Abstract
Synthesis of ribosomes in Escherichia coli requires an antitermination system that modifies RNA polymerase to achieve efficient transcription of the genes specifying 16S, 23S, and 5S rRNA. This modification requires nucleotide signals in the RNA and specific transcription factors, such as NusA and NusB. Transcription of rrn operons in strains lacking the ability to produce either NusA or NusB was examined by electron microscopy. The distribution and numbers of RNA polymerase molecules on rrn operons were determined for each mutant. Compared to the wild type, the 16S gene in the nusB mutant strain had an equivalent number of RNA polymerase molecules, but the number of RNA polymerase molecules was reduced 1.4-fold for the nusA mutant. For both mutant strains, there were twofold-fewer RNA polymerase molecules on the 23S RNA gene than for the wild type. Overall, the mutant strains each had 1.6-fold-fewer RNA polymerase molecules on their rrn operons than did the wild type. To determine if decreased transcription of the 23S gene observed by electron microscopy also affected the 30S/50S ribosomal subunit ratio, ribosome profiles were examined by sucrose gradient analysis. The 30S/50S ratio increased 2.5- to 3-fold for the nus mutant strains over that for wild-type cells. Thus, strains carrying either a nusA mutation or a nusB mutation have defects in transcription of 23S rRNA.
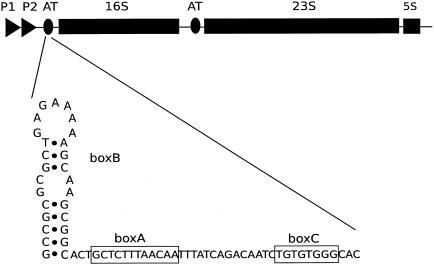
In the synthesis of rRNA in Escherichia coli, RNA polymerase undergoes several rrn operon-specific modifications that double the transcription elongation rate and allow read-through of Rho-dependent terminators (1, 7, 29, 30, 32). The modification events require specific nucleotide sequences in the leader and spacer regions of the rrn operons as well as interacting proteins that alter the properties of the transcribing polymerase. Thus, profound changes in the basic capabilities of RNA polymerase occur when rrn operons are transcribed, but these changes and their consequences have yet to be clearly described. The nucleotide sequence signals leading to the changes in RNA polymerase activity in rrn operon transcription are called BoxB, BoxA, and BoxC in the leader sequence and BoxB and BoxA in the spacer region between the 16S and 23S genes. BoxA is a conserved 12-nucleotide sequence (Fig. 1), BoxB is a conserved stem-loop structure, and BoxC is a GT-rich region (3, 19). These sequences are related to those used in the bacteriophage lambda N protein antitermination system (8, 12). Protein factors involved in rrn antitermination (rRNA-AT) are less well defined, but they likely include the N utilization substances (Nus) NusA, NusB, NusE (ribosomal protein S10), NusG, and ribosomal protein S4 (15, 23, 24, 27, 29, 30, 32). In vivo studies have documented the importance of NusB and NusG for rRNA-AT and, in conjunction with BoxA, for increasing the rate of elongation by RNA polymerase (28, 32). NusG, NusE (S10), and S4 are all essential proteins. It was assumed that NusB would also be an essential cellular protein, but in studies with transport protein mutants, two groups isolated suppressors of secretion mutations (secYts and secA5ts) that mapped to the nusB gene. One mutational change, ssyB, was an insertion of IS10 into the nusB gene (nusB::IS10), while another, ssaD1, introduced a nonsense codon early in the gene (17, 21, 25). The fact that cells containing a disrupted or dysfunctional nusB gene are viable suggests that NusB is not an essential protein. Cell physiology studies revealed that cells containing the nusB::IS10 mutation have a greatly increased doubling time and are cold sensitive. They also have a decreased peptide elongation rate at low temperatures, suggesting a potentially intriguing relationship between NusB and peptide synthesis (21). A point mutation in nusB, nusB5, has previously been examined for its effects on rRNA synthesis. A Y18D mutational change affecting a surface residue in the NusB5 protein results in defective lambda phage replication and defective rRNA transcription (2, 20). Compared to synthesis of the rrn leader region, 1.45-fold and 1.6-fold decreases in the amounts of 16S and 23S rRNA, respectively, have been documented. This drop-off in expression of 16S and 23S rRNA in nusB5 was interpreted as a loss of the rRNA-AT system leading to transcription termination within the rrn operon (20). Sucrose gradient profiles of the nusB5 mutant strain suggest that its 70S ribosomes are unstable and that it has a higher proportion of free ribosomal subunits than do wild-type strains when grown at 42°C (26). Another study of how antitermination features of rrn operons affect rRNA transcription focused on mutant BoxA sequences in the leader and spacer regions carried on plasmids encoding an rrn operon. The total ribosome content from strains carrying such plasmids was examined, and the proportion encoded by the mutant operon was determined. Quantitation of plasmid-encoded 16S and 23S rRNA in 30S and 50S subunits and 70S ribosomes showed that the leader BoxA mutation causes a 25% decrease in both 16S and 23S rRNA and a further 15% decrease in plasmid-encoded 23S in 50S subunits when a spacer BoxA mutation is added (11, 16). These in vivo results with nusB and BoxA mutants demonstrate the importance of each for efficient rRNA synthesis.
FIG. 1.
Schematic diagram of the rrnB operon showing the regulatory BoxBAC region and its position relative to the 16S gene in the leader and the 23S gene in the spacer region.
Although NusA seems by some tests to be an essential protein, a particular transposon insertion in nusA permits continued viability (4). In addition, Zheng and Friedman (33) were able to delete nusA in a strain that also carries a mutation in the Rho termination factor gene that causes Rho activity to be reduced eight- to ninefold (ΔnusA::cat rhoE134D). They were unable to delete nusA alone and hence postulated that the transcription pausing caused by binding of NusA to RNA polymerase is necessary to keep RNA polymerase from outpacing translation and revealing lethal Rho-binding and termination sites normally covered by translating ribosomes. The ΔnusA::cat rhoE134D mutant strain, like the nusB::IS10 mutant, has a slow-growth phenotype and is cold sensitive.
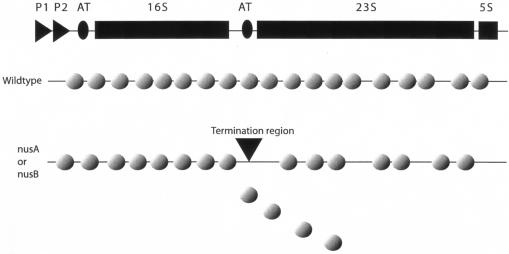
Because strains with mutations in nusA or nusB are viable but debilitated, we set out to determine directly what effect these mutations have on transcription of the rrn operons. Using electron microscopy, we visualized the number and position of transcribing RNA polymerase molecules on rrn operons in mutants that lack a fully functional antitermination system. Based on previous measurements of rrn expression in nusB5 and rRNA-AT mutants, we expected to find regions in the rrn operons where RNA polymerase molecules were falling off the DNA. We found that the number and spacing of RNA polymerase molecules on the 16S gene of the nusB::IS10 mutant were similar to those found on the rrn operons in a wild-type strain, but there was a twofold drop in the number of RNA polymerase molecules on the 23S gene. In the ΔnusA::cat rhoE134D strain, there was a 1.4-fold decrease in RNA polymerase molecules on the 16S gene as well as a twofold drop on the 23S gene. Sucrose gradient profiles of ribosomes from both of the nus mutant strains showed a 50S subunit peak that was decreased relative to the 30S peak, a result consistent with the decreased expression of 23S rRNA. Our data suggest that in the absence of Nus factors A and B, transcription of rrn operons is incomplete because RNA polymerase molecules leave the DNA near the 16S-23S junction. Apparently, there is a transcription block or terminator in this region that releases a portion of transcription complexes when a Nus factor is lacking.
MATERIALS AND METHODS
Strains.
Antitermination-defective mutant strains IQ527 (MC4100 nusB::IS10) (21, 25) and K7906 (ΔnusA::cat rhoE134D) (33) were grown in Luria broth at 37°C. The MC4100 and K4633 rhoE134D strains were used as reference strains for sucrose gradient analyses (6, 33).
Electron microscopy.
Chromatin spreads were prepared from mid-log-phase cultures as previously described by French and Miller (9). The dispersed chromatin was centrifuged onto carbon-coated copper electron microscope grids (Ted Pella, Redding, Calif.). The grids were negatively stained in uranyl acetate and phosphotungstic acid, rotary shadowed with platinum, and viewed with a Phillips CM10 transmission electron microscope. Electron-microscopic image negatives were digitized on an Epson 636 scanner at a resolution of 600 dpi. Analysis of these digitized images was performed with ImageJ software (18).
Sucrose gradients of ribosomes.
Ribosomes were prepared from mid-log-phase cells grown in Luria broth as previously described by Godson and Sinsheimer (10). Briefly, cell pellets were resuspended and incubated in 1/50th-volume TKM buffer (20 mM Tris HCl [pH 7.6], 60 mM KCl, 10 mM MgCl2, 20% sucrose) with lysozyme (150 μg/ml) for 20 min. Cells were collected by centrifugation and lysed during three freeze-thaw cycles. Additional lysis and clarification of the sample were achieved by resuspending the cells in 1/100th volume of a solution of TKMD buffer (Brij-35 [0.5%] and RQ1 RNase-free DNase I [0.05 U/μl] in TKM buffer). The mix was incubated on ice for 20 min. The lysate was centrifuged at 16,000 × g at 4°C for 10 min, and 12 A260 units of sample was layered onto a 10 to 40% sucrose gradient prepared in TKM buffer. Gradients were centrifuged in a Beckman SW41 rotor at 35,000 rpm for 2.5 h. The gradient was extracted with a Piston Gradient Fractionator (Biocomp, New Brunswick, Canada), and ribosome profiles were measured through a UV flow cell at 254 nm. Peaks were quantitated by using the fityk analysis package (http://www.unipress.waw.pl/∼wojdyr/fityk/).
RESULTS
Visualization of rRNA transcription in nusA and nusB mutant strains.
To observe rrn operon transcription directly, we used electron microscopy to examine chromosomal spreads of E. coli strains with mutations in the genes encoding NusA or NusB. From the image data collected, we counted the RNA polymerase molecules on the 16S and 23S genes, measured the distance between adjacent RNA polymerase molecules, and determined RNA polymerase distribution across the operon.
Electron microscopy provides a unique perspective into the transcriptional activity of individual genes that is difficult to obtain using conventional biochemical methods. The rrn operons are identified on the chromosome as sites of high transcriptional activity with a characteristic spatial arrangement of RNA polymerase molecules and nascent rRNA transcripts, suggestive of a double Christmas tree, that is easy to recognize (9).
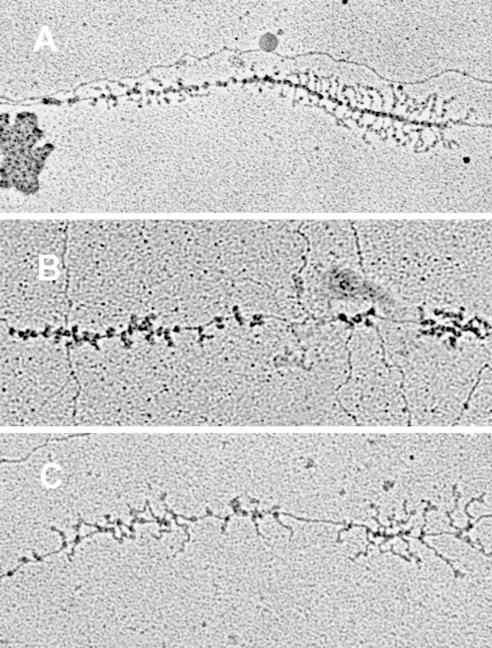
Figure 2 shows electron micrographs of an rRNA operon from each of the two nus mutant strains as well as a wild-type control for comparison. On each operon there are two gradients of increasing transcript length resulting from left-to-right transcription and RNase III cleavage of the nascent transcripts between the 16S and 23S cistrons. Based on the relative scarcity of RNA polymerases and nascent rRNA transcripts on the second gradient, it is evident that the mutant strains were deficient in transcription of the 23S gene. Total RNA polymerase numbers and their individual positions along each operon that could be unambiguously traced from the 5′ end to the 3′ end were recorded for the ΔnusA::cat rhoE134D and nusB::IS10 mutant strains and compared to those for a wild-type control (Table 1). The 3′ position of the 16S gene was determined as the point of transcript cleavage. RNA polymerase numbers were consistent between the wild type and the nusB::IS10 mutant on the 16S gene but were down 1.4-fold for the ΔnusA::cat rhoE134D strain. A twofold decrease in the number of RNA polymerase molecules engaged in 23S rRNA synthesis was found for both nus mutant strains, accounting for most of the 1.6-fold decrease in RNA polymerase numbers between the wild-type strain and two mutant strains over the entire operon (Table 1).
FIG. 2.
Electron micrographs of rrn operons. A. Chromosomal rrn operon from a wild-type strain. B. rrn operon from a nusA::cat rhoE134D mutant strain. C. rrn operon from a nusB::IS10 mutant strain. rrn operons are all 5.5 kb in length.
TABLE 1.
Electron-microscopic analysis of rrn operons in E. coli wild-type and nus mutant strains
| Genotype (na) | No. of polymerase molecules observed on rrn DNAb
|
Doubling time (min) | ||
|---|---|---|---|---|
| 16S | 23S | Total | ||
| Wild type (10) | 20.5 ± 5 | 31.0 ± 10 | 51.5 ± 12 | 24 |
| nusB::IS10 (70) | 16.4 ± 3 | 16.2 ± 4 | 32.6 ± 5 | 54 |
| ΔnusA::cat rhoE134D (14) | 14.6 ± 3 | 16.9 ± 3 | 31.5 ± 4 | 38 |
n, no. of operons.
Results are expressed as means ± standard deviations.
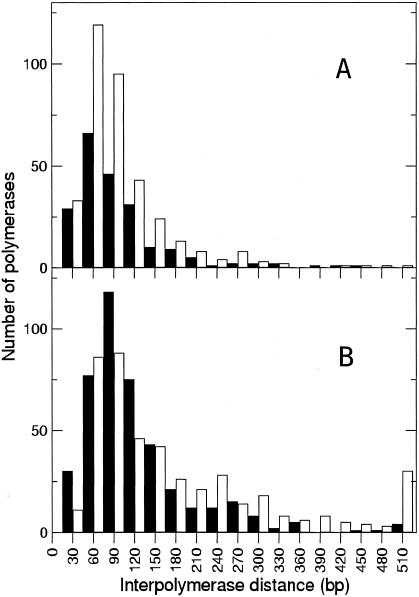
The distribution of RNA polymerase molecules on the rrn operons for the wild-type and nusB::IS10 strains used for Table 1 was analyzed by determining the center-to-center distance between adjacent RNA polymerases (Fig. 3). A subset (n = 27) of the nusB::IS10 rrn operons where RNA polymerases were clearly visible on a linear DNA contour were selected for further analysis. The average interpolymerase distance on the 16S gene was 85 and 111 bp for wild-type and nusB strains, respectively. In the wild-type strain, the interpolymerase distance on the 23S gene was 88 bp, similar to that along its 16S gene. In the nusB strain, however, the average interpolymerase distance on the 23S cistron increased to 176 bp. While the interpolymerase distance between the majority of RNA polymerases remained the same along the entire length of the rrn operons in nusB::IS10 (the mode of interpolymerase distances was the same for the 16S and 23S cistrons), the average distance between individual polymerases on the 23S cistron increased because of fewer polymerases. In the nusB mutant strain, 29% of polymerase molecules had interpolymerase gaps of more than 175 bp, while only 12% of polymerase molecules in the wild-type strain were separated by similarly sized gaps.
FIG. 3.
Determination of interpolymerase distance. The distance from polymerase center to polymerase center of all adjacent polymerases was measured. A. Interpolymerase spacing in the wild-type strain (n = 217 for 16S and n = 356 for 23S from 11 operons). B. Interpolymerase spacing in the nusB::IS10 mutant strain (n = 459 for 16S and n = 444 for 23S from 27 operons). Closed columns indicate polymerase molecules on the 16S genetic region, and open columns represent polymerase molecules on the 23S genetic region.
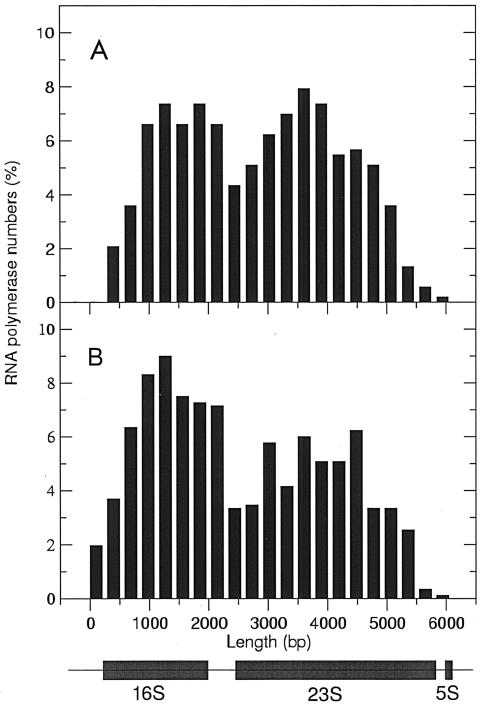
A plot of the relative positions of RNA polymerase molecules on the rrn operons in the nusB::IS10 strain showed no noticeable decrease in RNA polymerase molecules in the 16S-23S boundary region. This decreased density persisted throughout the 23S gene (Fig. 4). The wild-type operons also showed a slight decrease of the RNA polymerase population in the 16S-23S boundary, but the numbers once more increased throughout the rest of the 23S gene, suggesting that polymerase molecules were not lost but for some reason were more sparse in the boundary region.
FIG. 4.
Distribution of RNA polymerase molecules on rrn operons normalized to total polymerase numbers. A. Measurements of RNA polymerase numbers on rrn operons for the wild-type strain (n = 11). B. Measurements of RNA polymerase numbers for the nusB::IS10 mutant strain (n = 27).
Sucrose gradient analysis of ribosomes and ribosomal subunits in nusA and nusB mutant strains.
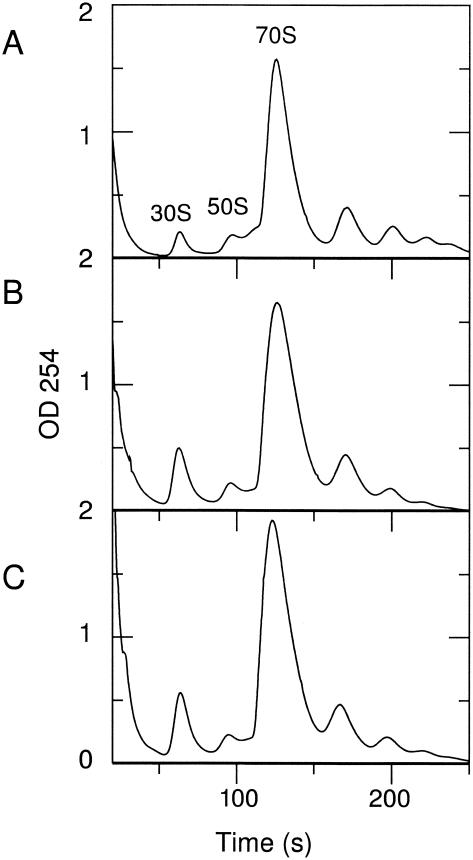
Since electron-microscopic analysis indicated a significant reduction in 23S rRNA transcription for both the nusA and nusB mutants, we examined whether this difference was also reflected in the relative amounts of 30S and 50S ribosomal subunits. Sucrose gradient profiles of ribosomes from the wild-type strain and two mutant strains were prepared, using a magnesium ion concentration that maintained the subunit association of 70S ribosomes (Fig. 5).
FIG. 5.
Sucrose gradient analysis of ribosomes prepared from the following: A. wild-type strain, MC4100; B. nusB::IS10 mutant strain; and C. nusA::cat rhoE134D mutant strain.
The ΔnusA::cat rhoE134D and nusB::IS10 mutant strains showed a significant change in the relative proportion of free 30S to 50S subunits compared to that for the wild-type strain (Table 2). Analysis of the rhoE134D mutant strain showed a pattern similar to that of wild-type cells, indicating that the rho mutation itself did not alter the proportion of 30S and 50S subunits in the ΔnusA::cat rhoE134D strain (data not shown). Thus, results obtained with both nus mutant strains supported the electron microscope analysis and confirmed that diminished transcription of the 23S gene resulted in a relative decrease in the 50S subunit pool (Table 2 and Fig. 5).
TABLE 2.
Sucrose gradient analysis of 70S ribosomes and their subunits from wild-type and nus mutant strains
| Strain or genotype | Peak areaa
|
% Relative to 70S
|
30S/50S ratio | |||
|---|---|---|---|---|---|---|
| 30S | 50S | 70S | 30S | 50S | ||
| MC4100 | 1.7 | 1.8 | 30.7 | 5.7 | 5.9 | 1 |
| nusB::IS10 | 4.8 | 1.8 | 37.7 | 12.7 | 4.8 | 2.5 |
| ΔnusA::cat rhoE134D | 5.9 | 2.1 | 38.8 | 14.4 | 5.1 | 3 |
Relative molar ratio.
DISCUSSION
The results of our electron microscope analysis of rrn transcription in strains lacking Nus factor A or B provided direct visual evidence that these proteins are needed for efficient expression of rrn operons. Because of their association with the transcription machinery, we assume that the two proteins act by direct or indirect interaction with RNA polymerase, RNA, and other proteins involved in the modifications of RNA polymerase required for rrn transcription. However, the specifics of these modifications of the rrn operon transcription machinery are not understood.
Both NusA and NusB have been previously implicated in rRNA transcription antitermination, but their precise role has not been determined. A study of rRNA-AT function in a nusA cold-sensitive strain showed that the increased rate of transcription elongation associated with BoxA-modified RNA polymerase complexes is abolished at the nonpermissive temperature, implicating NusA in this process (30). Likewise, in vivo studies of transcription rate and antitermination in the nusB::IS10 strain and in a strain depleted of NusG show that there is no elongation rate increase and no antitermination in these strains (28, 32). In vitro gel shift studies demonstrate that a complex of NusB and NusE binds to the rrn BoxA feature but that ribosomal protein S1 can compete for binding at this site (13, 14). Also, previous experiments suggest rRNA transcriptional polarity in a nusB5 mutant strain and BoxA substitution mutants (16, 20). Our finding of a relatively reduced number of RNA polymerase molecules transcribing the 23S gene in the nus strains (Table 1) supports the idea of a transcription defect and suggests that the mechanism of this defect in the absence of either cis or trans components of the antitermination complex is premature termination. Whether or not premature termination at the 16S leader is similarly increased is not apparent from the electron microscopy data. Sharrock et al. (20) suggested that because of premature termination at the rRNA operon in a nusB5 mutant, feedback control resulted in increased transcription initiation, compensating for reduced rRNA levels. Premature termination at the 16S leader region might be counteracted by increased transcription initiation, resulting in near-normal levels of 16S. Levels of 23S, however, are not maintained, since transcription at the 23S leader region is also subject to an increased termination frequency.
Previous studies looking at the effect of BoxA mutations on transcription have described a decrease in the stability of rRNA. Replacement of the leader BoxA leads to a decrease in 16S rRNA stability and to a relative reduction in 30S to 50S subunit ratio (26). Similarly, measurements of the relative proportion of plasmid-encoded rRNA in the 30S and 50S subunits from plasmids with a leader region BoxA mutation show a decrease of both plasmid-encoded 16S and 23S rRNA and a further drop in 23S rRNA when both leader and spacer BoxA mutations are present (11, 16). This is consistent with previous data that show that in the case of an unbalanced production of either 16S or 23S rRNA, excess RNA is rapidly degraded (22, 31). Combining these observations with the results we present here suggests that perturbation of the rRNA-AT system, whether at the level of the nucleotide sequence signaling the modifications (BoxA) or in the modifying proteins themselves (NusA and NusB), leads to a decrease in the stability or quantity of the rRNA transcript and to a relative decrease in the amount of 23S rRNA, probably as a result of premature termination at the 23S gene.
The excess of 30S free subunits over 50S free subunits in the nus mutant cells (Fig. 5) correlates with the electron microscopy results measuring RNA polymerase numbers on the 16S gene. Yamagishi and Nomura (31) show that a plasmid-encoded excess synthesis of either 16S or 23S rRNA does indeed lead to a corresponding imbalance in the relative molar ratios of ribosomal subunits. As with the nus mutants, a relative increase in 16S rRNA results in a corresponding relative increase in the 30S ribosome subunit because of the lack of partnering 50S subunits or because these “dead-end” 30S subunits are incompletely assembled and cannot proceed to form functional 70S ribosomes (31).
Our observation that the ΔnusA::cat rhoE134D strain was defective in rrn transcription was somewhat surprising (Fig. 2 and Tables 1 and 2). If overcoming of Rho-dependent terminators in the rrn operons were the only reason for an rRNA-AT system, the greatly diminished Rho activity in this strain would have prevented the 23S gene transcription polarity evident with the nusB::IS10 mutant strain. Instead, 23S gene transcription was decreased by at least twofold (Fig. 2 and Table 1). Because little is known about specific Rho-dependent terminators and their function, it is possible that such terminators in rrn operons have a high affinity for Rho protein and, while the mutant Rho protein's activity may be greatly diminished on some terminators, its activity on others might not be affected to the same extent (5). Alternatively, the antitermination system might play another role necessary for the maximal rate of rRNA synthesis.
Because the majority of RNA polymerase molecules on the 23S gene have the same relative spacing as on the 16S gene, the frequent gaps in RNA polymerase density present throughout the 23S gene in both nus mutant strains likely represent a significant malfunction of the transcription apparatus that occurs sporadically after transcription of the 16S gene. It may also be possible that whatever is releasing the hold of RNA polymerase on the DNA at the 16S-23S boundary is more readily overcome by groups of molecules than by single polymerases.
In summary, electron-microscopic examination of rrn operon transcription showed decreased transcription of the 23S rRNA gene and RNA polymerase molecules that were in more widely separated clusters in nusA and nusB mutant strains than in the wild type. Consistent with the electron microscopy data, the ratio of the concentration of free 50S subunit to that of free 30S subunits was decreased in the mutant strains. Our results suggest a mechanism for triggering transcription termination in rrn operons that works particularly effectively in the 16S-23S boundary region when factors necessary for increased transcription elongation rates and the antitermination system are missing (Fig. 6).
FIG. 6.
Diminished transcription of the 23S genetic region in a nusA or nusB mutant strain due to premature termination.
Acknowledgments
We thank C. Yanofsky and Linc Sonenshein for helpful comments on the manuscript, K. Ito for the IQ527 nusB::IS10 strain, and David Friedman for the K7096 ΔnusA::cat rhoE134D and K4633 rhoE134D strains.
This work was supported by NIH grant R01 GM24751 to C.L.S. S.F. was supported by NIH grant R01 GM63952 to Ann Beyer.
REFERENCES
- 1.Albrechtsen, B., C. L. Squires, S. Li, and C. Squires. 1990. Antitermination of characterized transcriptional terminators by the Escherichia coli rrnG leader region. J. Mol. Biol. 213:123-134. [DOI] [PubMed] [Google Scholar]
- 2.Altieri, A. S., M. J. Mazzulla, D. A. Horita, R. H. Coats, P. T. Wingfield, A. Das, D. L. Court, and R. A. Byrd. 2000. The structure of the transcriptional antiterminator NusB from Escherichia coli. Nat. Struct. Biol. 7:470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, K., C. Squires, and C. L. Squires. 1989. Ribosomal RNA operon antitermination. Function of the leader and spacer region boxB-boxA sequences and their conservation in diverse microorganisms. J. Mol. Biol. 209:345-358. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. 2002. E. coli genome project. [Online.] http://www.genome.wisc.edu.
- 5.Burns, C. M., and J. P. Richardson. 1995. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc. Natl. Acad. Sci. USA 92:4738-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 7.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeVito, J., and A. Das. 1994. Control of transcription processivity in phage: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc. Natl. Acad. Sci. USA 91:8660-8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French, S. L., and O. L. Miller, Jr. 1989. Transcription mapping of the Escherichia coli chromosome by electron microscopy. J. Bacteriol. 171:4207-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godson, G. N., and R. L. Sinsheimer. 1967. Use of Brij lysis as a general method to prepare polyribosomes from Escherichia coli. Biochim. Biophys. Acta 149:489-495. [DOI] [PubMed] [Google Scholar]
- 11.Heinrich, T., C. Condon, T. Pfeiffer, and R. K. Hartmann. 1995. Point mutations in the leader boxA of a plasmid-encoded Escherichia coli rrnB operon cause defective antitermination in vivo. J. Bacteriol. 177:3793-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason, S. W., and J. Greenblatt. 1991. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 5:1504-1512. [DOI] [PubMed] [Google Scholar]
- 13.Mason, S. W., J. Li, and J. Greenblatt. 1992. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J. Mol. Biol. 223:555-566. [DOI] [PubMed] [Google Scholar]
- 14.Mogridge, J., and J. Greenblatt. 1998. Specific binding of Escherichia coli ribosomal protein S1 to boxA transcriptional antiterminator RNA. J. Bacteriol. 180:2248-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nodwell, J. R., and J. Greenblatt. 1993. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell 72:261-268. [DOI] [PubMed] [Google Scholar]
- 16.Pfeiffer, T., and R. K. Hartmann. 1997. Role of the spacer boxA of Escherichia coli ribosomal RNA operons in efficient 23S rRNA synthesis in vivo. J. Mol. Biol. 265:385-393. [DOI] [PubMed] [Google Scholar]
- 17.Rajapandi, T., and D. Oliver. 1994. ssaD1, a suppressor of secA51(Ts) that renders growth of Escherichia coli cold sensitive, is an early amber mutation in the transcription factor gene nusB. J. Bacteriol. 176:4444-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasband, W. 2002. ImageJ, version 1.31. National Institutes of Health, Bethesda, Md.
- 19.Schaferkordt, J., and R. Wagner. 2001. Effects of base change mutations within an Escherichia coli ribosomal RNA leader region on rRNA maturation and ribosome formation. Nucleic Acids Res. 29:3394-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharrock, R. A., R. L. Gourse, and M. Nomura. 1985. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5275-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiba, K., K. Ito, and T. Yura. 1986. Suppressors of the secY24 mutation: identification and characterization of additional ssy genes in Escherichia coli. J. Bacteriol. 166:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siehnel, R. J., and E. A. Morgan. 1985. Unbalanced rRNA gene dosage and its effects on rRNA and ribosomal-protein synthesis. J. Bacteriol. 163:476-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Squires, C., J. Greenblatt, J. Li, C. Condon, and C. L. Squires. 1993. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. USA 90:970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squires, C. L., and D. Zaporojets. 2000. Proteins shared by the transcription and translation machines. Annu. Rev. Microbiol. 54:775-798. [DOI] [PubMed] [Google Scholar]
- 25.Taura, T., C. Ueguchi, K. Shiba, and K. Ito. 1992. Insertional disruption of the nusB (ssyB) gene leads to cold-sensitive growth of Escherichia coli and suppression of the secY24 mutation. Mol. Gen. Genet. 234:429-432. [DOI] [PubMed] [Google Scholar]
- 26.Theissen, G., S. E. Behrens, and R. Wagner. 1990. Functional importance of the Escherichia coli ribosomal RNA leader box A sequence for post-transcriptional events. Mol. Microbiol. 4:1667-1678. [DOI] [PubMed] [Google Scholar]
- 27.Torres, M., C. Condon, J.-M. Balada, C. Squires, and C. L. Squires. 2001. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in non-ribosomal and ribosomal RNA antitermination. EMBO J. 20:3811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres, M., J.-M. Balada, M. Zellars, C. Squires, and C. L. Squires. 2004. In vivo effect of NusB and NusG on rRNA transcription antitermination. J. Bacteriol. 186:1304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel, U., and K. F. Jensen. 1995. Effects of the antiterminator Box A on transcription elongation kinetics and ppGpp inhibition of transcription elongation in Escherichia coli. J. Biol. Chem. 270:18335-18340. [DOI] [PubMed] [Google Scholar]
- 30.Vogel, U., and K. F. Jensen. 1997. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J. Biol. Chem. 272:12265-12271. [DOI] [PubMed] [Google Scholar]
- 31.Yamagishi, M., and M. Nomura. 1988. Effects of induction of rRNA overproduction on ribosomal protein synthesis and ribosome subunit assembly in Escherichia coli. J. Bacteriol. 170:5042-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zellars, M., and C. L. Squires. 1999. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol. 32:1296-1304. [DOI] [PubMed] [Google Scholar]
- 33.Zheng, C., and D. I. Friedman. 1994. Reduced Rho-dependent transcription termination permits NusA-independent growth of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:7543-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]