Abstract
We determined the transcriptional start site of the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by using a PCR-based method and identified the promoter. Mutagenic analysis indicated that the promoter-bearing region rather than its downstream inverted repeat sequence was responsible for the low-revel of trh transcription.
Vibrio parahaemolyticus is a marine bacterium and virulent strains can cause gastroenteritis in humans through consumption of contaminated seafood. The strain capable of producing thermostable direct hemolysin (TDH) or TDH-related hemolysin (TRH) is considered the virulent strain and the tdh and trh genes encode TDH and TRH, respectively (17). Our group has reported five sequence variants of the tdh gene (tdh1 to tdh5), and their sequences are >97% identical (2, 17). The trh sequence varies from strain to strain, and various trh genes could be clustered into two subgroups represented by the trh1 and trh2 genes. The trh1 and trh2 genes share 84% sequence identity, and both are approximately 68% identical to the tdh2 gene (9, 19). The tdh1 and tdh2 genes were present in a strain exhibiting Kanagawa phenomenon (KP), a clear beta-type hemolysis in a special blood agar (Wagatsuma agar) (16). The KP is due to the high-level transcription of the tdh2 gene but not the tdh1 gene (18, 21). The tdh3 to tdh5 genes were cloned from KP-negative strains and transcriptional levels of the tdh3 to tdh5 genes were as low as that of the tdh1 gene (21). The high-level tdh2 transcription responsible for KP is mostly due to a strong promoter that was characterized by having a single-base difference from other tdh genes at a particular position in the −35 region (21). The trh1 and trh2 genes were cloned from KP-negative stains. The amounts of TRH produced from trh1-positive strains of V. parahaemolyticus under an in vitro condition were at least 50-fold less than that of TDH from KP-positive strains as examined by an enzyme-linked immunosorbent assay (24). No quantitative detection method has been available for the trh2 gene product, but it was produced in a smaller amount than was the trh1 gene product when compared by a maxicell method in an Escherichia coli background (9). Assays of the lacZ transcriptional fusions integrated into the chromosome showed that trh1 and trh2 transcriptions were 127- and 117-fold lower, respectively, than tdh2 transcription in the E. coli MC1061 background and 6- and 4-fold lower, respectively, than that in the V. parahaemolyticus AQ3815 background (15). We wished to elucidate why the levels of trh transcription are low. Unlike the tdh genes, the trh1 and trh2 genes both contain a 12-bp inverted repeat sequence (IRS) immediately upstream from the coding region (9, 19). This IRS may hinder transcription originating in the trh promoters. Alternatively, the trh promoters may be weak. In order to investigate these possibilities, it was imperative to identify the trh promoter. For this purpose, we attempted to determine the transcriptional start sites of the trh1 and trh2 genes by a conventional primer extension analysis method. As templates, we used the total RNA extracted from the V. parahaemolyticus strains, AQ4037 carrying the trh1 gene (19) and AT4 carrying the trh2 gene (9), and E. coli strains each carrying the plasmid-borne trh1 or trh2 gene. However, none of the attempts was successful, probably because of low yield of the trh mRNAs (data not shown). In this study, we successfully applied a technique called the cRACE method (defined below) that has been used to analyze eukaryotic mRNAs (13; explained below) to identify the transcriptional start sites of the trh1, trh2, and tdh2 genes. This allowed us to identify the trh promoters and thus to examine the effect of the promoter strength and the effect of the neighboring inverted repeat sequence on trh transcription.
All bacterial strains were grown in Luria-Bertani (LB) broth or on LB agar-based medium containing 1% NaCl (14) unless otherwise specified. When E. coli strains carrying the plasmid were grown, an appropriate antibiotic was added to maintain the plasmid. Detailed information on the plasmids and oligonucleotides used in this study is available upon request from the corresponding author.
Determination of the transcriptional start sites of the trh1, trh2, and tdh2 genes by the cRACE method.
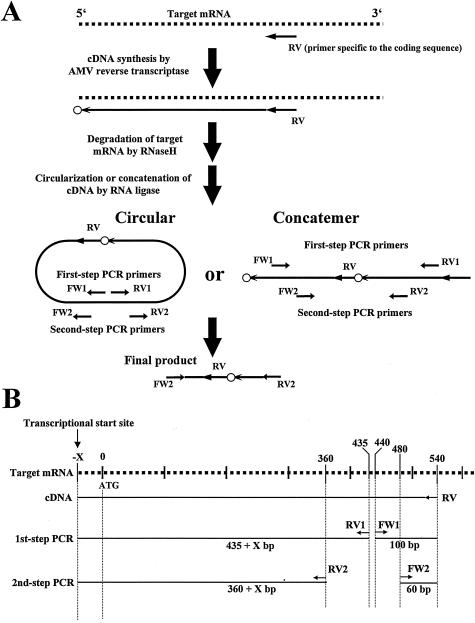
The protocol of the cRACE method used here is illustrated in Fig. 1A. Briefly, a partial cDNA sequence is reverse-transcripted from a known sequence of mRNA, is circularized (or concatenated), and rapidly amplified. The abbreviation “cRACE” stands for circular (or concatemeric) and rapid amplification of the cDNA end (5, 13, 20, 25). DNA sequencing of the second PCR product allows precise mapping of the transcriptional start site (the small open circle in Fig. 1A).
FIG. 1.
Outline chart showing the determination of the transcriptional start sites of the trh1 and trh2 genes by the cRACE method. (A) Schematic representation of the principles of the cRACE method (a slight modification of the method as reported by Maruyama et al. [13]). Oligonucleotide primers for cDNA synthesis and PCR are indicated by short horizontal arrows. Dotted lines and a long arrow represent the target mRNA and the first strand cDNA, respectively. The small open circle represents the nucleotide complementary to the transcriptional start site. (B) Locations of the primers and expected sizes of the first-strand cDNA and amplicons of the PCR. The dotted line and short arrows represent the target mRNA and primers, respectively. The straight lines depict cDNA or amplicons.
Derivatives of E. coli MC1061 (3) carrying a multicopy plasmid containing the trh1 or trh2 gene were cloned from V. parahaemolyticus AQ4037 (11) or strain AT4 (9). The strains were grown in LB broth at 37°C with shaking (160 rpm) to early log phase. The bacterial cells were harvested by centrifugation, and the total RNA was extracted with the Wizard SV Total RNA Isolation kit (Promega Corp., Madison, Wis.). The first-strand cDNA synthesis began with 6.75 μl of a reaction mixture containing 3.0 μg of total RNA, 0.5 μl of RV primer phosphorylated at the 5′ end (20 pmol/μl), 0.75 μl of 10× reverse transcriptase (RT) buffer (avian myeloblastosis virus [AMV] RT buffer; Life Sciences, Inc., St. Petersburg, Fla.), and distilled water. The reaction mixture was incubated at 70°C for 3 min; 0.25 μl of RNase inhibitor (40 U/μl; TaKaRa Bio, Inc., Otsu, Shiga, Japan) and 0.5 μl of AMV Reverse Transcriptase XL (35 U/μl; Life Sciences) were added; and the mixture was incubated at 30°C for 10 min, 50°C for 1 h, and finally at 80°C for 3 min. The total volume of the reaction mixture was increased to 29 μl by adding 21.5 μl of distilled water and 1 μl of RNase H (60 U/μl; TaKaRa) and incubated at 37°C for 30 min. Single-strand cDNA was precipitated with ethanol and subjected to a standard ligation reaction in a 20-μl volume.
The first-step PCR amplification was performed in a total volume of 25 μl containing 0.5 μl of the 10-fold-diluted ligation mixture (in TE buffer), 0.5 μl each of primers FW1 and RV1 (10 pmol/μl), 2 μl of 2.5 mM deoxynucleoside triphosphates (dNTPs), 2.5 μl of 10× buffer (Ex Taq buffer; TaKaRa), 0.25 μl of 5-U/μl Taq polymerase (Ex Taq; TaKaRa), and distilled water. The conditions for thermal cycling were denaturation at 94°C for 3 min followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. The expected amplicons should be larger than 535 (>435 + 100) bp (Fig. 1B). Amplicons of these sizes were isolated and served as the templates for the second-step (nested) amplification. The second-step amplification was performed with a total volume of 25 μl containing 0.5 μl of the template solution (ca. 10 ng of DNA/μl), 0.5 μl of each primer FW2 and RV2 (10 pmol/μl), 2 μl of 2.5 mM dNTPs, 2.5 μl of 10× buffer (Ex Taq buffer), 0.25 μl of 5-U/μl Taq polymerase (Ex Taq), and distilled water. The conditions for thermal cycling were denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. The expected amplicons in the second PCR should be larger than 420 (>360 + 60) bp (Fig. 1B). Only 460-bp amplicons were detected in this size range. The 460-bp amplicons were cloned into a plasmid vector, and their nucleotide sequences were determined. The transcriptional start site was linked to the 5′ end of RV primer (Fig. 1A) and was judged to be the C residue for both the trh1 and trh2 genes. The G residues complementary to the C residues were located 36 bases upstream from the coding region (Fig. 2).
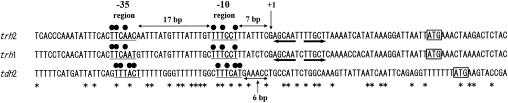
FIG. 2.
Alignment of the nucleotide sequences around the transcriptional start sites and the promoter regions of the trh1, trh2, and tdh2 genes. The base indicated by the boldface character and +1 is the transcriptional start site of each gene. The underlines depict the −10 and −35 regions of each promoter. The dots denote the bases showing identity with the consensus sequences of the E. coli promoter. The distances between the −35 and −10 regions and between the −10 region and the transcriptional start site are indicated. The bold arrows indicate IRS. The start (ATG) of the coding region is indicated by the box. The asterisk depicts the identical base among the three aligned sequences.
Similarly, we determined the transcriptional start site of the tdh2 gene, using the cRACE method. The total RNA extracted from an E. coli MC1061 derivative carrying a multicopy plasmid contained the tdh2 gene cloned from V. parahaemolyticus WP1 (16) and was used as the material for first-strand cDNA synthesis. The transcriptional start site of the tdh2 gene was found to be the T residue located 42 bases upstream from the coding sequence (Fig. 2).
It is easier to examine V. parahaemolyticus promoters by using the E. coli background rather than V. parahaemolyticus because many genetic tools are available for E. coli. To confirm the results obtained in the E. coli background, we also analyzed the total RNAs extracted from the V. parahaemolyticus strains, using the same cRACE method. In this E. coli-background cRACE experiment, we used the tdh2 gene previously cloned from V. parahaemolyticus WP1 (16). However, for the V. parahaemolyticus background, this strain is old and now grows more slowly than previously for an unknown reason. Therefore, we used strain AQ3815 carrying the tdh2 coding sequence and its upstream sequence, which are identical to those of the WP1 strain (16; unpublished data), and determined the tdh2 transcriptional start site. The transcriptional start site was identical to that of an E. coli derivative containing the tdh2 gene cloned from WP1. We used V. parahaemolyticus strains AQ4037 (trh1+) and AT4 (trh2+) to determine the transcriptional start sites of the trh1 and trh2 genes, respectively. However, we could not determine the transcriptional start sites by using the V. parahaemolyticus backgrounds because the amplicons of the expected sizes were not detected after the first-step amplification by the inverse PCR. This indicates that there is a limit to the sensitivity of the cRACE method for the analysis of bacterial mRNA, although it is more sensitive than primer extension analysis. However, using the E. coli background, we successfully determined the transcriptional start sites for the trh1 and trh2 genes by the cRACE method. This can be attributed to an increase in the relative amount of the target mRNA because we cloned the gene into a multicopy plasmid.
The tdh2 transcriptional start site of WP1 determined by the cRACE method was located 2 bases upstream from that determined by the primer extension analysis in our previous study (21). This discrepancy may be due to the resolution of the respective analytical methods. We used RNA extracted from E. coli carrying the cloned tdh2 gene and the same AMV reverse transcriptase in both studies. The transcriptional start site is empirically visually aligned in the conventional sequencing gel by the primer extension analysis, whereas it is located to the exact base by the advanced sequencing technique of the cRACE method. Therefore, we consider that the transcriptional start site determined by the cRACE method is more precise than that found by the primer extension analysis.
Identification of the promoters of the trh1, trh2, and tdh2 genes.
The nucleotide sequences upstream of the trh1, trh2, and tdh2 coding regions are compared in Fig. 2. The transcriptional start sites of the three genes determined by the cRACE method as described above are indicated (position +1) in this figure. Considering the location of the transcriptional start site, in homology with the consensus sequences of the −10 and −35 regions of the E. coli promoter (10), we identified the −10 and −35 regions of the trh1, trh2, and tdh2 promoters as shown in Fig. 2. The relative locations of the promoter regions and the transcriptional start sites and −10 and −35 sequences of the three genes matched well, except that the distances between the −10 region and the transcriptional start site were 7 bp for the trh1 and trh2 genes and 6 bp for the tdh2 gene. Consensus −35 and −10 sequences constructed from three promoters would be T100T100C67A100A67T67 and T100T100T100C100C67T100, respectively, where the subscript denotes the percent occurrence of the most frequently found base. The trh1 promoter sequence was identical with the consensus promoter sequence. The space between the −35 and −10 sequences was 17 bases without exception. The high similarities among the three promoters support the hypothesis that these genes were derived from a common ancestor (17). To the best of our knowledge, other than the tdh2, trh1, and trh2 promoters, promoters were described for only 10 of the V. parahaemolyticus gene sequences deposited in GenBank. Two of these promoters were identified by primer extension analysis (6, 7), but the others are putative promoters determined from homologies with consensus sequences. We have compared the sequence of the trh1 promoter with those of the 10 non-tdh/trh promoters. The identities were 60, 50, 0, 50, 30, and 50% for the bases in the −35 sequence; 80, 0, 70, 10, 10, and 80% for those in the −10 sequence; and 50% for the 17-base spacing. The low identity values might support the hypothesis that the tdh and trh genes were acquired by V. parahaemolytiucs from another organism (17). This interpretation needs to be supported by identification of the promoter sequences of other V. parahaemolytiucs genes.
Effect of the IRSs on trh1 and trh2 transcriptions.
The 12-bp IRSs of the trh1 and trh2 genes were located immediately downstream from the transcriptional start sites (Fig. 2). To examined whether the IRSs influence the transcription of these genes, we mutated (deleted or made substitutions in) the IRS of the trh1 and trh2 genes and the corresponding region of the tdh2 gene by using the PCR-based tail-to-tail mutagenesis method of Imai et al. (8) (Fig. 3A). We then measured the transcriptional levels of the genes with or without the mutation, using a lacZ transcriptional fusion vector, pUJ8 (4). The fusion genes were placed in a multicopy plasmid in the E. coli MC1061 background. They were also subcloned into a suicide vector, pUTminiTn5Sm/Sp (4), and integrated into the chromosome of V. parahaemolyticus AQ3815 by conjugation on the membrane filter as described previously (15, 18). Three or more colonies were randomly selected for each construction, and their β-galactosidase activities were measured as described previously (23).
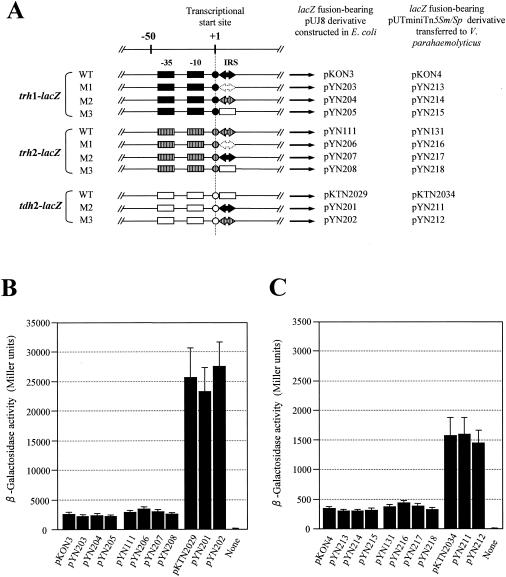
FIG. 3.
Transcriptional levels of the trh1-, trh2-, and tdh2-lacZ fusion genes with or without mutations in the IRS. (A) Schematic representation of the promoter and IRS regions of the trh1, trh2, and tdh2 genes with or without mutations in the IRS that were fused to the lacZ gene. Two boxes, a circle, and large arrows (solid, open, or hatched) indicate the −35 and −10 regions of the promoter, the transcriptional start site, and the IRS, respectively. WT, wild-type sequence without mutation; M1, deletion of the IRS; M2 and M3, replacement of the IRS with the other IRS or corresponding tdh2 sequence. The long arrows indicate the location and the transcriptional direction of the lacZ gene, but the vector sequence is not shown. The designations of the pUJ8 derivatives or pUTminiTn5Sm/Sp derivatives containing the respective lacZ fusion constructs are indicated. β-Galactosidase activities of the E. coli MC1061 derivatives and V. parahaemolyticus AQ3815 derivatives carrying the plasmids indicated in panel A are shown in panels B and C, respectively. “None” indicates the parent strain without a lacZ fusion (a negative control). Each result represents the mean ± standard deviation determined from three or more randomly selected colonies.
There was no significant difference in the transcriptional level between the wild-type and mutants of each of the trh1-, trh2-, and tdh2-lacZ fusions in the E. coli background (P > 0.05) (Fig. 3B) and in the V. parahaemolyticus background (P > 0.05) (Fig. 3C). The results clearly indicate that the IRSs of the trh1 and trh2 have no influence on the transcription of not only the trh1 and trh2 genes but also the tdh2 gene.
We searched the whole genome sequence of V. parahaemolyticus RIMD2210633 (12; http://genome.gen-info.osaka-u.ac.jp/bacteria/vpara) for homology with the IRS of the trh1 and trh2 genes. The trh1 IRS homologues with 100% identity were detected within the coding sequences of four genes encoding proteins. The trh2 IRS homologues with 100% identity were detected within the coding sequences of 13 genes coding for proteins, putative proteins, or hypothetical proteins and in two other locations outside the genes (noncoding regions). Detection of the IRS homologues mostly inside the coding regions is in favor of the conclusion that these IRSs are not involved in transcriptional control of V. parahaemolyticus genes.
Effect of the promoter-bearing region on trh1 and trh2 transcriptions.
Since high-level transcription of the tdh2 gene was attributable mostly to its promoter strength (21), we examined the effect of the promoter-bearing region located upstream of the transcriptional start site on the trh1 and trh2 transcriptions. We replaced the upstream region among the trh1-, trh2-, and tdh2-lacZ fusions, using the PCR-ligation-PCR mutagenesis method of Ali et al. (1), and monitored their transcriptional levels in the E. coli and V. parahaemolyticus backgrounds, using the lacZ fusion system as described for the mutation in the IRS (Fig. 4).
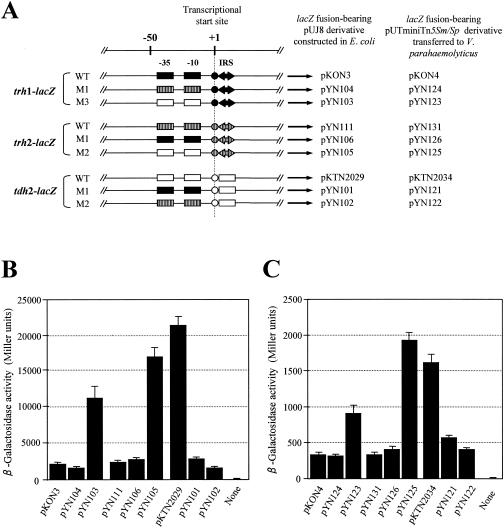
FIG. 4.
Transcriptional levels of the trh1-, trh2-, and tdh2-lacZ fusion genes with or without replacement in the promoter-bearing region located upstream from the transcriptional start site. (A) Schematic representation of the promoter and IRS regions of the trh1, trh2, and tdh2 genes with or without replacement in the promoter region that were fused to the lacZ gene. Two boxes, a circle, and large arrows (solid or hatched) indicate the −35 and −10 regions of the promoter and IRS, respectively. WT, wild-type sequence without replacement; M1, M2, and M3, replacement of the promoter-bearing region. The long arrows indicate the location and transcriptional direction of the lacZ gene, but the vector sequence is not shown. The designations of the pUJ8 derivatives or pUTminiTn5Sm/Sp derivatives containing the respective lacZ fusion constructs are indicated. β-Galactosidase activities of the E. coli MC1061 derivatives and V. parahaemolyticus AQ3815 derivatives carrying the plasmid indicated in panel A are shown in panels B and C, respectively. “None” indicates the parent strain without a lacZ fusion (a negative control). Each result represents the mean ± standard deviation determined from three or more randomly selected colonies.
A similar tendency was observed in the results obtained with the E. coli (Fig. 4B) and V. parahaemolyticus (Fig. 4C) backgrounds. The transcription of the trh1- and trh2-lacZ fusions increased five- to sevenfold in the E. coli background and three- to fivefold in the V. parahaemolyticus background when their upstream sequences were replaced with the tdh2 upstream sequence, but not very significantly (P > 0.05) when replaced with the other trh upstream sequence. On the other hand, the tdh2-lacZ transcription was reduced 7- to 12-fold in the E. coli background and 3- to 4-fold in the V. parahaemolyticus background when the upstream region was replaced with that of the trh1 and trh2 fusion genes. The above results show that the low transcriptional levels of the trh1 and trh2 genes are ascribable to the nucleotide sequences of the promoter-bearing upstream region.
The trh-bearing strains are unique in that they produce urease. The trh gene is located in close proximity to the urease gene cluster but this gene cluster has no influence on trh expression (15, 22). Others have reported that there exists the ureR gene, which is a presumed regulator of the urease gene cluster, approximately 0.4 kb upstream of the trh1 gene (22). We previously determined only the 38- and 252-bp nucleotide sequences upstream of the trh1 and trh2 coding regions, respectively (9, 19). We determined 741- and 1,927-bp nucleotide sequences upstream of the trh1 and trh2 coding regions that were contained in the recombinant plasmids used in this study. The 3′ end of the ureR gene was detected in the 438- and 434-bp upstream regions of the trh1 and trh2 coding regions, respectively; but there were no other unique structures including an inverted repeat sequence or a possible gene in the upstream sequences. It is likely therefore that the trh promoters contained in the upstream regions are responsible for low-level transcriptions of the trh1 and trh2 genes. The significant difference in the transcription level between the tdh2 and other tdh1 genes was most likely due to a single-base difference in the −35 region of the tdh promoter (21). Interestingly, the A residue unique to the −35 region of the tdh2 promoter and is essential for high-level transcription of the tdh2 gene was also found in the trh promoters (Fig. 2) but the homology of the −35 and −10 regions of the trh promoters with the consensus sequence of the E. coli promoter (10) were lower than that of the tdh2 promoter (Fig. 2). One or more of the bases in the −35 or −10 regions that are not completely conserved among the trh1, trh2, and tdh2 genes or the 7-base distance between the −10 region and the transcriptional start site (Fig. 2) may be responsible for the low level of the trh transcription. Our hypothesis on the weak trh promoter needs to be supported by a future study including site-directed mutagenesis experiments as previously carried out for the tdh genes (21).
Nucleotide sequence accession number.
The nucleotide sequences of the coding regions and upstream regions of the trh1, trh2, and tdh2 genes determined in this study have been deposited into the DDBJ/EMBL/GenBank databases under accession no. AB112353, AB112354, and AB112355, respectively.
Acknowledgments
This research was supported in part by a Grant-in Aid for Scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan; by the US-Japan Cooperative Medical Science Program, Cholera and Related Diarrheal Diseases, Japanese Panel; and by the “Research for the Future” Program from The Japan Society for the Promotion of Science (JSPS-RFTF97L00706).
REFERENCES
- 1.Ali, S. A., and A. Steinkasserer. 1995. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques 18:746-750. [PubMed] [Google Scholar]
- 2.Baba, K., H. Shirai, A. Terai, Y. Takeda, and M. Nishibuchi. 1991. Analysis of the tdh gene cloned from a tdh gene- and trh gene-positive strain of Vibrio parahaemolyticus. Microbiol. Immunol. 35:253-258. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 4.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. MiniTn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative bacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNA from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funahashi, T., T. Moriya, S. Uemura, S.-I. Miyoshi, S. Shinoda, S. Narimatsu, and S. Yamamoto. 2002. Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus. J. Bacteriol. 184:936-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funahashi, T., T. Tanabe, H. Aso, H. Nakao, Y. Fujii, K. Okamoto, S. Narimatsu, and S. Yamamoto. 2003. An iron-regulated gene required for utilization of aerobactin as an exogenous siderophore in Vibrio parahaemolyticus. Microbiology 149:1217-1225. [DOI] [PubMed] [Google Scholar]
- 8.Imai, Y., Y. Matsushima, T. Sugiura, and M. Terada. 1991. A simple and rapid method for genetating a deletion by PCR. Nucleic Acid Res. 19:2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishishita, M., N. Matsuoka, K. Kumagai, S. Yamasaki, Y. Takeda, and M. Nishibuchi. 1992. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 58:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewin, B. 2000. Genes VII. Oxford University Press, New York, N.Y.
- 11.Lin, Z., K. Kumagai, K. Baba, J. J. Mekalanos, and M. Nishibuchi. 1993. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J. Bacteriol. 175:3844-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 13.Maruyama, I. N., and T. L. Rakow, and H. I. Maruyama. 1995. cRACE: a simple method for identification of the 5′ end of mRNA. Nucleic Acid Res. 23:3796-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, H. 1972. Experiments in molecular genetics. Cold Spring Harbor, Cold Spring Harbor Laboratory Press, N.Y.
- 15.Nakaguchi, Y., J. Okuda, T. Iida, and M. Nishibuchi. 2003. The urease gene cluster of Vibrio parahaemolyticus does not influence the expression of the thermostable direct hemolysin (TDH) gene or the TDH-related hemolysin gene. Microbiol. Immunol. 47:233-239. [DOI] [PubMed] [Google Scholar]
- 16.Nishibuchi, M., and J. B. Kaper. 1990. Duplication and variation of the thermostable direct haemolysin (tdh) gene in Vibrio parahaemolyticus. Mol. Microbiol. 4:87-99. [DOI] [PubMed] [Google Scholar]
- 17.Nishibuchi, M., and J. B. Kaper. 1995. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63:2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishibuchi, M., K. Kumagai, and J. B. Kaper. 1991. Contribution of the tdh1 gene of Kanagawa phenomenon-positive Vibrio parahaemolyticus to production of extracellular thermostable direct hemolysin. Microb. Pathog. 11:453-460. [DOI] [PubMed] [Google Scholar]
- 19.Nishibuchi, M., T. Taniguchi, T. Misawa, V. Khaeomanee-Iam, T. Honda, and T. Miwatani. 1989. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect. Immun. 57:2691-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic application of an inverse polymerase chain reaction. Genetics 120:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuda, J., and M. Nishibuchi. 1998. Manifestation of the Kanagawa phenomenon, the virulence-associated phenotype, of Vibrio parahaemolyticus depends on a particular single base change in the promoter of the thermostable direct haemolysin gene. Mol. Microbiol. 30:499-511. [DOI] [PubMed] [Google Scholar]
- 22.Park, K.-S., T. Iida, Y. Yamaichi, T. Oyagi, K. Yamamoto, and T. Honda. 2000. Genetic characterization of DNA region containing the trh and ure genes of Vibrio parahaemolyticus. Infect. Immun. 68:5742-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsot, C., and J. J. Mekalanos. 1990. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc. Natl. Acad. Sci. USA 87:9898-9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirai, H., H. Ito, T. Hirayama, Y. Nakamoto, N. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triglia, T., M. G. Peterson, and D. J. Kemp. 1988. A procedure for in vitro amplification of cDNA segments that lie outside the boundaries of known sequences. Nucleic Acid Res. 16:8186. [DOI] [PMC free article] [PubMed] [Google Scholar]