Abstract
Cupin-type phosphoglucose isomerases (cPGIs) were identified in some archaeal and bacterial genomes and the respective coding function of cpgi's from the euryarchaeota Archaeoglobus fulgidus and Methanosarcina mazei, as well as the bacteria Salmonella enterica serovar Typhimurium and Ensifer meliloti, was proven by functional overexpression. These cPGIs and the cPGIs from Pyrococcus and Thermococcus spp. represent the cPGI family and were compared with respect to kinetic, inhibitory, thermophilic, and metal-binding properties. cPGIs showed a high specificity for the substrates fructose-6-phosphate and glucose-6-phosphate and were inhibited by millimolar concentrations of sorbitol-6-phosphate, erythrose-4-phosphate, and 6-phosphogluconate. Treatment of cPGIs with EDTA resulted in a complete loss of catalytic activity, which could be regained by the addition of some divalent cations, most effectively by Fe2+ and Ni2+, indicating a metal dependence of cPGI activity. The motifs TX3PX3GXEX3TXGHXHX6-11EXY and PPX3HX3N were deduced as the two signature patterns of the novel cPGI family. Phylogenetic analysis suggests lateral gene transfer for the bacterial cPGIs from euryarchaeota.
Phosphoglucose isomerase (PGI; EC 5.3.1.9) catalyzes the reversible isomerization of glucose-6-phosphate to fructose-6-phosphate. PGI plays a central role in sugar metabolism of eukarya, bacteria, and archaea, both in glycolysis via the Embden-Meyerhof pathway in eukarya and bacteria and in its modified versions found in archaea. PGI is also involved in gluconeogenesis, where the enzyme operates in the reverse direction (see references 22, 26, and 40). PGIs have evolved convergently. Most PGIs belong to the PGI superfamily, which can be divided into the PGI family and the recently identifiedbifunctional phosphoglucose/phosphomannose isomerase(PGI/PMI) family (22). PGIs from the PGI superfamily, often referred to as conventional PGIs, are found in all domains of life and are well studied. Crystal structures have been determined for the eukaryotic PGIs from pigs, rabbits, humans and from the bacterium Bacillus stearothermophilus, and conserved amino acids proposed to be involved in substrate binding and/or catalysis have been identified (3, 6, 10-13, 28, 29, 42). Bifunctional PGI/PMIs, which have been characterized as a novel family within the PGI superfamily, were predominantly found in the crenarchaeotal branch of the archaea (26).
Recently, a novel type of PGI has been identified and characterized from the hyperthermophilic euryarchaeon Pyrococcus furiosus (22, 52) and later from the closely related Thermococcus litoralis (30). These PGIs belong to the cupin superfamily and thus represent a convergent line of PGI evolution. The cupin superfamily is present in all three domains of life—eukarya, bacteria, and archaea—and comprises a group of functionally diverse proteins that contain a central domain composed of β-strands forming a small β-barrel called “cupin.” Proteins from the cupin superfamily range, for example, from mannose-6-phosphate isomerases and epimerases involved in formation of bacterial cell wall carbohydrates to nonenzymatic storage proteins in plant seeds and transcription factors (16-19). The two euryarchaeal cPGIs were the first PGIs of the cupin family to be described. Sequence comparison suggested the presence of few putative cPGI homologs; however, none of them have been characterized (44, 53). Two consensus sequences (GX5HXHX3,4EX6G and GX5PXGX2HX3N) were established for the cupin superfamily (17, 18), which include a metal-binding site found in most cupin enzymes, including the PfcPGI and TlPGI (4, 30, 49). Two crystal structures are available for the PfcPGI; however, the role of the metal is discussed to be either involved in substrate binding and/or in catalysis (4, 30, 49). Berrisford et al. (4) showed that catalytic activity is not strongly influenced by the presence or absence of the bound metal, suggesting that the metal might be implicated in substrate recognition rather than being directly involved in catalysis. Loss of metal-binding ability in some TlcPGI mutants goes along with a loss of catalytic function, suggesting a role in catalysis (30). Thus far, conclusive biochemical studies analyzing the role of metals have not been performed.
We describe here the identification and functional characterization of cPGIs from the euryarchaeota Archaeoglobus fulgidus and Methanosarcina mazei, as well as the bacteria Salmonella enterica serovar Typhimurium and Ensifer meliloti. Molecular, kinetic, thermophilic, and inhibitory properties were analyzed and compared. In particular, metal depletion and reactivation experiments were performed with these cPGIs and with PfcPGI. The data strongly indicate an involvement of divalent cations in the catalytic mechanism. Two consensus sequences for the novel cPGI family were derived. Phylogenetic analyses suggest that bacterial cPGIs originated from euryarchaeota by lateral gene transfer
MATERIALS AND METHODS
Overexpression and purification of recombinant protein.
The open reading frame (ORFs) AF1494, STM2757, SMc02042, and MM1968, which encode putative cPGIs, have been identified by BLASTP searches with the PfcPGI sequence (22) in the genomes of A. fulgidus (AfcPGI), M. mazei (MmcPGI), S. enterica serovar Typhimurium (StcPGI), and E. meliloti (EmcPGIA) (15, 21, 33, 39). To prove their coding function as cpgi, these ORFs were cloned and functionally overexpressed in Escherichia coli as follows. The respective coding region of these cpgi's was amplified by PCR with Pwo or Taq polymerase (PEQLAB, Erlangen, Germany) from the respective genomic DNA as a template. The following primers were used to introduce unique restriction sites for NdeI and XhoI, respectively: pfcpgi, 5-′CTGGTGGTGCATATGTATAAGGAACCTTT-3′ (forward), 5-′TAACATTGTCCAGTTAACTACTTTTTCCACC-3′ (reverse); afcpgi, 5-′CCGGCTCTTTTTTTACATATGGAGTTTGTT-3′ (forward), 5-′GCTCGCTCTCTCGAGGTAAAAATTTTAA-3′ (reverse); mmcpgi 5-′CAAAGGGGAATCATATGGCAGTAACACTGAAG-3′ (forward), 5-′CCCATTATAAAATTCTCGAGCCTATTTCAGAGC-3′ (reverse); stcpgi, 5-′CAGGGGGCACATATGAAACAGCTACACC-3′ (forward), 5-′GCAACGCCTGCTCGAGATTGCGTTTCC-3′ (reverse); emcpgiA, 5-′CGAGACATACGAGGAATCCATATGACAAAGCT-3′ (forward), and 5-′GAGTTGCGGCTTGCTCGAGCGGAGCA-3′ (reverse). After amplification and NdeI/XhoI double digestion, the PCR products were inserted by T4 DNA ligase (Roche Diagnostics) into pET (Novagen) expression vectors linearized by NdeI/XhoI double digestion. The resulting plasmids pfcpgi-pET17b, afcpgi-pET19b, afcpgi-pET17b, stcpgi-pET19b, and emcpgi-pET19b were introduced into E. coli JM109 and BL21(DE3) or BL21-CodonPlus(DE3)-RIL (pfcpgi-pET-17b, mmcpgi-pET19b) (Stratagene) via transformation. The inserted gene sequences were confirmed on each strand by the method of Sanger et al. (45). E. coli BL21(DE3) and BL21-CodonPlus(DE3)-RIL cells transformed with different cpgi-pET constructs were grown in two flasks with 400 ml of Luria-Bertani medium containing 100 μg of carbenicillin/ml and 34 μg of chloramphenicol/ml (only BL21-CodonPlus(DE3)-RIL cells) at 37°C to an optical density at 600 nm (OD600) of 0.8, and cpgi expressions were initiated by induction with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (pfcpgi) or 1 mM IPTG (afcpgi, mmcpgi, stcpgi, and emcpgiA). After 4 h of further growth (OD600 of ∼2.5 to 3.2), the cells were harvested by centrifugation at 4°C and washed in 50 mM Tris-HCl (pH 7.0) containing 50 mM NaCl. Recombinant proteins were purified as follows. PfcPGI was purified as previously described (22). For AfcPGI (obtained from afcpgi-pET17b transformed E. coli), cell extracts were prepared by French press treatment (1.3 × 108 Pa) of cell suspensions in buffer A (50 mM Tris-HCl pH 8.4). After ultracentrifugation (100,000 × g for 60 min), the solution was heat precipitated at 80°C for 45 min, centrifuged again (15,000 × g for 15 min), extensively dialyzed against buffer B (50 mM Tris-HCl [pH 8.0]), and applied to DEAE-Sepharose (63 ml) that had been equilibrated with buffer B. After the column was washed with 60 ml of buffer B and 150 ml of buffer C (50 mM piperazine [pH 5.6]), protein was desorbed with a linear NaCl-gradient (0 to 2 M) in buffer C. AfcPGI obtained from 0.1 to 0.2 M NaCl was essentially pure. For His-tagged AfcPGI, StcPGI, MmcPGI, and EmcPGIA (due to expression via pET19b vectors, these PGIs contain an additional 23-amino-acid N-terminal sequence [MGHHHHHHHHHHSSGHIDDDDKH]), cell extracts were prepared from cell suspensions in buffer D (300 mM NaCl, 1 mM imidazole, 50 mM Tris-HCl [pH 8.2]) as described above and ultracentrifuged (100,000 × g for 60 min), and the solutions were applied on a 3-ml Ni-NTA-Superflow (Qiagen, Hilden, Germany) flowthrough column previously equilibrated with buffer A. After the column was washed with 20 ml of buffer A, protein was eluted by four steps with increasing imidazole concentrations in buffer A of 20 mM (10 ml), 50 mM (10 ml), and 500 mM (2 × 10 ml). PGI activity containing fractions were recovered from the last three elution fractions, the one obtained from the last two were essentially pure. Pure cPGIs were dialyzed against 10 mM Tris-HCl (pH 8.0) and stored at −20°C. Under these conditions activity remained about constant. The respective His tag of AfcPGI, StcPGI, MmcPGI could not be removed by enterokinase treatment.
Analytical assays.
The purity of the respective cPGI preparation was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% gels, followed by staining with Coomassie brilliant blue R 250 according to standard procedures (35). Protein concentrations were determined by the method of Bradford (5) with bovine serum albumin as a standard. Using the ExPASy ProtParam Tool (http://us.expasy.org/tools/protparam.html), the molar extinction coefficients at 280 nm (ɛ280) were calculated and used for the determination of protein concentrations. ɛ280 values were as follows: PfcPGI, 35,560 M−1 cm−1; AfcPGI, 38,830 M−1 cm−1; MmcPGI 29,300 M−1 cm−1; StcPGI, 26,880 M−1 cm−1; and EmcPGIA, 36,840 M−1 cm−1. Gel filtration chromatography was carried out at ambient temperature on a Bio-Prep SE1000/17 column (Bio-Rad, Munich, Germany) in 50 mM Tris-HCl-150 mM NaCl (pH 7.4) at a flow rate of 0.5 ml min−1. For analytical ultracentrifugation, the AfcPGI (protein without His tag was used) was dialyzed against 20 mM Tris-HCl (pH 7.0). Ultracentrifugation was done at 20°C in a Beckman Optima XL-A analytical ultracentrifuge equipped with a Titan AN 50 rotor and absorption optics. Sedimentation velocity experiments were done for an approximation of the molecular mass at 22,000 rpm, and sedimentation-diffusion equilibrium runs were done at 60,000 rpm for determining the molecular mass in a 150-μl volume. In sedimentation velocity experiments the apparent sedimentation coefficient sapp20,W and an approximated molecular mass were evaluated from the velocity and shape of the sedimenting boundary by fitting the time-dependent concentration profiles calculated with Lamm's differential equation for a single sedimenting species to the measured data by using the program package AKKUPROG (32). When the measured concentration profile of sedimentation-diffusion equilibrium runs remained unchanged for 12 h, we assumed equilibrium to be attained. AKKUPROG was used to calculate the apparent molecular masses by fitting the ideal distribution for a single species to the measured concentration profiles. The partial specific volume of the protein was calculated from the amino acid sequence.
Determination of metal content.
After purification, the cPGIs (PfcPGI, AfcPGI, AfcPGI with His tag, MmcPGI, and StcPGI) were dialyzed with a 100-fold excess volume of 10 mM Tris-HCl-10 mM EDTA (pH 7.0) and subsequently with an 100-fold excess volume of H2O at 25°C. Dialysis with EDTA was necessary to remove unspecific binding of Fe2+ or Ni2+ to cPGIs. Without EDTA treatment the metal contents gave unreliable, significantly higher values (more than fivefold). The following controls were used. For His-tagged cPGIs, to correct for unspecific metal binding by the respective His tags as well as for H2O impurities, His-tagged glucose-6-phosphate dehydrogenase from T. maritima (23) and His-tagged PGI from P. aerophilum (25) were dialyzed and prepared as described above for the cPGIs, and the respective metal contents of the proteins were measured. The controls contained on average 0.02 Ni2+ per enzyme molecule, whereas no iron could be detected in any of the control samples. For non-His-tagged cPGIs, to correct for H2O impurities, H2O used for dialysis was used as a control. Determination of iron and nickel was performed by atom absorption spectroscopy as described recently (38).
Enzyme assays and determination of kinetic parameters.
Since the enzyme activities were not sensitive to oxygen, all assays were performed in the presence of oxygen. pH optima were determined by using acetate, Tris-HCl, ethanolamine, CHES (N-cyclohexyl-2-aminoethanesulfonic acid), citrate, phosphate, and CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) at 50 or 100 mM each. PGI activity (G6P ↔ F6P) was determined in both directions. The formation of G6P and F6P was measured by using glucose-6-phosphate dehydrogenase from T. maritima (23) and mannitol-1-phosphate dehydrogenase from Klebsiella pneumoniae (41) as recently described (26). The following standard conditions were used: for PfcPGI, 80°C (pH 7.0) and 2 μg of enzyme; for AfcPGI, 50°C or 70°C (pH 7.4) and 5 μg of enzyme; for MmcPGI, 37°C (pH 7.4) and 20 μg of enzyme; for StcPGI, 37°C (pH 7.2) and 33 μg of enzyme; for EmcPGIA, 50°C (pH 7.4) and 80 μg of enzyme. Standard assay mixture 1 (F6P formation) contained 100 mM Tris-HCl, 0.3 mM NADH, 0.1 to 15 mM G6P, and 0.6 U of mannitol-1-phosphate dehydrogenase; standard assay mixture 2 (G6P formation) contained 100 mM Tris-HCl, 0.5 mM NADP+, 5 mM F6P, and 1.1 U of glucose-6-phosphate dehydrogenase. The latter assay was routinely used for most PGI activity determinations. (The given pH values correspond to the respective temperature used.) The following substances were tested for potential inhibitory effects on cPGI activity: EDTA, erythrose-4-phosphate, 6-phosphogluconate, sorbitol-6-phosphate, phosphoenolpyruvate, ADP, fructose-1-phosphate, and fructose-1,6-bisphosphate. Kinetic data were fit to the Michaelis-Menten equation with the MicroCal software by using the Levenberg-Marquard algorithm.
Effect of metals on PGI activity.
In order to obtain metal-free enzyme, 2 mg of enzyme was incubated in the presence of 0.5 M EDTA for 30 min under the following indicated conditions: PfcPGI, 80°C (pH 7.0); AfcPGI (enzyme without His tag), 70°C (pH 7.4); MmcPGI, 37°C (pH 7.6); StcPGI, 37°C (pH 7.2); and EmcPGIA, 50°C (pH 7.4). The proteins were subsequently dialyzed five times against a 100-fold excess of H2O. The effect of metals on PGI activity was tested by adding 5 to 3,000 μM concentrations of Fe2+, Ni2+, Mn2+, or Zn2+ to the respective standard assay mixtures, which was modified by using 10 mM Tris-HCl instead of 100 mM Tris-HCl (at the respective pH). Low buffer concentrations (10 mM) and Ultrapure Milli-Q water had to be used for all substances in the assays. When higher buffer concentrations or deionized H2O was used, the residual catalytic activity was still measured (data not shown) after EDTA treatment, which was presumably due to soaking of metal impurities into the metal-binding site of the enzymes. cPGI activity in the presence of Co2+, Ca2+, Cd2+, and Mg2+ was determined at 100 μM concentrations of the respective metal. For the determination of EDTA Ki values, cPGIs were preincubated at the respective assay temperature for 30 min (except for MmcPGI, which was preincubated for 2 h) before the respective cPGI activity was determined and compared to the control (cPGI preincubated without EDTA).
Temperature dependence and thermal stability.
The temperature dependence of the enzyme activity was measured at between 20 and 96°C by using the respective standard assay for G6P formation except that 50 mM sodium phosphate was used instead of 100 mM Tris-HCl. The thermostability of the purified enzymes (125 μg of AfcPG or MmcPGI/ml in 50 mM sodium phosphate [pH 7.0], 400 μg of StcPGI/ml in 50 mM Tris-HCl [pH 7.0], and 600 μg of EmcPGIA/ml in Tris-HCl [pH 7.4]) were tested in sealed vials, which were incubated at temperatures between 40 and 100°C for 2 to 120 min. The vials were then cooled on ice for 5 min, and the residual enzyme activity was tested and compared to the controls (unheated samples).
CD spectroscopy.
Circular dichroism (CD) spectroscopy analyses were performed on JASCO J-715 CD spectrometer. Spectra were recorded in 2-mm cuvettes and corrected for the signal of the solvent (10 mM sodium phosphate [pH 7.0]). Secondary structure analysis and assignment to different secondary structure types were performed by the experimentally established spectra-structure correlation by using the Varselec option of Dicroprot (14). Heat-induced unfolding of the respective cPGIs was analyzed in temperature gradient experiments by using closed cuvettes (0.5 cm). The protein samples were dialyzed against 10 mM sodium phosphate buffer (pH 7.0), and the protein concentrations were set to 200 μg/ml. The temperature of the respective samples was raised at a rate of 1°C/min (depending on the protein in a range from 20 to 100°C). Protein unfolding was followed by changes of the CD ellipticity at 226 nm (PfcPGI, StcPGI, and EmcPGIA) or 232 nm (AfcPGI and MmcPGI). The ellipticity at a given temperature (Φ) was corrected for the temperature-dependent baseline giving Φ′. The corrected values of the folded state Φ′f and the unfolded state Φ′uf were used for calculating the fraction of folded protein (xf) as follows: (Φ′ − Φ′uf)/(Φ′ − Φ′f). Spectra were recorded before and after each temperature gradient. The spectra, as well as the ellipticities of the unfolded state, were obtained at pH 2 and pH 12 when complete thermal unfolding was not obtained up to 100°C.
Sequence handling.
Sequence alignments were constructed with the neighbor-joining (NJ) method of CLUSTAL X (50) by using the GONNET matrix. Phylogenetic trees were constructed by using both the NJ option of CLUSTAL X and the maximum-likelihood (ML) method of PROML (PHYLIP, version 3.6). Confidence limits were estimated by 100 bootstrapping replicates.
RESULTS AND DISCUSSION
In this report we give a comprehensive characterization of cPGIs from archaea and bacteria which represent a novel PGI family.
Identification, cloning and functional overexpression of cPGIs.
Based on the PfcPGI sequence, the ORFs AF1494, MA0821, Mb1339, MM1968, STM2757, SMc02042, and SMb20857, which have been previously annotated as hypothetical proteins, were identified by BLASTP searches as putative PGIs in the complete genomes of A. fulgidus, Methanosarcina species, S. enterica serovar Typhimurium, and E. meliloti (formerly Sinorhizobium meliloti), respectively (15, 20, 21, 33, 39). Meanwhile, AF1494 and MM1968 have been proposed as putative cPGIs (44, 53). The coding function of the ORFs AF1494, MM1968, STM2757, and SMc02042, as cpgi's were proven by cloning into pET19b and functional overexpression in E. coli (see below). To test the in vivo transcription of cPGIs (ORFs AF1494, MM1968, and SMc02042), total RNA was extracted from A. fulgidus, M. mazei, and E. meliloti cells. cpgi-specific mRNAs were amplified as cDNA by reverse transcription-PCR, and cpgi-specific cDNAs of the expected respective lengths were detected, indicating in vivo transcription of cpgi's in these three organisms.
Cloning and functional overexpression.
ORF AF1494 contains 708 bp, ORF MM1968 contains 741 bp, ORF STM2757 contains 843 bp, and ORF SMc02042 contains 639 bp, coding for polypeptides of 235, 246, 280, and 212 amino acids, respectively. The recombinant His-tagged AfcPGI, StcPGI, and EmcPGIA were purified to apparent homogeneity by a single chromatographic step on Ni-NTA agarose. In addition, AfcPGI and the pfcpgi gene were cloned into pET17b. Recombinant AfcPGI and PfcPGI were purified by a heat treatment and two chromatographic steps on phenyl Sepharose and gel filtration. Expression of the pfcpgi from the T7 promoter was more than five times higher than from the araC promoter (see reference 22). With the exception of the PfcPGI, all cPGIs could be purified to yield more than 10 mg of pure protein per g of transformed E. coli (wet weight).
Molecular properties.
Proteins from the diverse cupin superfamily have been described as monomers or oligomers with either a monocupin or a bicupin fold (16-19) (Table 1). PfcPGI was previously described as a dimer of 21.4-kDa subunits (22). Using size-exclusion chromatography, the purified cPGIs resulted in a single elution peak, and apparent molecular masses of 29 kDa (AfcPGI), 28 kDa (MmcPGI), 67 kDa (StcPGI), and 43 kDa (EmcPGIA), respectively, were deduced. SDS-PAGE revealed one subunit each with apparent molecular masses of 27, 28, 32, and 27 kDa, respectively, indicating a dimeric structure for the StcPGI and the EmcPGIA and monomeric structures for the AfcPGI and MmcPGI. The monomeric structure of the AfcPGI was verified by analytical ultracentrifugation. The measured concentration profile of sedimentation-diffusion equilibrium runs were best fit by using a molecular mass of 27 kDa, which is about the calculated mass of the AfcPGI (27.424 kDa). In accordance with this, the intersubunit hydrophilic cluster as described for PfcPGI (Lys3, Tyr129, Gln59′, and Asp65′) (4) is partially conserved in the dimeric EmcPGIA but is not present in the monomeric AfcPGI and MmcPGI (see alignment in Fig. 5). Thus, cPGIs are either monomeric or dimeric monocupins. In comparison, a dimeric arrangement [α2 or (α2)2] is a prerequisite for catalytic activity for conventional PGIs, as well as for PGI/PMIs, which also have significantly larger subunits ranging from 296 amino acids to more than 600 amino acids.
TABLE 1.
Comparison of cPGIs with selected members of the cupin superfamily
| Protein | Source | Fold type | Quaternary structure | Metalc | Source or reference |
|---|---|---|---|---|---|
| Archaea | |||||
| PGI | Pyrococcus furiosus, Salmonella enterica serovar Typhimurium, and Ensifer meliloti | Monocupin | α2 | Me | 2; This work |
| PGI | Archaeoglobus fulgidus and Methanosarcina mazei | Monocupin | α | Me | This work |
| PGI | Thermococcus litoralis | Monocupin | α2 | Fe and Zn | 30 |
| dTDP-6-deoxy-d-xylo-4-hexulose 3,5-epimerase | Methanothermobacter thermoautotrophicus | Monocupin | α2 | None | 7 |
| Bacteria | |||||
| AraC | Escherichia coli | Monocupin | α2 | None | 47 |
| Proline 3-hydroxylase | Streptomyces sp. | Monocupin | α2 | Fe | 9 |
| dTDP-6-deoxy-d-xylo-4-hexulose 3,5-epimerase | Salmonella enterica serovar Typhimurium | Monocupin | α2 | None | 1 |
| Oxalate decarboxylase | Bacillus subtilis | Bicupin | α6 | Mn | 2 |
| Eukarya | |||||
| Oxalate oxidase | Barley, Hordeum vulgare | Monocupin | α6 | Mn | 54 |
| Homogentisate dioxygenase | Human | Monocupin | α6 | Fea | 51 |
| Phosphomannose isomerase | Candida albicans | Bicupin | α | Zn | 8 |
| Isopenicillin N-synthase | Emericella nidulans | Monocupin | α | Fe or Mnb | 43 |
| Canavalin | Jack bean | Bicupin | α3 | None | 34 |
| Phaseolin | Phaseolus vulgaris | Bicupin | α3 | None | 36 |
Iron is not associated with the cupin domain.
Mn was found in the structure.
cMe, divalent metal.
FIG. 5.
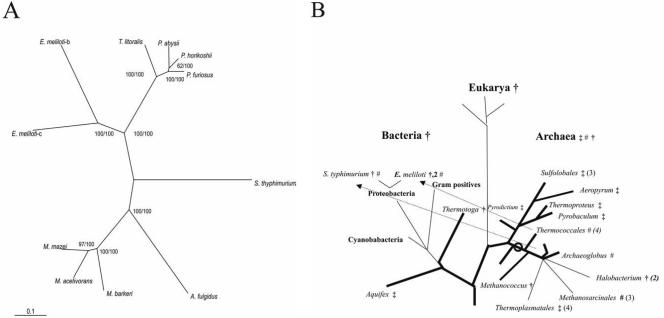
(A and B) Phylogenetic relationship of enzymes from the cPGI family (A) and distribution of PGIs in the universal phylogenetic tree (B). In panel A, the tree was constructed by the NJ algorithm of CLUSTAL X (50). The numbers at the nodes are bootstrapping values according to NJ (first) or ML (second). For NCBI accession numbers or SwissProt identifiers, see Fig. 4. In panel B, a phylogenetic tree according to 16S rRNA and 18S rRNA sequences (48) is shown. Organisms or groups of organisms are marked to contain PGIs belonging to the cPGI family (#) or to the PGI superfamily including the PGI family (†) and PGI/PMI family (‡). The proposed origin of the cPGI family in the euryarchaeal evolution is indicated by an open circle, and the postulated lateral gene transfers are indicated by arrows.
CD spectra.
In order to compare the overall fold of the cupin-type PGIs with PGIs and PGI/PMI, CD spectra were recorded. All cPGIs showed very distinct CD spectra compared to PGIs from the PGI superfamily, e.g., to the PGI/PMI from Thermoplasma acidophilum (26). In comparison to conventional PGIs, which consist of two αβα-sandwich domains, cPGIs are characterized by a central β-barrel forming the unique central cupin core and show lower ellipticities due to their higher β-sheet content. In accordance with this, a lower α-helical content and higher β-sheets were estimated for cPGIs from the CD spectrum.
Catalytic properties.
Kinetic properties of purified cPGIs were determined for both reaction directions (G6P and F6P) and are summarized in Table 2. The pH optima of the PGIs analyzed were quite similar, i.e., between pH 7.2 and 7.6. For all cPGIs the respective rate dependence on both substrates of the enzymes followed Michaelis-Menten kinetics, and the respective Km and Vmax values are given in Table 2. All Vmax values given correspond to the purification procedure described. The catalytic rates of the three mesophilic cPGIs analyzed (MmcPGI, EmcPGIA, and StcPGI) were significantly lower compared to both the hyperthermophilic cPGIs and the conventional PGIs or PGI/PMIs (26, 27, 44, 46). It should be noted that S. enterica serovar Typhimurium and E. meliloti contain several PGI isoenzymes in addition to the cPGI described here; S. enterica serovar Typhimurium also contains a conventional PGI (Q8ZKI4) and E. meliloti contains a conventional PGI (StPGI, Q92SC4), as well as a second cPGI (EmcPGIB, Q92UI1). The recombinant StPGI and EmcPGIB encode for active PGIs as well (unpublished data). In E. meliloti all three pgi genes are transcribed in vivo during growth on complex medium (unpublished data). In M. mazei no other pgi gene could be identified. cPGIs exhibited a high specificity for the substrates G6P and F6P. They do not accept M6P at significant rates (<2%). Thus, cPGIs do not show PMI activity and differ from the enzymes of the bifunctional PGI/PMI family. It should be noted that pmi genes are present in all organisms containing a cupin-type PGI.
TABLE 2.
Comparison of molecular and kinetic properties of cPGIs from P. furiosus, A. fulgidus, M. mazei, S. enterica serovar Typhimurium, E. meliloti, and T. litoralis
| Parameter | Propertya of:
|
|||||
|---|---|---|---|---|---|---|
| PfcPGI* (22) | AfcPGI (This work) | MmcPGI (This work) | StcPGI (This work) | EmcPGIA (This work) | TlcPGI* (30) | |
| Standard assay temp (°C) | 80 | 50 | 37 | 37 | 50 | 50 |
| Apparent molecular mass (kDa) | ||||||
| Native enzyme | ||||||
| Gel filtration | 43 | 28 | 28 | 67 | 43 | 39 |
| Analytical ultracentrifugation | - | 27 | - | - | - | - |
| Subunit (SDS-PAGE) | 20 | 28 | 28 | 32 | 27 | 26.7 |
| Subunit (calculated) | 21.476 | 27.424 | 30.209 | 26.834 | 34.684 | 26.7 |
| Oligomeric structure | α2 | α | α | α2 | α2 | α2 |
| pH optimum | 7.0 | 7.4 | 7.6 | 7.2 | 7.4 | - |
| Temp optimum (°C) | >99 | 82 | 57 | 55 | 57 | - |
| Arrhenius activation energy (kJ/mol) | 50 | 71 ± 3 | 72 | 88 | 41 | - |
| Linear part of Arrhenius plot (°C) | 20-96 | 30-83 | 30-55 | 35-55 | 27-40 | - |
| Tm (°C) | >100* | 97 | 66 | 64 | 58 | - |
| Km (mM) | ||||||
| F6P | 1.0 | 0.6 | 0.7 | 3 | 0.8 | 1.7 |
| G6P | 2.9* | 0.6 | 1.0 | 1.0 | 0.4 | 11.7 |
| Vmax (U/mg) | ||||||
| F6P | 800 | 30 | 2.6 | 0.5 | 0.3 | 72.6 |
| G6P | 130 | 8.5 | 2.4 | 1.5 | 0.5 | 45.5 |
| Ki (mM) | ||||||
| Erythrose-4-phosphate | 3.9* | ND | 1.8 | ND | 1.8 | - |
| 6-Phosphogluconate | 0.30* | 0.67 | 0.15 | 3.0 | ND | 0.38 |
| Sorbitol-6-phosphate | 0.19* | 0.7 | 0.03 | 0.36 | 0.54 | - |
| Mannose-6-phosphate | ND* | ND | ND | ND | ND | 2.3 |
*, New data; ND, not detectable; -, not measured. The reference or source is indicated in parentheses in the subheading. F6P, fructose-6-phosphate; G6P, glucose-6-phosphate.
Inhibitors.
The potential inhibitory effects of several substances, including inhibitors of conventional PGIs, on cPGIs were analyzed (Table 2). Among the substances tested, only sorbitol-6-phosphate had a significant effect on the cPGIs with Ki values ranging from 0.003 to 0.7 mM. Erythrose-4-phosphate and 6-phosphogluconate, classical inhibitors of the conventional PGIs with Ki values in the micromolar range (e.g., (26, 44), could hardly inhibit cPGIs. Erythrose-4-phosphate had no effect on AfcPGI and StcPGI, and 6-phosphogluconate did not inhibit EmcPGIA (up to 5 mM). The distinct response with respect to these inhibitors might be due to the different catalytic mechanisms proposed, i.e., hydride transfer in cPGIs (49) versus proton transfer via cis-enediol intermediate reported for conventional PGIs (13, 37, 42). All other substances tested (ATP, Pi, fructose-1,6-bisphosphate, and fructose-1-phosphate) had no significant effect on the cPGIs used in the present study.
Metal dependence of cPGI activity.
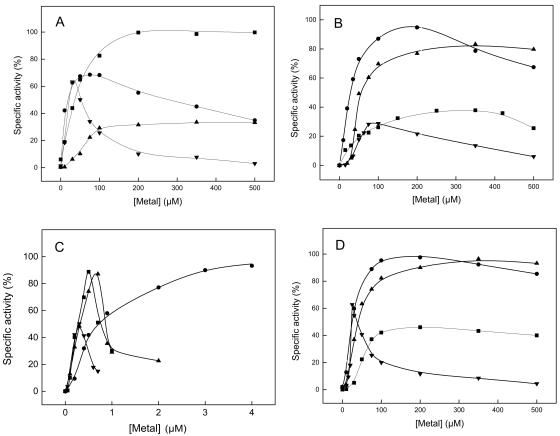
The majority of proteins with a cupin domain contain a metal-binding site for divalent cations that are essential for function (Table 3) (for a review, see reference 18). We analyzed here the role of metals on five cPGIs, including PfcPGI. (i) As analyzed by atomic absorption spectroscopy, all PGIs contained both iron and nickel, the respective metal contents (expressed as mole/mole subunits) are given in Table 3. None of the cPGIs were fully occupied by metals, indicating an underestimation of metal content, which is probably due to EDTA treatment of the proteins used to reduce unspecific binding of metals, e.g., to the His tag, prior to iron and nickel measurements (see Materials and Methods). The metal content measured depended on both the source of the protein and the purification procedure used due to different (contaminating) metal contents of the buffer systems used. (ii) All cPGIs were inhibited by millimolar concentrations of EDTA; the high Ki values obtained for EDTA (55 to 210 mM) (Table 3) indicate a high affinity for metals. (iii) Metal depletion of cPGIs was achieved by long-term incubation with high EDTA concentrations (500 mM) 10 to 20°C below the temperature optimum of the respective enzyme and extensive subsequent dialysis with H2O. After this treatment, all cPGIs in the present study, except for EmcPGIA, completely lost catalytic activity. The residual activity of EmcPGIA measured after EDTA treatment is probably due to an extreme tight binding of metals, which is also reflected by the highest Ki value for EDTA compared to the other cPGIs. However, catalytic activity of PfcPGI, AfcPGI, MmcPGI, and StcPGI could be restored by the addition of several divalent cations. The respective reactivation curves of PfcPGI, AfcPGI, MmcPGI, StcPGI, and EmcPGIA with Fe2+, Ni2+, Mn2+, and Zn2+ are given in Fig. 1, and the derived apparent Vmax values and apparent metal affinities as reflected by the metal concentrations at half-maximal velocity (KMe,0.5V values) are summarized in Table 3. The measured KMe,0.5V values for Fe2+, Ni2+, Mn2+, or Zn2+ varied between 7 and 1,180 μM, and the enzymes showed the lowest apparent KMe,0.5V values, which indicate a high affinity to either Fe2+ (AfcPGI), Ni2+ (PfcPGI and StcPGI), or Zn2+ (MmcPGI and StcPGI). Thus, under physiological conditions cPGIs presumably represent a heterogeneous mixture with respect to their metal content depending on both affinity and availability of the respective divalent metals. The highest catalytic activity was measured in the presence of Fe2+ (PfcPGI and MmcPGI), Ni2+ (AfcPGI, MmcPGI, StcPGI, and EmcPGIA), or Mn2+ (StcPGI). Generally, divalent metals were inhibitory at higher millimolar concentrations. This was especially true for Zn2+, which reactivates the cPGIs at low concentrations but was inhibitory at concentrations of >200 μM. In addition, when added at 100 μM, Co2+ and Cd2+ could be used by PfcPGI, AfcPGI, and StcPGI, whereas Mg2+ and Ca2+ were to some extent efficient only with PfcPGI and Cu2+ was rather inhibitory for all cPGIs.
TABLE 3.
Metal content, EDTA inhibition, and apparent affinities and activities with divalent cations of cPGIs from P. furiosus, A. fulgidus, M. mazei, S. enterica serovar Typhimurium, and E. melilotia
| Parameter | Propertyb of:
|
||||
|---|---|---|---|---|---|
| PfPGI* | AfcPGI* | MmcPGI | StcPGI | EmcPGIA | |
| Assay temp (°C) used | 80 | 50 | 37 | 37 | 50 |
| Metal content (mol/mol subunit) | |||||
| Ni2+ | 0.39*/0.12† | 0.41‡/0.06* | - | 0.20 | 0.07 |
| Fe2+ | 0.04*/0.07† | 0.03‡/0.01* | - | 0.17 | 0.13 |
| Ki EDTA (mM) | 55 | 97 | 101 | 120 | 210 |
| Metal reactivation apparent affinities Kaff (μM) | |||||
| Ni2+ | 7 | 37 | 1,180 | 22 | ND |
| Fe2+ | 40 | 7 | 200 | 50 | ND |
| Mn2+ | 61 | 81 | 220 | 38 | ND |
| Zn2+ | 17c | 46 | 130 | 22 | ND |
| Apparent Vmax (U/mg) | |||||
| Ni2+ | 61 | 7.0 | 2.3 | 1.2 | ND |
| Fe2+ | 92 | 2.8 | 2.3 | 0.5 | ND |
| Mn2+ | 26 | 6.2 | 2.3 | 1.2 | ND |
| Zn2+ | 51 | 1.8 | 1.3 | 0.7 | ND |
| Velocity at a 100 μM concn of metal | |||||
| Co2+ | 44 | 2.2 | 1.2 | ||
| Cd2+ | 23 | 2.1 | 0.4 | ND | |
| Ca2+ | 33 | 0.1 | 0.03 | ND | |
| Cu2+ | 0.5 | ND | - | 0.02 | ND |
| Mg2+ | 28 | 0.01 | - | 0.02 | ND |
The metal content was measured by atomic absorption spectroscopy. Apparent KMe,0.5V values and apparent Vmax values (substrate fructose-6-phosphate) were derived from reactivation curves of cPGIs (Fig. 2). All metals were used in the form of chlorides.
*, Recombinant protein without His tag; †, purified from P. furiosus; ‡, recombinant protein with His tag; ND, not detectable; -, not measured.
FIG. 1.
Reactivation of cPGIs from P. furiosus, A. fulgidus, M. mazei, and S. enterica serovar Typhimurium by metals after EDTA treatment and subsequent H2O dialyses. (A) PfcPGI; (B) AfcPGI; (C) MmcPGI; (D) StcPGI. Reactivation obtained with Fe2+ (▪), Ni2+ (•), Mn2+ (▴), and Zn2+ (▾) is shown. A total of 100% of the specific activity corresponds to 91 U/mg (PfcPGI), 7 U/mg (AfcPGI), 2.3 U/mg (MmcPGI), or 1.2 U/mg (StcPGI).
Thus, the data presented here, and in particular the reactivation experiments, indicate an involvement of metals in catalysis. Moreover, the presence of a divalent metal, most likely iron, as deduced from the crystal structure of PfcPGI bound with 6-phosphogluconate is a prerequisite in the recently proposed hydride transfer mechanism of PfcPGI (49).
Temperature optimum and thermostability.
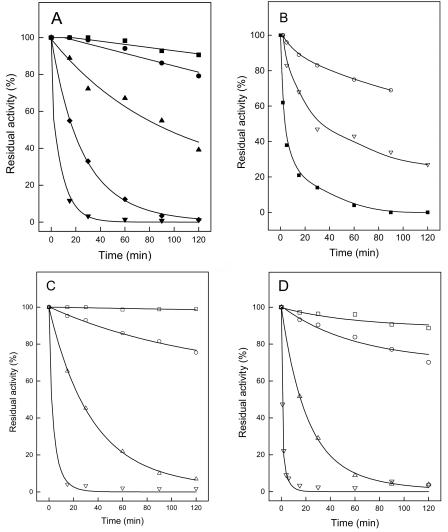
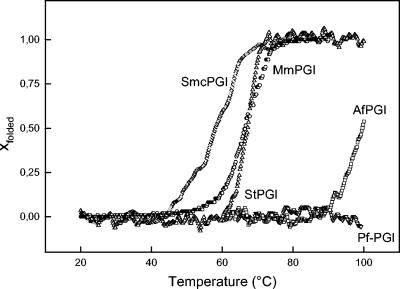
The thermophilic properties of the cPGIs were analyzed by determining the temperature dependence of the specific activity by time-dependent heat inactivation of the proteins (Fig. 2), as well as by heat-induced unfolding as detected by CD spectroscopy (Fig. 3). Not surprisingly, PfcPGI showed the highest thermostability; its temperature optimum has been shown to be >99°C (22), and no thermal unfolding could be detected at up to 100°C. AfcPGI had a temperature optimum at 82°C and an apparent melting temperature (Tm) of 97°C; however, complete unfolding could not be detected at temperatures up to 100°C. The enzyme was highly thermostable, retained >80% of the activity upon 120 min incubation at 80°C, and showed a half-life of ∼100 min at 90°C. At 100°C an almost complete loss of activity was observed after 60 min. MmcPGI and StcPGI showed temperature optima of 57 and 55°C, respectively, and Tm values of 66 and 64°C, respectively, were deduced from the respective monophasic and sigmoidal melting curve. MmcPGI and StcPGI did not lose activity at 40°C and had half-lives of ca. 25 min at 60 and 55°C, respectively. The EmcPGIA exhibited a temperature optimum of 57°C and a Tm of 58°C. The deduced activation energy of the latter was significant lower than the corresponding values of the other cPGIs in the present study. The EmcPGIA was quite heat resistant up to 50°C, with a half-life of >240 min, whereas at 55°C the half-life was ca. 20 min. The cPGIs from the mesophiles M. mazei, S. enterica serovar Typhimurium, and E. meliloti showed heat resistance and temperature optima 10 to 15°C above their respective growth temperature optima of 37, 37, and 28°C. One might speculate that the proteobacterial cPGIs might be more relevant at higher temperatures in the respective organisms.
FIG. 2.
Thermostability of cPGIs from A. fulgidus, M. mazei, S. enterica serovar Typhimurium, and E. meliloti. (A) AfcPGI; (B) MmcPGI; (C) StcPGI; (D) EmcPGIA. Symbols indicate thermostability at 100°C (▾), 95°C (⧫), 90°C (▴), 80°C (•), 70°C (▪), 60°C (▿), 55°C (▵), 50°C (○), and 40°C (□).
FIG. 3.
Thermal unfolding of cPGIs from P. furiosus, A. fulgidus, M. mazei, S. enterica serovar Typhimurium, and E. meliloti as monitored by CD spectroscopy. The values for PfcPGI (▿), AfcPGI (□), MmcPGI (○), StcPGI (▵), and EmcPGIA (◊) are indicated. Thermal unfolding was either detected at 226 nm (PfcPGI, MmcPGI, StcPGI, and EmcPGIA) or detected at 232 nm (AfcPGI and MmcPGI).
Sequence comparison and phylogenetic affiliation.
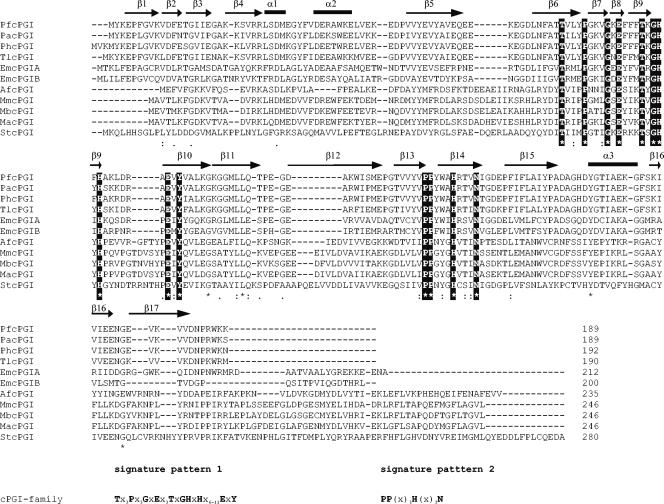
cPGIs show neither homology (ca. 5%) nor structural similarity to the PGI superfamily and represent a convergent line of PGI evolution (4, 22, 49, 52). Their function as cPGIs has been described either recently (22, 30, 52) or in the present study. Additional putative homologs are currently not present in protein databases. An alignment of all known cPGIs is given in Fig. 4 and revealed significant amino acid sequence identity (11 to 89%) among these sequences. A total of 17 residues are completely conserved among these 11 sequences, including the metal-binding site (His88, His90, His136, and Glu97; PfcPGI numbering) (4, 30, 49). In most cases the corresponding metal-binding site of other cupin superfamily proteins is coordinated by three histidines and a glutamate as well (18). In addition, all residues except for Tyr52, which is substituted by a His in StcPGI, i.e., residues Thr71, Gly87, Tyr99, and Tyr160, that have been proposed to be involved in substrate binding as deduced from the X-ray structure of the PfcPGI in the presence of inhibitors are conserved as well (49). Tyr152, Ala69, and Ala150, which have been suggested to contribute to the hydrophobicity of the active site of PfcPGI, are not conserved. Based on the alignment, two specific signature patterns of the cPGI family were deduced: TX3PX3GXEX3TXGHXHX6-11EXY and PPX3HX3N. The region between the two cupin motifs, which varies from a minimum of 11 amino acids in some microbial enzymes to more than 100 amino acids in certain eukaryotic transcription factors and dioxygenases (17, 18), comprises 20 to 31 amino acids in cPGIs.
FIG. 4.
Multiple sequence alignment of amino acid sequences of the cPGI family. The alignment was generated by using CLUSTAL X (50). Accession numbers were as follows: PfcPGI, NP577925; AfcPGI, NP070323; MmcPGI, NP633992; StcPGI, NP461684; EmcPGIA, Q92MQ8; EmcPGIB, Q92UI1 (and its putative homologs from P. horikoshi [O59618], P. abyssi [Q9UXW3], M. barkeri [ZP00076760], and M. acetivorans [NP615780]). The two derived signature patterns of the cPGI family are given below the alignment; invariable residues of these patterns are in boldface and are also highlighted in the alignment. Secondary structure assignments as derived from the X-ray structure of the PfcPGIs (49) are given above the sequences; α-helices are indicated as bars, and β-sheets are indicated as arrows.
Phylogenetic analyses and evolution of cPGIs.
In order to analyze the phylogenetic relation of cPGIs within the cupin superfamily, phylogenetic analyses were performed by two tree construction methods: NJ and ML. Indeed, in a phylogram including both cPGIs and several putative members of the cupin superfamily from eukaryal, bacterial, and archaeal cPGIs form a separate cluster within the cupin superfamily (not shown) and thus represents its own cPGI family. Thus far this family comprises nine euryarchaeal and two proteobacterial sequences. All other bacterial genomes thus far appear to contain only a conventional PGI. This includes all proteobacteria, especially Agrobacterium tumefaciens and S. enterica, which are the most closely related species from E. meliloti and S. enterica serovar Typhimurium, respectively, with sequenced genomes. Thus, for the respective proteobacterial cpgi genes, a lateral gene transfer from euryarchaeal species has to be assumed, since a loss of this gene in almost all bacteria appears to be unlikely. To test this hypothesis, phylogenetic analyses of the cPGI family were performed by the NJ and ML tree construction methods. A phylogram of the cPGI family based on an NJ tree is given in Fig. 5A. The topology was achieved by both methods (NJ and ML) and is supported by very good bootstrapping values. Not surprisingly, cPGI family sequences cannot be divided into a bacterial and an euryarchaeal cluster but rather into two or three subfamilies: Thermococcales and Methanosarcinales (Salmonella). The identity between these subfamilies is ca. 11 to 22% and members of a subfamily share molecular and spectral properties. Subfamily II comprises the monomeric cPGIs from Methanosarcinales and A. fulgidus. The Thermococcales subfamily includes the dimeric cPGIs from Thermococcus and Pyrococcales spp., as well as the two cPGI isoenzymes from E. meliloti. EmcPGIA and EmcPGIB share 51% amino acid identity and cluster closest to the TlcPGI, which also shows a high homology to these proteobacterial cPGIs (38 and 35% identity, respectively). Their presence is presumably the result of a lateral gene transfer from a Thermococcus-like species after the lines of E. meliloti and A. tumefaciens divided, followed by a late gene duplication in the evolution of E. meliloti. For the stcpgi gene, a lateral gene transfer has to be assumed after the lines of S. enterica serovar Typhimurium and S. enterica divided; however, the source of this transfer remains unknown.
Lateral gene transfers have been suggested for several conventional pgi genes (26, 31, 44), e.g., from eukarya to their symbionts and parasites or between (hyper)thermophilic bacteria and archaea (26, 44). Thus, the evolution and distribution of PGIs in the phylogenetic tree is complex and rather puzzling (Fig. 5B). The PGI superfamily is found in all three domains—archaea, bacteria, and eukarya—and probably has been present in the last common ancestor as well. Whereas PGIs from the PGI superfamily are found in all bacteria and the euryarchaea Methanococcus and halobacterial species, PGI/PMIs presumably have originated in the early crenarchaeal evolution by functional diversification and are found mainly in crenarchaea and also in two (hyper)thermophilic bacteria and in Thermoplasmatales (26). The assumption of lateral gene transfers from a euryarchaeal source to some proteobacteria defines the cPGI family as an euryarchaeal family that originated during early euryarchaeal evolution. A scenario would be plausible in which these novel cPGIs have replaced an ancestral conventional euryarchaeal PGI before the Thermococcales evolved. In the following euryarchaeal evolution this cpgi gene might have been replaced by a pgi or a pgi/pmi gene in Methanococcales and Halobacteriales or Thermoplasmatales, respectively.
In summary, we give an elaborate characterization of a novel protein family, the cPGI family, which represents a convergent line of PGIs. Cupin-type PGI represents a euryarchaeal PGI family and have spread to some bacteria by lateral gene transfer. They differ from conventional PGIs by their fold, a lower molecular mass, their metal dependence, and their inhibitory potential.
Acknowledgments
We thank C. Urbanke (Hanover, Germany) for analytical ultracentrifugation and R. Hedderich and J. Koch for determination of the metal content. The expert technical assistance of K. Lutter-Mohr, M. Kusche, and A. Brandenburger is also gratefully acknowledged.
This study was supported by grants of the Deutsche Forschungsgemeinschaft (SCHO 316/9-1) and the Fonds der Chemischen Industrie to P.S.
REFERENCES
- 1.Allard, S. T., M. F. Giraud, and J. H. Naismith. 2001. Epimerases: structure, function, and mechanism. Cell Mol. Life Sci. 58:1650-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, R., P. C. Dorrestein, C. Kinsland, T. P. Begley, and S. E. Ealick. 2002. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution. Biochemistry 41:7659-7669. [DOI] [PubMed] [Google Scholar]
- 3.Arsenieva, D., and C. J. Jeffery. 2002. Conformational changes in phosphoglucose isomerase induced by ligand binding. J. Mol. Biol. 323:77-84. [DOI] [PubMed] [Google Scholar]
- 4.Berrisford, J. M., J. Akerboom, A. P. Turnbull, D. DeGeus, S. E. Sedelnikova, I. Staton, C. W. McLeod, C. H. Verhees, O. J. van der, D. W. Rice, and P. J. Baker. 2003. Crystal structure of Pyrococcus furiosus phosphoglucose isomerase: implications for substrate binding and catalysis. J. Biol. Chem. 278:33290-33297. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chou, C. C., Y. J. Sun, M. Meng, and C. D. Hsiao. 2000. The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition. J. Biol. Chem. 275:23154-23160. [DOI] [PubMed] [Google Scholar]
- 7.Christendat, D., V. Saridakis, A. Dharamsi, A. Bochkarev, E. F. Pai, C. H. Arrowsmith, and A. M. Edwards. 2000. Crystal structure of dTDP-4-keto-6-deoxy-d-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP. J. Biol. Chem. 275:24608-24612. [DOI] [PubMed] [Google Scholar]
- 8.Cleasby, A., A. Wonacott, T. Skarzynski, R. E. Hubbard, G. J. Davies, A. E. Proudfoot, A. R. Bernard, M. A. Payton, and T. N. Wells. 1996. The X-ray crystal structure of phosphomannose isomerase from Candida albicans at 1.7 angstrom resolution. Nat. Struct. Biol. 3:470-479. [DOI] [PubMed] [Google Scholar]
- 9.Clifton, I. J., L. C. Hsueh, J. E. Baldwin, K. Harlos, and C. J. Schofield. 2001. Structure of proline 3-hydroxylase: evolution of the family of 2-oxoglutarate dependent oxygenases. Eur. J. Biochem. 268:6625-6636. [DOI] [PubMed] [Google Scholar]
- 10.Cordeiro, A. T., P. H. Godoi, C. H. Silva, R. C. Garratt, G. Oliva, and O. H. Thiemann. 2003. Crystal structure of human phosphoglucose isomerase and analysis of the initial catalytic steps. Biochim. Biophys. Acta 1645:117-122. [DOI] [PubMed] [Google Scholar]
- 11.Davies, C., and H. Muirhead. 2002. Crystal structure of phosphoglucose isomerase from pig muscle and its complex with 5-phosphoarabinonate. Proteins 49:577-579. [DOI] [PubMed] [Google Scholar]
- 12.Davies, C., and H. Muirhead. 2003. Structure of native phosphoglucose isomerase from rabbit: conformational changes associated with catalytic function. Acta Crystallogr. D Biol. Crystallogr. 59:453-465. [DOI] [PubMed] [Google Scholar]
- 13.Davies, C., H. Muirhead, and J. Chirgwin. 2003. The structure of human phosphoglucose isomerase complexed with a transition-state analogue. Acta Crystallogr. D Biol. Crystallogr. 59:1111-1113. [DOI] [PubMed] [Google Scholar]
- 14.Deleage, G., and C. Geourjon. 1993. An interactive graphic program for calculating the secondary structure content of proteins from circular dichroism spectrum. Comput. Appl. Biosci. 9:197-199. [DOI] [PubMed] [Google Scholar]
- 15.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 16.Dunwell, J. M. 1998. Sequence analysis of the cupin gene family in Synechocystis PCC6803. Microb. Comp. Genomics 3:141-148. [DOI] [PubMed] [Google Scholar]
- 17.Dunwell, J. M., A. Culham, C. E. Carter, C. R. Sosa-Aguirre, and P. W. Goodenough. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26:740-746. [DOI] [PubMed] [Google Scholar]
- 18.Dunwell, J. M., S. Khuri, and P. J. Gane. 2000. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64:153-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunwell, J. M., A. Purvis, and S. Khuri. 2004. Cupins: the most functionally diverse protein superfamily? Phytochemistry 65:7-17. [DOI] [PubMed] [Google Scholar]
- 20.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. Fitzhugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, D. M. Conway, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 22.Hansen, T., M. Oehlmann, and P. Schönheit. 2001. Novel type of glucose-6-phosphate isomerase in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:3428-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, T., B. Schlichting, and P. Schönheit. 2002. Glucose-6-phosphate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima: expression of the g6pd gene and characterization of an extremely thermophilic enzyme. FEMS Microbiol. Lett. 216:249-253. [DOI] [PubMed] [Google Scholar]
- 24.Hansen, T., and P. Schönheit. 2000. Purification and properties of the first-identified, archaeal, ATP-dependent 6-phosphofructokinase, an extremely thermophilic non-allosteric enzyme, from the hyperthermophile Desulfurococcus amylolyticus. Arch. Microbiol. 173:103-109. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, T., Urbanke, C., and P. Schönheit. 2004. Bifunctional phosphoglucose/phosphomannose isomerase from the hyperthermophilic archaeon Pyrobaculum aerophilum. Extremophiles 8:507-512. [DOI] [PubMed]
- 26.Hansen, T., D. Wendorff, and P. Schonheit. 2004. Bifunctional phosphoglucose/phosphomannose isomerases from the Archaea Aeropyrum pernix and Thermoplasma acidophilum constitute a novel enzyme family within the phosphoglucose isomerase superfamily. J. Biol. Chem. 279:2262-2272. [DOI] [PubMed] [Google Scholar]
- 27.Hesman, T. L., W. O. Barnell, and T. Conway. 1991. Cloning, characterization, and nucleotide sequence analysis of a Zymomonas mobilis phosphoglucose isomerase gene that is subject to carbon source-dependent regulation. J. Bacteriol. 173:3215-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeffery, C. J., B. J. Bahnson, W. Chien, D. Ringe, and G. A. Petsko. 2000. Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator. Biochemistry 39:955-964. [DOI] [PubMed] [Google Scholar]
- 29.Jeffery, C. J., R. Hardre, and L. Salmon. 2001. Crystal structure of rabbit phosphoglucose isomerase complexed with 5-phospho-d-arabinonate identifies the role of Glu357 in catalysis. Biochemistry 40:1560-1566. [DOI] [PubMed] [Google Scholar]
- 30.Jeong, J. J., S. Fushinobu, S. Ito, B. S. Jeon, H. Shoun, and T. Wakagi. 2003. Characterization of the cupin-type phosphoglucose isomerase from the hyperthermophilic archaeon Thermococcus litoralis. FEBS Lett. 535:200-204. [DOI] [PubMed] [Google Scholar]
- 31.Katz, L. A. 1996. Transkingdom transfer of the phosphoglucose isomerase gene. J. Mol. Evol. 43:453-459. [DOI] [PubMed] [Google Scholar]
- 32.Kindler, B. 1997. AKKUPROG. Ph.D. thesis. University of Hanover, Hanover, Germany.
- 33.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 34.Ko, T. P., Y. G. Kuznetsov, A. J. Malkin, J. Day, A. McPherson, T. P. Ko, J. Day, A. McPherson. 2001. X-ray diffraction and atomic force microscopy analysis of twinned crystals: rhombohedral canavalin. Acta Crystallogr. D Biol. Crystallogr. 57:829-839. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence, M. C., E. Suzuki, J. N. Varghese, P. C. Davis, A. Van Donkelaar, P. A. Tulloch, and P. M. Colman. 1990. The three-dimensional structure of the seed storage protein phaseolin at 3 Å resolution. EMBO J. 9:9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, J. H., K. Z. Chang, V. Patel, and C. J. Jeffery. 2001. Crystal structure of rabbit phosphoglucose isomerase complexed with its substrate d-fructose 6-phosphate. Biochemistry 40:7799-7805. [DOI] [PubMed] [Google Scholar]
- 38.Mander, G. J., A. J. Pierik, H. Huber, and R. Hedderich. 2004. Two distinct heterodisulfide reductase-like enzymes in the sulfate-reducing archaeon Archaeoglobus profundus. Eur. J. Biochem. 271:1106-1116. [DOI] [PubMed] [Google Scholar]
- 39.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 40.Noltmann, E. A. 1972. Aldose-ketose isomerases, p. 271-354. In P. D. Boyer (ed.), The enzymes. Academic Press, Inc., New York, N.Y.
- 41.Otte, S., and J. W. Lengeler. 2001. The mtl genes and the mannitol-1-phosphate dehydrogenase from Klebsiella pneumoniae KAY2026. FEMS Microbiol. Lett. 194:221-227. [DOI] [PubMed] [Google Scholar]
- 42.Read, J., J. Pearce, X. Li, H. Muirhead, J. Chirgwin, and C. Davies. 2001. The crystal structure of human phosphoglucose isomerase at 1.6 Å resolution: implications for catalytic mechanism, cytokine activity and hemolytic anaemia. J. Mol. Biol. 309:447-463. [DOI] [PubMed] [Google Scholar]
- 43.Roach, P. L., I. J. Clifton, C. M. Hensgens, N. Shibata, C. J. Schofield, J. Hajdu, and J. E. Baldwin. 1997. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature 387:827-830. [DOI] [PubMed] [Google Scholar]
- 44.Rudolph, B., T. Hansen, and P. Schönheit. 2004. Glucose-6-phosphate isomerase from the hyperthermophilic archaeon Methanococcus jannaschii: characterization of the first archaeal member of the phosphoglucose isomerase superfamily. Arch. Microbiol. 181:82-87. [DOI] [PubMed] [Google Scholar]
- 45.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreyer, R., and A. Bock. 1980. Phosphoglucose isomerase from Escherichia coli K-10: purification, properties, and formation under aerobic and anaerobic condition. Arch. Microbiol. 127:289-298. [DOI] [PubMed] [Google Scholar]
- 47.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, C. Wolberger, S. M. Soisson, B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. The 1.6 Å crystal structure of the AraC sugar-binding and dimerization domain complexed with d-fucose: structural basis for ligand-regulated oligomerization of AraC. J. Mol. Biol. 273:226-237. [DOI] [PubMed] [Google Scholar]
- 48.Stetter, K. O. 1996. Hyperthermophilic procaryotes. FEMS Microbiol. Rev. 18:149-158. [Google Scholar]
- 49.Swan, M. K., T. Hansen, P. Schönheit, and C. Davies. 2003. Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus. J. Biol. Chem. 278:47261-47268. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Titus, G. P., H. A. Mueller, J. Burgner, D. C. Rodriguez, M. A. Penalva, and D. E. Timm. 2000. Crystal structure of human homogentisate dioxygenase. Nat. Struct. Biol. 7:542-546. [DOI] [PubMed] [Google Scholar]
- 52.Verhees, C. H., M. A. Huynen, D. E. Ward, E. Schiltz, W. M. De Vos, and J. Van der Oost. 2001. The phosphoglucose isomerase from the hyperthermophilic archaeon Pyrococcus furiosus is a unique glycolytic enzyme that belongs to the cupin superfamily. J. Biol. Chem. 276:40926-40932. [DOI] [PubMed] [Google Scholar]
- 53.Verhees, C. H., S. W. Kengen, J. E. Tuininga, G. J. Schut, M. W. Adams, W. M. De Vos, and O. J. van der. 2003. The unique features of glycolytic pathways in Archaea. Biochem. J. 375:231-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woo, E. J., J. M. Dunwell, P. W. Goodenough, A. C. Marvier, and R. W. Pickersgill. 2000. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7:1036-1040. [DOI] [PubMed] [Google Scholar]