garlic extracts did not influence growth when used at concentrations below 1 to 2% (vol/vol). This indicates that garlic extract at this concentration interferes with the QS-controlled induction of lasB (Fig. 3A). We noted that differences in age (since picking), origin, and possible subspecies of garlic affect the dose-response relationship. Such batch variations make calibration of each extract obligatory. For 4-NPO, we found 100 μM to be the optimum concentration without affecting growth (Fig. 3B).
FIG. 3.
Dose-response curves of garlic extract (A) and 4-NPO (B). The QS monitor PAO1 (lasB-gfp) (23) was incubated with 2% (solid squares), 1% (solid triangles), 0.5% (solid circles), 0.25% (X), 0.13% (open circles), 0.06% (open triangles), or 0% (open circles) (vol/vol) garlic extract. Likewise, the QS monitor was treated with 100 μM (solid circles), 50 μM (X), 25 μM (open squares), 13 μM (open triangles), or 0 μM (open circles) 4-NPO. Growth and fluorescence were followed over time. RFU, relative fluorescence units.
Several QSI compounds are present in crude garlic extract.
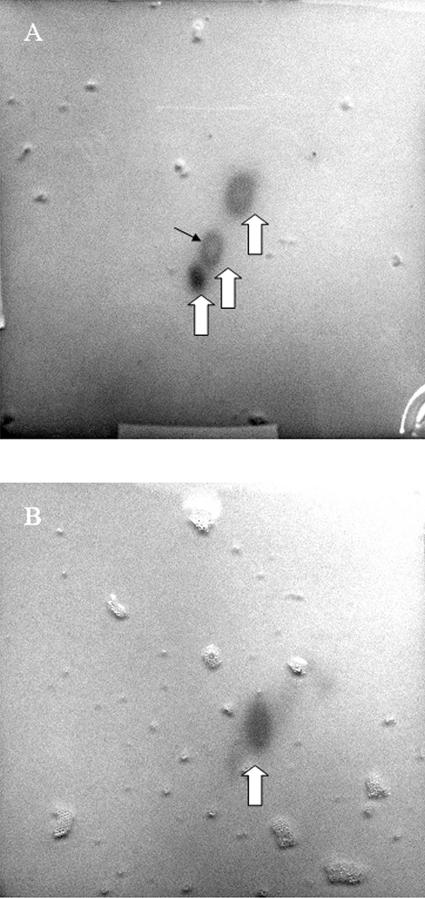
Crude toluene extract of garlic was spotted onto a reversed-phase TLC plate and developed in two dimensions, after which the TLC plates were cast into agar containing QSIS1. After incubation, three spots of QSI activity were observed, indicating that the extract contained more than one biologically active QSI compound (Fig. 4A). This assay was performed with the crude extract retaining the growth inhibitory compound(s). The no-growth area suggested that this QSI activity comigrated with growth inhibitory activity. TLC separation performed on the aqueous phase revealed the presence of only one compound with QSI activity (Fig. 4B). This compound did not block growth in the tested concentration.
FIG. 4.
Two-dimensional TLC of crude toluene extract of garlic (A) and an aqueous phase separated from the toluene (B). Compounds having QSI effect were visualized by overlaying the TLC plate with agar containing QSIS1, X-Gal, and 3-oxo-C6-HSL. Large arrows indicate areas with growth of strain QSIS1, and the small black arrow points to the center where growth inhibitory activity comigrates with QSI activity.
DNA microarray analysis.
With QSIS and promoter fusions, the QSI compounds were only tested with a few QS-regulated genes. To gain more comprehensive information about the target specificity, the effect of two different garlic extracts (G1 and G6) as well as the pure compound 4-NPO on the transcriptome of P. aeruginosa was assessed by Affymetrix DNA arrays. Cultures of PAO1 were brought into exponential growth and at OD600 of 0.7, cultures were treated with either 2% (vol/vol) garlic or 100 μM 4-NPO. Although induction and repression of QS-controlled genes formed a continuum throughout the growth cycle (40), a majority of the genes are maximally regulated at the transition to stationary phase (25); hence, growth was followed to OD600 of 2.0, at which point samples for array analysis were taken. Absolute expression values from treated cultures were compared to the corresponding values of the untreated control. Expression values from genes estimated not to be present by using the Micro Array suite software, version 5.0, were not included in the analysis. Changes in gene expression were reported as simple fold changes; a change in expression of less than fivefold has proved to be insignificant in previous studies (25, 41). Genes that were significantly (>5-fold) downregulated are listed in Table 2, whereas significantly upregulated genes are listed in Table 3. Performed under these restrictions, the analysis demonstrated that the two garlic extracts affected the expression of 167 (3%) genes in total (Tables 2 and 3). According to our previous mapping (25), 34% of the QS regulon was downregulated by garlic treatment. Affected genes were found among LasR-controlled genes (8% of total number of LasR-controlled genes (25), RhlR-controlled genes (34% of total RhlR-controlled genes) (25), and LasR- plus RhlR-controlled genes (62% of total LasR- and RhlR-controlled genes) (25), indicating that garlic treatment was less efficient against LasR-controlled gene expression. 4-NPO showed a broader spectrum of target genes, as it affected expression of 337 (6%) genes in total; 323 were downregulated and 14 were upregulated (Tables 2 and 3). A total of 37% of the QS regulon was found to be downregulated by this treatment. Affected genes were found among LasR (13%), RhlR (42%), and LasR plus RhlR (67%) genes. This indicates that the garlic compounds and 4-NPO preferentially target the RhlR receptor. When analyzed under similar restrictions, the well-established QSI, furanone compound 30, was found to inhibit 46% of the QS-regulated genes in P. aeruginosa PAO1. Affected genes were found among LasR (23%), RhlR (84%) and LasR plus RhlR (39%) genes. Of the genes regulated by garlic extract and 4-NPO, 53 and 49%, respectively, are of unknown function. This seems reasonable, as about 44% of the genes in the PAO1 genome have unknown functions (25).
TABLE 2.
Genes downregulated by two different garlic extracts (G1 and G6) and 4-NPO
| PA genea | Gene | Regulation | Descriptionb | Fold changec
|
||
|---|---|---|---|---|---|---|
| G1 | G6 | 4-NPO | ||||
| PA0012 | Hypothetical protein | 1.2 | −1.0 | −6.8 | ||
| PA0049 | Hypothetical protein | −31.7 | −12.6 | −76.3 | ||
| PA0059 | osmC | las + rhl | Osmotically inducible protein OsmC | −22.2 | −23.1 | −51.2 |
| PA0063 | Hypothetical protein | 1.3 | 1.3 | −14.5 | ||
| PA0105 | coxB | Cytochrome c oxidase, subunit II | −2.3 | −1.0 | −7.9 | |
| PA0106 | coxA | Cytochrome c oxidase, subunit I | −2.1 | −1.2 | −5.9 | |
| PA0107 | Conserved hypothetical protein | −14.4 | −6.2 | −23.3 | ||
| PA0108 | colII | Cytochrome c oxidase, subunit III | −10.3 | −4.3 | −5.7 | |
| PA0112 | Hypothetical protein | −6.9 | −2.1 | −15.2 | ||
| PA0113 | Probable cytochrome c oxidase assembly factor | −1.7 | −1.8 | −19.3 | ||
| PA0119 | Probable dicarboxylate transporter | −2.8 | −1.5 | −7.8 | ||
| PA0121 | Hypothetical protein | −1.4 | −1.3 | −5.6 | ||
| PA0122 | las + rhl | Conserved hypothetical protein | −4.5 | −4.5 | −14.6 | |
| PA0133 | Probable transcriptional regulator | −1.5 | 1.4 | −14.5 | ||
| PA0177 | Probable purine binding chemotaxis protein | −2.7 | −3.0 | −35.6 | ||
| PA0187 | Hypothetical protein | −3.8 | −4.1 | −16.6 | ||
| PA0188 | Hypothetical protein | −28.0 | −3.5 | −2.4 | ||
| PA0215 | Probable transporter | −1.0 | 1.1 | −7.6 | ||
| PA0216 | Probable transporter | −1,5 | −1.3 | −16.4 | ||
| PA0224 | Probable aldolase | −1.9 | −1.2 | −8.5 | ||
| PA0229 | pcaT | Dicarboxylic acid transporter PcaT | −5.8 | −3.1 | −1.7 | |
| PA0276 | Hypothetical protein | 0.0 | 0.0 | −7.1 | ||
| PA0279 | Probable transcriptional regulator | −2.1 | −1.4 | −12.7 | ||
| PA0355 | pfpI | Protease PfpI | −14.2 | −8.4 | −8.0 | |
| PA0397 | Probable cation efflux system protein | −2.5 | −1.5 | −10.4 | ||
| PA0534 | Conserved hypothetical protein | −3.8 | −4.0 | −13.2 | ||
| PA0567 | las + rhl | Conserved hypothetical protein | −21.9 | −14.9 | −11.4 | |
| PA0699 | Probable peptidyl-prolyl cis-trans isomerase, PpiC type | 0.0 | 0.0 | −11.4 | ||
| PA0755 | Probable porin | −2.0 | 1.3 | −5.6 | ||
| PA0797 | Probable transcriptional regulator | 2.3 | 1.1 | −9.0 | ||
| PA0798 | pmtA | Phospholipid methyltransferase | −1.7 | −1.3 | −15.3 | |
| PA0800 | Hypothetical protein | 0.0 | 0.0 | −35.9 | ||
| PA0803 | Hypothetical protein | −2.3 | −2.1 | −17.2 | ||
| PA0812 | Hypothetical protein | −5.1 | −1.1 | 1.2 | ||
| PA0828 | Probable transcriptional regulator | −1.8 | −1.1 | −8.0 | ||
| PA0846 | Probable sulfate uptake protein | 1.8 | −1.2 | −17.3 | ||
| PA0852 | cpbD | las + rhl | Chitin binding protein CbpD precursor | −14.4 | −6.2 | −16.9 |
| PA0873 | phhR | Transcriptional regulator PhhR | 0.0 | 0.0 | −5.0 | |
| PA0887 | acsA | Acetyl-coenzyme A synthetase | 3.6 | 1.9 | −5.4 | |
| PA0990 | Conserved hypothetical protein | −3.5 | −5.3 | −9.9 | ||
| PA0996 | pqsA | las | Probable coenzyme A ligase | −1.3 | 1,3 | −7.6 |
| PA1111 | Hypothetical protein | −5.2 | −5.0 | −6.9 | ||
| PA1190 | Conserved hypothetical protein | −2.1 | −1.6 | −5.2 | ||
| PA1202 | Probable hydrolase | −1.9 | −1.4 | −5.7 | ||
| PA1216 | Hypothetical protein | −3.4 | −2.2 | −13.3 | ||
| PA1242 | Hypothetical protein | −6.9 | −3.8 | −38.9 | ||
| PA1249 | aprA | las + rhl | Alkaline metalloproteinase precursor | −3.4 | −5.1 | −16.6 |
| PA1255 | Hypothetical protein | −1.1 | −1.6 | −9.8 | ||
| PA1290 | Probable transcriptional regulator | −1.8 | −1.5 | −8.6 | ||
| PA1297 | Probable metal transporter | −9.8 | −8.9 | −3.2 | ||
| PA1298 | Conserved hypothetical protein | −13.3 | −2.7 | −2.8 | ||
| PA1323 | las + rhl | Hypothetical protein | −26.5 | −19.5 | −32.3 | |
| PA1324 | las + rhl | Hypothetical protein | −22.5 | −19.1 | −21.6 | |
| PA1349 | Conserved hypothetical protein | −1.9 | −2.2 | −5.4 | ||
| PA1351 | Probable sigma-70 factor, ECF subfamily | −1.8 | −2.5 | −21.7 | ||
| PA1404 | Hypothetical protein | −2.7 | −3.2 | −8.8 | ||
| PA1412 | Hypothetical protein | −6.1 | −3.7 | −3.6 | ||
| PA1415 | Hypothetical protein | −2.3 | −2.6 | −12.6 | ||
| PA1418 | Probable sodium:solute symport protein | −3.4 | −2.8 | −5.2 | ||
| PA1419 | Probable transporter | 0.0 | 0.0 | −9.5 | ||
| PA1421 | gbuA | Guanidinobutyrase | 0.0 | 0.0 | −5.3 | |
| PA1470 | Probable short-chain dehydrogenase | −1.9 | −1.6 | −5.9 | ||
| PA1471 | Hypothetical protein | −6.5 | −4.7 | −4.4 | ||
| PA1475 | ccmA | Heme exporter protein CcmA | 1.4 | −1.0 | −13.0 | |
| PA1485 | Probable amino acid permease | 0.0 | 0.0 | −11.5 | ||
| PA1486 | Hypothetical protein | 0.0 | 0.0 | −9.0 | ||
| PA1514 | Conserved hypothetical protein | −3.2 | −3.4 | −15.5 | ||
| PA1523 | xdhB | Xanthine dehydrogenase | −2.2 | −2.2 | −24.4 | |
| PA1562 | acnA | Aconitate hydratase 1 | −4.3 | −3.3 | −6.2 | |
| PA1617 | Probable AMP-binding enzyme | −1.6 | −2.2 | −7.0 | ||
| PA1651 | Probable transporter | −1.4 | −1.3 | −7.8 | ||
| PA1652 | Hypothetical protein | −1.2 | 1.1 | −5.7 | ||
| PA1660 | las + rhl | Hypothetical protein | −8.5 | −2.5 | −10.6 | |
| PA1664 | las + rhl | Hypothetical protein | −8.6 | −2.1 | −4.6 | |
| PA1665 | las + rhl | Hypothetical protein | −5.2 | −3.8 | −1.8 | |
| PA1667 | las + rhl | Hypothetical protein | −5.0 | −2.2 | −3.4 | |
| PA1668 | Hypothetical protein | −2.7 | −1.7 | −13.9 | ||
| PA1669 | las + rhl | Hypothetical protein | −7.7 | −2.4 | −4.6 | |
| PA1670 | stp1 | Serine/threonine phosphoprotein phosphatase Stp1 | −4.9 | −3.3 | −54.3 | |
| PA1700 | Conserved hypothetical protein in type III secretion | 1.4 | 1.1 | −14.4 | ||
| PA1730 | Conserved hypothetical protein | −2.8 | −2.6 | −5.6 | ||
| PA1763 | Hypothetical protein | −1.7 | −1.5 | −13.7 | ||
| PA1782 | Probable serine/threonine-protein kinase | 0.0 | 0.0 | −27.6 | ||
| PA1784 | las | Hypothetical protein | −5.8 | −4.1 | −5.2 | |
| PA1840 | Hypothetical protein | 1.4 | 1.0 | −33.2 | ||
| PA1864 | Probable transcriptional regulator | −1.7 | −2.0 | −13.0 | ||
| PA1866 | Hypothetical protein | −2.1 | −2.1 | −36.0 | ||
| PA1869 | las + rhl | Probable acyl carrier protein | −3.1 | −1.8 | −8.8 | |
| PA1870 | Hypothetical protein | −7.1 | −2.9 | −5.7 | ||
| PA1871 | lasA | las + rhl | Las A protease precursor | −12.9 | −8.1 | −65.2 |
| PA1874 | Hypothetical protein | −5.5 | −8.0 | −10.1 | ||
| PA1875 | las | Probable outer membrane protein precursor | −7.5 | −7.5 | −45.8 | |
| PA1876 | Probable ATP binding/permease fusion ABC transporter | −4.1 | −5.8 | −38.3 | ||
| PA1881 | Probable oxidoreductase | −2.1 | −2.8 | −12.6 | ||
| PA1887 | Hypothetical protein | −2.2 | −6.2 | −3.8 | ||
| PA1888 | Hypothetical protein | −3.2 | −3.4 | −5.0 | ||
| PA1895 | Hypothetical protein | −1.3 | −1.3 | −7.9 | ||
| PA1901 | phzC2 | rhl | Phenazine biosynthesis protein PhzC | −7.2 | −3.9 | −16.1 |
| PA1902 | phzD2 | rhl | Phenazine biosynthesis protein PhzD | −7.4 | −5.1 | −17.4 |
| PA1903 | phzE2 | rhl | Phenazine biosynthesis protein PhzE | −10.0 | −4.4 | −11.5 |
| PA1904 | phzF2 | rhl | Probable phenazine biosynthesis protein | −7.5 | −5.3 | −221.1 |
| PA1905 | phzG2 | rhl | Probable pyridoxamine 5′-phosphate oxidase | −8.2 | −4.7 | −26.2 |
| PA1914 | Conserved hypothetical protein | −15.8 | −11.9 | −33.1 | ||
| PA1921 | Hypothetical protein | −34.0 | −3.0 | −18.4 | ||
| PA1956 | Hypothetical protein | 0.0 | 0.0 | −8.6 | ||
| PA1970 | Hypothetical protein | −2.3 | −1.3 | −6.4 | ||
| PA1978 | Probable transcriptional regulator | 0.0 | 0.0 | −23.7 | ||
| PA1985 | pqqA | Pyrroloquinoline quinone biosynthesis protein A | −1.8 | −2.1 | −7.8 | |
| PA1988 | pqqD | Pyrroloquinoline quinone biosynthesis protein D | 1.1 | −1.6 | −11.5 | |
| PA1989 | pqqE | Pyrroloquinoline quinone biosynthesis protein E | −1.3 | −1.9 | −5.3 | |
| PA1999 | Probable CoA transferase, subunit A | 1.1 | −2.3 | −8.6 | ||
| PA2000 | Probable CoA transferase, subunit B | 1.5 | −1.7 | −8.0 | ||
| PA2001 | atoB | Acetyl-CoA acetyltransferase | 1.5 | −1.9 | −9.1 | |
| PA2002 | Conserved hypothetical protein | −2.4 | −11.3 | −39.5 | ||
| PA2013 | Probable enoyl-CoA hydratase/isomerase | −1.2 | 1.3 | −8.6 | ||
| PA2021 | Hypothetical protein | −13.0 | −10.6 | −60.1 | ||
| PA2030 | las + rhl | Hypothetical protein | −6.8 | −4.7 | −18.4 | |
| PA2031 | las + rhl | Hypothetical protein | −10.8 | −4.4 | −17.3 | |
| PA2035 | Probable decarboxylase | 0.0 | 0.0 | −27.6 | ||
| PA2046 | Hypothetical protein | −47.2 | −16.3 | −31.2 | ||
| PA2050 | Probable sigma-70 factor, ECF subfamily | 2.4 | −2.2 | −6.7 | ||
| PA2066 | Hypothetical protein | −3.9 | −5.2 | −9.5 | ||
| PA2067 | Probable hydrolase | −6.1 | −5.7 | −6.9 | ||
| PA2068 | rhl | Probable MFS transporter | −8.8 | −5.7 | −18.6 | |
| PA2069 | rhl | Probable carbamoyl transferase | −24.8 | −11.3 | −34.1 | |
| PA2084 | Probable asparagine synthetase | 1.5 | −3.2 | −5.3 | ||
| PA2086 | Probable epoxide hydrolase | 1.1 | −3.3 | −5.1 | ||
| PA2088 | las + rhl | Hypothetical protein | 0.0 | 0.0 | −12.2 | |
| PA2090 | Hypothetical protein | −1.2 | −4.8 | −8.2 | ||
| PA2093 | Probable sigma-70 factor, ECF subfamily | −1.6 | −5.4 | −2.0 | ||
| PA2110 | Hypothetical protein | −13.4 | −15.7 | −70.7 | ||
| PA2111 | Hypothetical protein | −6.9 | −13.6 | −14.2 | ||
| PA2112 | Conserved hypothetical protein | −18.8 | −16.1 | −25.5 | ||
| PA2113 | Probable porin | −10.8 | −6.8 | −36.0 | ||
| PA2114 | Probable MFS transporter | −73.2 | −15.0 | −33.5 | ||
| PA2116 | Conserved hypothetical protein | −153.1 | −62.4 | −87.4 | ||
| PA2134 | Hypothetical protein | −2.3 | −2.4 | −29.8 | ||
| PA2137 | Hypothetical protein | 0.0 | 0.0 | −33.5 | ||
| PA2139 | Hypothetical protein | 0.0 | 0.0 | −5.5 | ||
| PA2142 | Probable short-chain dehydrogenase | −8.8 | −6.5 | −4.1 | ||
| PA2143 | Hypothetical protein | −30.8 | −8.1 | −67.1 | ||
| PA2144 | glgP | Glycogen phosphorylase | 0.0 | 0.0 | −40.2 | |
| PA2146 | Conserved hypothetical protein | −7.2 | −30.9 | −24.5 | ||
| PA2149 | Hypothetical protein | −4.8 | −3.7 | −8.3 | ||
| PA2151 | Conserved hypothetical protein | −92.9 | −78.8 | −24.2 | ||
| PA2153 | glgB | 1,4 α-Glucan branching enzyme | −9.5 | −6.6 | −26.0 | |
| PA2158 | Probable alcohol dehydrogenase (Zn dependent) | −4.5 | −2.7 | −17.7 | ||
| PA2159 | Conserved hypothetical protein | −1.1 | −1.8 | −7.9 | ||
| PA2162 | Probable glycosyl hydrolase | −2.8 | −4.3 | −12.4 | ||
| PA2163 | Hypothetical protein | −3.1 | −4.7 | −17.9 | ||
| PA2165 | Probable glycogen synthase | −1.5 | −2.1 | −5.6 | ||
| PA2166 | Hypothetical protein | −5.3 | −6.3 | −10.6 | ||
| PA2167 | Hypothetical protein | −12.2 | −6.8 | −126.9 | ||
| PA2168 | Hypothetical protein | −2.5 | −4.3 | −12.4 | ||
| PA2169 | Hypothetical protein | −4.1 | −5.9 | −3.0 | ||
| PA2170 | Hypothetical protein | −9.3 | −7.6 | −2.1 | ||
| PA2171 | Hypothetical protein | −3.0 | −2.2 | −9.4 | ||
| PA2172 | Hypothetical protein | −3.3 | −5.5 | −3.0 | ||
| PA2174 | Hypothetical protein | −3.7 | −3.1 | −5.9 | ||
| PA2175 | Hypothetical protein | −3.2 | −3.8 | −25.5 | ||
| PA2176 | Hypothetical protein | −13.2 | −6.5 | −143.6 | ||
| PA2184 | Conserved hypothetical protein | −3.5 | −3.1 | −65.9 | ||
| PA2187 | Hypothetical protein | −3.2 | −1.3 | −42.8 | ||
| PA2190 | Conserved hypothetical protein | −5.1 | −4.6 | −18.0 | ||
| PA2191 | exoY | Adenylate cyclase ExoY | −1.2 | −1.1 | −5.6 | |
| PA2198 | Hypothetical protein | −1.9 | −1.7 | −21.2 | ||
| PA2225 | Hypothetical protein | −2.2 | −1.3 | −10.2 | ||
| PA2226 | Hypothetical protein | −1.6 | −1.8 | −5.5 | ||
| PA2244 | Hypothetical protein | −3.5 | −5.4 | −3.3 | ||
| PA2245 | Hypothetical protein | −14.1 | −3.8 | −15.6 | ||
| PA2250 | lpdV | Lipoamide dehydrogenase-Val | −1.0 | 1.1 | −5.7 | |
| PA2251 | Hypothetical protein | −1.2 | 1.0 | −8.6 | ||
| PA2272 | pbpC | Penicillin binding protein 3A | 0.0 | 0.0 | −19.9 | |
| PA2277 | arsR | ArsR protein | 1.2 | −1.0 | −12.1 | |
| PA2278 | arsB | ArsB protein | −2.0 | −1.2 | −13.1 | |
| PA2296 | Hypothetical protein | 1.1 | −1.6 | −10.1 | ||
| PA2300 | chiC | rhl | Chitinase | −37.1 | −20.5 | −32.4 |
| PA2317 | Probable oxidoreductase | −1.3 | −1.4 | −10.6 | ||
| PA2322 | Gluconate permease | −2.9 | −2.4 | −49.5 | ||
| PA2414 | las + rhl | l-sorbosone dehydrogenase | −14.8 | −8.7 | −7.0 | |
| PA2415 | las + rhl | Hypothetical protein | −13.2 | −5.7 | −9.4 | |
| PA2433 | las + rhl | Hypothetical protein | −41.8 | −28.9 | −30.8 | |
| PA2448 | Hypothetical protein | −3.5 | −3.3 | −35.8 | ||
| PA2457 | Hypothetical protein | −1.4 | −1.3 | −9.8 | ||
| PA2485 | Hypothetical protein | −8.9 | −10.4 | −6.8 | ||
| PA2486 | Hypothetical protein | −6.9 | −6.4 | −5.0 | ||
| PA2505 | Probable porin | −1.5 | −1.2 | −8.1 | ||
| PA2528 | Probable RND efflux membrane fusion protein precursor | −1.1 | −1.5 | −5.3 | ||
| PA2534 | Probable transcriptional regulator | −1.4 | −1.7 | −6.3 | ||
| PA2557 | Probable AMP binding enzyme | −9.0 | −6.6 | −6.9 | ||
| PA2564 | las + rhl | Hypothetical protein | −2.5 | −18.8 | −1.9 | |
| PA2570 | palL | rhl | PA-I galactophilic lectin | −50.6 | −16.9 | −19.0 |
| PA2598 | Hypothetical protein | 2.9 | −1.8 | −20.7 | ||
| PA2602 | Hypothetical protein | 1.5 | −1.2 | −10.8 | ||
| PA2692 | Probable transcriptional regulator | −2.2 | 1.3 | −5.3 | ||
| PA2696 | Probable transcriptional regulator | −1.5 | −1.1 | −10.4 | ||
| PA2698 | Probable hydrolase | −1.5 | −1.1 | −12.6 | ||
| PA2701 | Probable MFS transporter | 1.2 | −1.9 | −6.2 | ||
| PA2708 | Hypothetical protein | −2.3 | −3.4 | −98.8 | ||
| PA2717 | cpo | Chloroperoxidase precursor | −4.2 | −4.9 | −39.8 | |
| PA2722 | Hypothetical protein | −2.7 | −2.2 | −7.9 | ||
| PA2746 | Hypothetical protein | −4.2 | −2.7 | −18.6 | ||
| PA2747 | las + rhl | Hypothetical protein | −18.8 | −16.6 | −11.5 | |
| PA2751 | Conserved hypothetical protein | −5.0 | −3.4 | −10.8 | ||
| PA2754 | Conserved hypothetical protein | −10.7 | −8.9 | −4.8 | ||
| PA2777 | Conserved hypothetical protein | −6.7 | −6.9 | −14.1 | ||
| PA2819 | Hypothetical protein | −0.0 | 0.0 | −5.2 | ||
| PA2862 | lipA | Lactonizing lipase precursor | −1.4 | −1.6 | −6.3 | |
| PA2893 | Probable very-long-chain acyl-CoA synthetase | −1.6 | −1.2 | −5.8 | ||
| PA2895 | Hypothetical protein | −2.3 | −3.8 | −11.1 | ||
| PA2909 | Hypothetical protein | 1.0 | −1.3 | −11.1 | ||
| PA2937 | Hypothetical protein | −3.8 | −3.9 | −13.7 | ||
| PA2939 | las | Probable aminopeptidase | −4.7 | −3.2 | −38.9 | |
| PA3019 | Probable ATP binding component of ABC transporter | 1.2 | −1.2 | −6.8 | ||
| PA3025 | Probable FAD dependent glycerol-3-phosphate dehydrogenase | −1.5 | −1.1 | −8.0 | ||
| PA3042 | Hypothetical protein | −1.8 | −4.0 | −8.2 | ||
| PA3181 | 2-Keto-3-deoxy-6-phosphogluconate aldolase | −1.4 | −1.6 | −8.8 | ||
| PA3183 | zwf | Glucose-6-phosphate 1-dehydrogenase | −2.2 | −3.2 | −5.1 | |
| PA3231 | Hypothetical protein | −11.8 | −12.0 | −24.7 | ||
| PA3234 | Probable sodium:solute symporter | −13.2 | −13.9 | −5.3 | ||
| PA3235 | Conserved hypothetical protein | 0.0 | 0.0 | −13.4 | ||
| PA3250 | Hypothetical protein | −1.7 | −1.7 | −6.2 | ||
| PA3251 | Hypothetical protein | −2.4 | −2.3 | −6.0 | ||
| PA3271 | Probable two-component sensor | −1.3 | −1.6 | −21.5 | ||
| PA3273 | Hypothetical protein | −4.5 | −5.9 | −14.4 | ||
| PA3274 | las + rhl | Hypothetical protein | −15.6 | −7.3 | −58.6 | |
| PA3316 | Probable permease of ABC transporter | −1.7 | −1.2 | −12.5 | ||
| PA3324 | Probable short-chain dehydrogenase | 0.0 | 0.0 | −9.9 | ||
| PA3332 | rhl | Conserved hypothetical protein | −1.7 | 1.6 | −5.7 | |
| PA3333 | fabH2 | rhl | 3-Oxoacyl-[acyl-carrier-protein] synthase III | −1.5 | 1.3 | −10.5 |
| PA3336 | rhl | Probable MFS transporter | −1.7 | 1.5 | −6.7 | |
| PA3361 | rhl | Hypothetical protein | −8.7 | −7.0 | −46.9 | |
| PA3365 | Probable chaperone | 0.0 | 0.0 | −18.0 | ||
| PA3366 | amiE | Aliphatic amidase | −1.7 | −2.0 | −6.8 | |
| PA3369 | Hypothetical protein | −5.4 | −6.4 | −41.8 | ||
| PA3370 | Hypothetical protein | −10.1 | −12.3 | −23.7 | ||
| PA3371 | las + rhl | Hypothetical protein | −9.8 | −9.5 | −84.9 | |
| PA3416 | Probable pyruvate dehydrogenase E1 component, beta chain | −4.1 | −6.3 | −5.8 | ||
| PA3451 | Hypothetical protein | −2.7 | −3.7 | −9.1 | ||
| PA3459 | Probable glutamine amidotransferase | −9.7 | −5.3 | −4.3 | ||
| PA3460 | Probable acetyltransferase | −9.9 | −5.7 | −14.0 | ||
| PA3461 | Conserved hypothetical protein | −6.0 | −7.3 | −8.6 | ||
| PA3466 | Probable ATP-dependent RNA helicase | 1.1 | 1.1 | −7.9 | ||
| PA3478 | rhlB | las + rhl | Rhamnosyltransferase chain B | −10.5 | −7.1 | −44.0 |
| PA3479 | rhlA | las + rhl | Rhamnosyltransferase chain A | −9.6 | −7.2 | −29.1 |
| PA3492 | Conserved hypothetical protein | −3.0 | −1.7 | −5.5 | ||
| PA3500 | Conserved hypothetical protein | −1.8 | −1.8 | −7.6 | ||
| PA3520 | las + rhl | Hypothetical protein | −8.5 | −9.1 | −87.3 | |
| PA3562 | Probable phosphotransferase system enzyme I | −1.7 | −1.8 | −13.2 | ||
| PA3581 | glpF | Glycerol uptake facilitator protein | 0.0 | 0.0 | −76.5 | |
| PA3584 | glpD | Glycerol-3-phosphate dehydrogenase | −1.9 | −1.4 | −9.2 | |
| PA3691 | las + rhl | Hypothetical protein | −13.4 | −9.4 | −12.8 | |
| PA3692 | las + rhl | Probable outer-membrane protein precursor | −22.3 | −14.2 | −14.0 | |
| PA3706 | Probable protein methyltransferase | −1.4 | −1.7 | −5.3 | ||
| PA3710 | Probable GMC-type oxidoreductase | 0.0 | 0.0 | −10.2 | ||
| PA3724 | lasB | las + rhl | Elastase LasB | −6.8 | −3.1 | −22.6 |
| PA3734 | Hypothetical protein | −7.3 | −5.0 | −32.9 | ||
| PA3739 | Probable sodium/hydrogen antiporter | −1.5 | −1.4 | −22.3 | ||
| PA3758 | Probable N-acetylglucosamine-6-phosphate deacetylase | −3.1 | −2.1 | −6.0 | ||
| PA3779 | Hypothetical protein | −1.7 | −2.0 | −5.1 | ||
| PA3788 | Hypothetical protein | −8.4 | −7.6 | −6.5 | ||
| PA3815 | Conserved hypothetical protein | −2.1 | −2.1 | −9.8 | ||
| PA3819 | Conserved hypothetical protein | −5.2 | −5.3 | −3.1 | ||
| PA3845 | Probable transcriptional regulator | −1.7 | −1.6 | −17.1 | ||
| PA3887 | nhaP | Na+/H+ antiporter NhaP | 1.0 | 1.1 | −17.2 | |
| PA3888 | Probable permease of ABC transporter | −7.1 | −8.6 | −8.7 | ||
| PA3889 | Probable binding protein component of ABC transporter | −4.8 | −6.7 | −10.5 | ||
| PA3890 | las + rhl | Probable permease of ABC transporter | −5.6 | −5.7 | −95.6 | |
| PA3898 | Probable transcriptional regulator | 1.2 | −1.4 | −18.3 | ||
| PA3920 | Probable metal transporting P-type ATPase | −1.3 | −1.8 | −28.4 | ||
| PA3923 | Hypothetical protein | −1.6 | −1.0 | −9.0 | ||
| PA3957 | Probable short-chain dehydrogenase | −3.6 | −1.8 | −5.9 | ||
| PA4078 | Probable nonribosomal peptide synthetase | −7.2 | −5.2 | −8.0 | ||
| PA4086 | cupB1 | Probable fimbrial subunit CupB1 | −1.6 | 1.1 | −11.2 | |
| PA4106 | Conserved hypothetical protein | −1.7 | −1.1 | −5.5 | ||
| PA4129 | las + rhl | Hypothetical protein | −2.6 | −6.5 | −7.2 | |
| PA4130 | las + rhl | Probable sulfite or nitrite reductase | −2.1 | −8.3 | −13.2 | |
| PA4133 | las + rhl | Cytochrome c oxidase subunit (cbb3 type) | −5.6 | −11.5 | −123.5 | |
| PA4134 | las + rhl | Hypothetical protein | −3.4 | −3.5 | −5.4 | |
| PA4138 | tyrS | Tyrosyl-tRNA synthetase | 0.0 | 0.0 | −11.7 | |
| PA4141 | las + rhl | Hypothetical protein | −6.6 | −4.4 | −23.7 | |
| PA4142 | las + rhl | Probable secretion protein | −7.2 | −5.6 | −33.9 | |
| PA4143 | Probable toxin transporter | −4.7 | −3.5 | −148.3 | ||
| PA4171 | Probable protease | −4.7 | −2.5 | −16.8 | ||
| PA4172 | Probable nuclease | −6.4 | −3.4 | −7.0 | ||
| PA4175 | prpL | las | Pvds-regulated endoprotease, lysyl class | −6.9 | −4.2 | −20.7 |
| PA4199 | Probable acyl-CoA dehydrogenase | −1.7 | −1.2 | −9.2 | ||
| PA4204 | Conserved hypothetical protein | −7.6 | −3.2 | −4.5 | ||
| PA4205 | mexG | Hypothetical protein | −4.1 | −3.7 | −6.0 | |
| PA4207 | mexl | Probable RND efflux transporter | −4.1 | −5.3 | −3.3 | |
| PA4209 | phzM | rhl | Probable phenazine-specific methyltransferase | −4.2 | −3.1 | −17.1 |
| PA4210 | phzA1 | rhl | Probable phenazine biosynthesis protein | −5.6 | −2.7 | −15.17 |
| PA4211 | phzB1 | rhl | Probable phenazine biosynthesis protein | −6.4 | −3.5 | −25.0 |
| PA4217 | phzS | rhl | Flavin-containing monooxygenase | −5.3 | −2.7 | −15.4 |
| PA4290 | Probable chemotaxis transducer | −4.4 | −8.1 | −27.1 | ||
| PA4294 | Hypothetical protein | −2.0 | −2.7 | −5.5 | ||
| PA4300 | Hypothetical protein | −2.8 | −3.5 | −5.7 | ||
| PA4302 | Probable type II secretion system protein | −3.6 | −8.5 | −3.2 | ||
| PA4303 | Hypothetical protein | −2.5 | −6.0 | −6.0 | ||
| PA4304 | Probable type II secretion system protein | −2.6 | −3.5 | −5.2 | ||
| PA4305 | Hypothetical protein | −3.4 | −4.3 | −21.1 | ||
| PA4306 | las + rhl | Hypothetical protein | −5.7 | −10.1 | −20.1 | |
| PA4342 | Probable amidase | 1.0 | −1.0 | −25.8 | ||
| PA4345 | Hypothetical protein | −4.9 | −5.7 | −10.8 | ||
| PA4346 | Hypothetical protein | −1.4 | −2.3 | −9.0 | ||
| PA4354 | Conserved hypothetical protein | −1.7 | −1.4 | −28.4 | ||
| PA4377 | Hypothetical protein | −4.7 | −6.1 | −15.9 | ||
| PA4391 | Hypothetical protein | −1.5 | −1.6 | −7.1 | ||
| PA4397 | panE | Ketopantoate reductase | −1.4 | −1.5 | −6.1 | |
| PA4435 | Probable acyl-CoA dehydrogenase | −1.0 | 1.4 | −5.4 | ||
| PA4456 | Probable ATP-binding component of ABC transporter | 1.5 | 1.3 | −5.9 | ||
| PA4573 | Hypothetical protein | −4.1 | −5.6 | −4.4 | ||
| PA4648 | Hypothetical protein | −5.2 | −8.8 | −11.9 | ||
| PA4649 | Hypothetical protein | −3.7 | −8.7 | −20.9 | ||
| PA4650 | Hypothetical protein | −5.4 | −4.3 | −31.2 | ||
| PA4651 | Probable pilus assembly chaperone | −3.4 | −5.7 | −4.3 | ||
| PA4653 | Hypothetical protein | −2.0 | −1.8 | −8.5 | ||
| PA4738 | las + rhl | Conserved hypothetical protein | −5.4 | −7.2 | −76.7 | |
| PA4739 | las + rhl | Conserved hypothetical protein | −11.4 | −11.3 | −28.1 | |
| PA4785 | Probable acyl-CoA thiolase | −4.8 | −4.5 | −10.1 | ||
| PA4786 | Probable short-chain dehydrogenase | −3.2 | −3.3 | −12.0 | ||
| PA4788 | Hypothetical protein | −1.7 | −1.7 | −13.9 | ||
| PA4816 | Hypothetical protein | −4.1 | −1.8 | −11.4 | ||
| PA4828 | Conserved hypothetical protein | −1.3 | 1.1 | −8.6 | ||
| PA4829 | lpd3 | Dihydrolipoamide dehydrogenase 3 | −1.3 | −1.7 | −14.3 | |
| PA4835 | Hypothetical protein | −11.9 | −2.3 | −2.1 | ||
| PA4869 | Hypothetical protein | −2.5 | −2.2 | −15.9 | ||
| PA4876 | osmE | Osmotically inducible lipoprotein OsmE | −13.5 | −12.9 | −10.2 | |
| PA4877 | Hypothetical protein | −4.0 | −5.7 | −3.9 | ||
| PA4880 | Probable bacterioferritin | −3.7 | −4.2 | −14.2 | ||
| PA4908 | Hypothetical protein | 0.0 | 0.0 | −8.4 | ||
| PA4910 | Probable ATP-binding component of ABC transporter | −1.5 | −1.2 | −18.7 | ||
| PA4980 | Probable enoyl-CoA hydratase/isomerase | 1.0 | −1.0 | −14.4 | ||
| PA5034 | hemE | Uroporphyrinogen decarboxylase | 0.0 | 0.0 | −6.1 | |
| PA5061 | Conserved hypothetical protein | −4.1 | −3.7 | −7.2 | ||
| PA5093 | Probable histidine/phenylalanine ammonia-lyase | 0.0 | 0.0 | −5.5 | ||
| PA5096 | Probable binding protein component of ABC transporter | 0.0 | 0.0 | −12.8 | ||
| PA5098 | hutH | Histidine ammonia-lyase | −2.9 | −6.6 | −2.6 | |
| PA5099 | Probable transporter | 0.0 | 0.0 | −21.9 | ||
| PA5100 | hutU | Urocanase | 1.6 | −3.2 | −5.2 | |
| PA5155 | Probable permease of ABC transporter | 1.0 | 1.5 | −8.3 | ||
| PA5171 | arcA | Arginine deiminase | −5.7 | −2.6 | 1.5 | |
| PA5172 | arcB | Omithine carbamoyltransferase, catabolic | −6.4 | −2.7 | 1.4 | |
| PA5173 | arcC | Carbamate kinase | −7.6 | −4.5 | −1.3 | |
| PA5212 | Hypothetical protein | −8.4 | −8.7 | −5.8 | ||
| PA5235 | glpT | Glycerol-3-phosphate transporter | −5.7 | −2.1 | −13.3 | |
| PA5297 | paxB | Pyruvate dehydrogenase (cytochrome) | −4.7 | −3.7 | −6.6 | |
| PA5352 | Conserved hypothetical protein | −1.3 | −1.3 | −5.1 | ||
| PA5383 | Conserved hypothetical protein | −5.7 | −3.1 | −5.9 | ||
| PA5457 | Hypothetical protein | 1.0 | 1.0 | −17.0 | ||
| PA5460 | Hypothetical protein | −6.8 | −2.8 | −2.0 | ||
| PA5468 | Probable citrate transporter | −1.9 | 1.2 | −6.7 | ||
| PA5481 | las + rhl | Hypothetical protein | −5.8 | −12.2 | −79.3 | |
| PA5482 | las + rhl | Hypothetical protein | −22.7 | −16.6 | −77.0 | |
| PA5503 | Probable ATP binding component of ABC transporter | 1.8 | −1.9 | −5.4 | ||
| PA5506 | Hypothetical protein | −5.0 | −3.1 | −4.5 | ||
| PA5517 | Conserved hypothetical protein | −1.1 | −1.1 | −13.6 | ||
| PA5541 | Probable dihydroorotase | −2.4 | −1.1 | −11.6 | ||
| PA5544 | Conserved hypothetical protein | −5.5 | −2.5 | −1.3 | ||
Genes marked with boldface type were found to be QS regulated (25).
Description from the Pseudomonas genome project (http://www.pseudomonas.com). CoA, coenzyme A; MFS, major facilitator superfamily.
Values are fold change in hybridization signal comparing a treated and untreated planktonic culture at OD600 = 2.0. Boldface type indicates a change of fivefold or higher. LasR/RhlR regulation is defined by Hentzer et al. (25).
TABLE 3.
Genes upregulated by two different garlic extracts (G1 and G6) and 4-NPO
| PA genea | Gene | Regulation | Descriptionb | Fold Changec
|
||
|---|---|---|---|---|---|---|
| G1 | G6 | 4-NPO | ||||
| PA0047 | Hypothetical protein | 3.0 | 2.6 | 5.8 | ||
| PA0083 | Conserved hypothetical protein | 2.5 | 3.3 | 5.8 | ||
| PA0085 | Conserved hypothetical protein | 3.1 | 3.6 | 8.8 | ||
| PA0197 | Hypothetical protein | 8.1 | −1.6 | −1.9 | ||
| PA0198 | exbB1 | Transport protein ExbB | 6.2 | −2.1 | −2.5 | |
| PA1556 | Probable cytochrome c oxidase subunit | 5.4 | 1.8 | 11.0 | ||
| PA1557 | Probable cytochrome oxidase subunit (cbb3 type) | 3.3 | 1.6 | 8.3 | ||
| PA1673 | Hypothetical protein | 2.0 | 1.8 | 18.1 | ||
| PA1715 | pscB | Type III export apparatus protein | 7.4 | 5.4 | 4.6 | |
| PA1718 | pscE | Type III export protein PscE | 4.4 | 2.7 | 6.5 | |
| PA1746 | Hypothetical protein | 1.1 | −1.1 | 7.1 | ||
| PA2285 | Hypothetical protein | −1.1 | −1.1 | 5.4 | ||
| PA2310 | Hypothetical protein | 5.9 | −3.2 | 1.0 | ||
| PA3283 | Conserved hypothetical protein | 2.4 | 2.4 | 25.4 | ||
| PA3284 | Hypothetical protein | 2.0 | 2.0 | 12.1 | ||
| PA3441 | las + rhl | Probable molybdopterin binding protein | 5.9 | −2.5 | −2.5 | |
| PA3442 | Probable ATP-binding component of ABC transporter | 9.2 | −1.9 | −2.5 | ||
| PA3444 | Conserved hypothetical protein | 8.3 | 1.1 | −1.1 | ||
| PA3445 | Conserved hypothetical protein | 5.3 | −1.1 | 1.3 | ||
| PA3570 | mmsA | Methylmalonate-semialdehyde dehydrogenase | 7.6 | 2.4 | −3.0 | |
| PA3720 | Hypothetical protein | 10.2 | 3.2 | 1.1 | ||
| PA3935 | tauD | Murine dioxygenase | 8.4 | −1.3 | 1.4 | |
| PA3938 | Probable periplasmic taurine binding protein precursor | 6.2 | −1.3 | −1.2 | ||
| PA4067 | oprG | Outer membrane protein OprG precursor | 2.3 | 1.6 | 5.9 | |
| PA4191 | Probable iron-ascorbate oxidoreductase | 5.9 | 1.2 | 1.0 | ||
| PA4195 | Probable binding protein component of ABC transporter | 7.1 | 1.7 | −1.1 | ||
| PA4710 | phuR | Hemoglobin uptake outer membrane receptor PhuR precursor | 4.6 | 5.5 | 2.7 | |
| PA5027 | Hypothetical protein | 1.3 | 1.8 | 6.3 | ||
| PA5054 | hslU | Heat shock protein HslU | 4.4 | 5.4 | 2.3 | |
| PA5083 | Conserved hypothetical protein | 5.8 | 2.3 | 1.2 | ||
| PA5475 | Hypothetical protein | −1.0 | −1.2 | 5.4 | ||
Gene marked with boldface were found to be QS regulated (25).
Description from the Pseudomonas genome project (http://www.pseudomonas.com).
Values are fold changes in hybridization signal comparing a treated and untreated planktonic culture at OD600 = 2.0; data in boldface indicate a change of fivefold or higher. LasR-RhlR regulation is defined by Hentzer et al. (25).
The two garlic extracts and 4-NPO have 111 target genes in common. Genes such as lasA and lasB (encoding elastase protease), rhlAB (encoding rhamolipids), and chiC (encoding chitinase), as well as aprA, phzA1B1, phzS, phzC2D2E2F2G2, and PA1L, were downregulated by the garlic extracts and 4-NPO. All these are involved in virulence and pathogenesis of P. aeruginosa.
The genes of P. aeruginosa have been divided into several functional classes (by PseudoCAP) (25). Garlic extract has a preference for genes belonging to the secreted factors (toxins, enzymes, and alginate) group, targeting 11 genes (22%) of that group. Similar results are seen with 4-NPO, as it targets 13 genes from that group. This correlates with the fraction of secreted factors (toxins, enzymes, alginate) genes regulated by QS. QS regulates 11 genes (22%) from this group (25), 10 of which match the genes targeted by garlic and 4-NPO. As rhamnolipid, phenazine, and other virulence factors are members of this group, it suggests that the garlic extracts can downregulate virulence of PAO1.
The garlic extract used in this transcriptome analysis is still a relatively crude extract containing a mixture of compounds. This may account for the effects on non-QS-regulated genes. However, it is fair to assume that the pure compound in the optimum concentration will exert a much more pronounced effect and have a higher specificity on the QS regulon. 4-NPO (which is a pure compound) downregulated a significant portion of the QS-regulated genes. However, it also affected expression of a number of non-QS-regulated genes. Together with the fact that 4-NPO is required at a 10-fold-higher concentration than furanone compound 30 (25), it indicates that 4-NPO is less efficient in affecting the QS regulon than the furanone compound.
Effect on biofilm tolerance to tobramycin.
It has previously been shown that P. aeruginosa biofilm cells are highly tolerant to antibiotic treatment (3). Davies et al. (9) demonstrated that a QS mutant of PAO1 is more susceptible to sodium dodecyl sulfate (SDS) treatment than the wild-type counterpart. Further, Hentzer et al. (25) showed that biofilms treated with a QSI became susceptible to both SDS and tobramycin. We tested if garlic extract exhibited a similar effect. Two sets of PAO1 biofilms were allowed to form for 3 days in flow chambers in which the medium contained either no garlic or 1% garlic. Following this, each biofilm was challenged with 340 μg of tobramycin/ml for 24 h. The effect of the antibiotic treatment was assessed by live/dead staining. In Fig. 5D, it can be seen that most of the cells in the biofilm treated with both garlic and tobramycin died (Fig. 5D). In striking contrast, only the cells in the top layer of the biofilm treated with tobramycin alone were killed (Fig. 5B). Further, Fig. 5C shows that treatment with garlic extract alone had no effect on biofilm viability. Comparing the untreated biofilm (Fig. 5A) with a garlic-treated one (Fig. 5C), a difference in architecture is observed. The untreated biofilm exhibited the classical mushrooms of a P. aeruginosa biofilm grown in glucose-containing medium. A flatter, undifferentiated biofilm was formed when the medium contained 1% garlic extract. This effect would be expected by QSI treatment, since a QS-defective lasI mutant of PAO1 has been found to form flat, undifferentiated biofilms (9).
FIG. 5.
Three-dimensonal images of 3-day-old PAO1 biofilms grown in the presence (bottom) or absence (top) of garlic extract. The biofilms are either untreated (left) or treated with tobramycin for 24 h (right). Bacterial viability was assessed with the BacLight viability stain: green (live)/red (dead).
C. elegans nematode model.
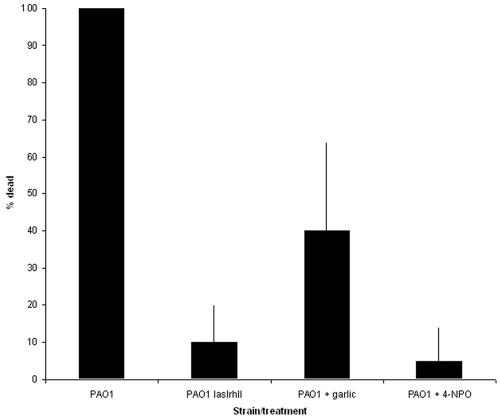
Previous work has shown that P. aeruginosa PAO1 kills C. elegans when the strain is grown on brain heart infusion (BHI) agar (17). However, a lasR mutant is strongly attenuated in this virulence model (17). Thus, the presence of a QS blocker in the medium is expected to abolish the ability of P. aeruginosa PAO1 to kill C. elegans. In agreement with a previous study (17), we observed that when C. elegans was placed on a lawn of P. aeruginosa PAO1 grown on BHI plates, the nematodes were killed within a few hours (Fig. 6). However, when the medium was supplemented with either 2% garlic extract or 100 μM 4-NPO, only 40 or 5% of the worms were killed within 5 h, respectively. As a comparison, a QS-deficient lasI rhlI double mutant killed 10% of the nematodes. These results provide strong evidence that garlic extract and 4-NPO reduce expression of pathogenic traits in PAO1.
FIG. 6.
Mortality of C. elegans nematodes living on a lawn of P. aeruginosa. Bars show an average of five experiments, and errors bars indicate the standard deviation between experiments.
In conclusion, we have developed recombinant bacteria which can be employed as rapid QSI screens. These QSIS systems have been used to screen a library of pure compounds and a selection of extracts from food sources and herbal medicine. We have identified a number of active QSI (Table 1) and further investigated the effects of the two most active: garlic extracts, which contain at least three different QSIs, and 4-NPO. Both were able to inhibit QS in a concentration-dependent manner. GeneChip analysis revealed that garlic extract and 4-NPO had a profound effect on genes, especially virulence genes, regulated by QS. Both garlic extract and 4-NPO significantly lowered the pathogenicity of P. aeruginosa PAO1 in a C. elegans nematode model. In a clinical context, drugs based on QSI compounds might become interesting, due both to the ability to downregulate expression of virulence factors and to the ability to make biofilms much more susceptible to conventional antibiotic treatment.
Work is currently in progress to identify the active QSI component in garlic extract, as well as to investigate the effect of garlic extract in a pulmonary mouse model.
Acknowledgments
Our work has received financial support from the Danish Technical Research Council, the Villum Kann-Rasmussen Foundation, the Cystic Fibrosis Foundation Therapeutics, Inc., and the Mukoviszidose e.V.
REFERENCES
- 1.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geisenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anwar, H., M. K. Dasgupta, and J. W. Costerton. 1990. Testing the susceptibility of bacteria in biofilms to antibacterial agents. Antimicrob. Agents Chemother. 34:2043-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlier, A., S. Uroz, B. Smadja, R. Fray, X. Latour, Y. Dessaux, and D. Faure. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 69:4989-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, A. B., K. Riedel, L. Eberl, L. R. Flodgaard, S. Molin, L. Gram, and M. Givskov. 2003. Quorum-sensing-directed protein expression in Serratia proteamaculans B5a. Microbiology 149:471-483. [DOI] [PubMed] [Google Scholar]
- 7.Clark, D. J., and O. Maaløe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 8.Collier, D. N., L. Anderson, S. L. McKnight, T. L. Noah, M. Knowles, R. Boucher, U. Schwab, P. Gilligan, and E. C. Pesci. 2002. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol. Lett. 215:41-46. [DOI] [PubMed] [Google Scholar]
- 9.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V, M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMango, E., H. J. Zar, R. Bryan, and A. Prince. 1995. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J. Clin. Investig. 96:2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, Y.-H., A. R. Gusti, Q. Zhang, J.-L. Xu, and L.-H. Zhang. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, Y.-H., L.-H. Wang, J.-L. Xu, H.-B. Zhang, X.-F. Zhang, and L.-H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 14.Dong, Y.-H., J.-L. Xu, X.-Z. Li, and L.-H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson, D. L., R. Endersby, A. Kirkham, K. Stuber, D. D. Vollman, H. R. Rabin, I. Mitchell, and D. G. Storey. 2002. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 70:1783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Givskov, M., R. de Nys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. D. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 178:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Givskov, M., L. Eberl, G. Christiansen, M. J. Benedik, and S. Molin. 1995. Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol. Microbiol. 15:445-454. [DOI] [PubMed] [Google Scholar]
- 20.Givskov, M., and S. Molin. 1993. Secretion of Serratia liquefaciens phospholipase from Escherichia coli. Mol. Microbiol. 8:229-242. [DOI] [PubMed] [Google Scholar]
- 21.Givskov, M., L. Olsen, and S. Molin. 1988. Cloning and expression in Escherichia coli of the gene for extracellular phospholipase A1 from Serratia liquefaciens. J. Bacteriol. 170:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardman, A. M., G. S. Stewart, and P. Williams. 1998. Quorum sensing and the cell-cell communication dependent regulation of gene expression in pathogenic and non-pathogenic bacteria. Antonie Leeuwenhoek 74:199-210. [DOI] [PubMed] [Google Scholar]
- 23.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 24.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, S. J., S. Y. Park, J. J. Lee, D. Y. Yum, B. T. Koo, and J. K. Lee. 2002. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl. Environ. Microbiol. 68:3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manefield, M., R. de Nys, N. Kumar, R. Read, M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145:283-291. [DOI] [PubMed] [Google Scholar]
- 30.Manefield, M., L. Harris, S. A. Rice, R. de Nys, and S. Kjelleberg. 2000. Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl. Environ. Microbiol. 66:2079-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manefield, M., T. B. Rasmussen, M. Henzter, J. B. Andersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2002. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148:1119-1127. [DOI] [PubMed] [Google Scholar]
- 32.Martinez, E., B. Bartolome, and F. de la Cruz. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159-162. [DOI] [PubMed] [Google Scholar]
- 33.Park, S. Y., S. J. Lee, T. K. Oh, J. W. Oh, B. T. Koo, D. Y. Yum, and J. K. Lee. 2003. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149:1541-1550. [DOI] [PubMed] [Google Scholar]
- 34.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen, T. B., M. Manefield, J. B. Andersen, L. Eberl, U. Anthoni, C. Christophersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2000. How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146:3237-3244. [DOI] [PubMed] [Google Scholar]
- 39.Rumbaugh, K. P., J. A. Griswold, B. H. Iglewski, and A. N. Hamood. 1999. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 67:5854-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 42.Smith, R. S., R. Kelly, B. H. Iglewski, and R. P. Phipps. 2002. The Pseudomonas autoinducer N-(3-oxododecanoyl) homoserine lactone induces cyclooxygenase-2 and prostaglandin E2 production in human lung fibroblasts: implications for inflammation. J. Immunol. 169:2636-2642. [DOI] [PubMed] [Google Scholar]
- 43.Steinmetz, M., D. Le Coq, H. B. Djemia, and P. Gay. 1983. Genetic analysis of sacB, the structural gene of a secreted enzyme, levansucrase of Bacillus subtilis Marburg. Mol. Gen. Genet. 191:138-144. (In French.) [DOI] [PubMed] [Google Scholar]
- 44.Telford, G., D. Wheeler, P. Williams, P. T. Tomkins, P. Appleby, H. Sewell, G. S. Stewart, B. W. Bycroft, and D. I. Pritchard. 1998. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect. Immun. 66:36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uroz, S., C. D'Angelo-Picard, A. Carlier, M. Elasri, C. Sicot, A. Petit, P. Oger, D. Faure, and Y. Dessaux. 2003. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology 149:1981-1989. [DOI] [PubMed] [Google Scholar]
- 46.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]