Abstract
The σS (or RpoS) subunit of RNA polymerase is the master regulator of the general stress response in Escherichia coli. While nearly absent in rapidly growing cells, σS is strongly induced during entry into stationary phase and/or many other stress conditions and is essential for the expression of multiple stress resistances. Genome-wide expression profiling data presented here indicate that up to 10% of the E. coli genes are under direct or indirect control of σS and that σS should be considered a second vegetative sigma factor with a major impact not only on stress tolerance but on the entire cell physiology under nonoptimal growth conditions. This large data set allowed us to unequivocally identify a σS consensus promoter in silico. Moreover, our results suggest that σS-dependent genes represent a regulatory network with complex internal control (as exemplified by the acid resistance genes). This network also exhibits extensive regulatory overlaps with other global regulons (e.g., the cyclic AMP receptor protein regulon). In addition, the global regulatory protein Lrp was found to affect σS and/or σ70 selectivity of many promoters. These observations indicate that certain modules of the σS-dependent general stress response can be temporarily recruited by stress-specific regulons, which are controlled by other stress-responsive regulators that act together with σ70 RNA polymerase. Thus, not only the expression of genes within a regulatory network but also the architecture of the network itself can be subject to regulation.
The general stress sigma factor σS (or RpoS) is strongly induced when Escherichia coli cells are exposed to various stress conditions, which include starvation, hyperosmolarity, pH downshift, or nonoptimal high or low temperature (for a review of σS regulation, see reference 24). By standard genetic and molecular biology methods, more than 80 σS-controlled genes have been identified to date, indicating that σS is the master regulator of a rather large regulon which represents the genetic basis of the E. coli general stress response (for summaries, see references 23 and 41).
In their regulatory patterns, many σS-controlled genes just follow the cellular σS level; i.e., they are activated whenever σS and therefore σS-containing RNA polymerase (EσS) accumulate in the cell. Other σS-dependent genes, however, exhibit highly specific regulation, with a narrow window of expression only under some sort of stress condition. The best-studied example of this type of σS-controlled gene is the csiD gene, which is mainly induced by carbon starvation because the cyclic AMP (cAMP)-cAMP receptor protein (CRP) acts as an essential activator for σS-containing RNA polymerase at the csiD promoter (21, 46, 49). Also, the leucine-responsive regulatory protein (Lrp) is involved in the regulation of certain σS-dependent genes (9, 13, 33, 64). These findings indicate that the σS-containing RNA polymerase holoenzyme has the ability to cooperate with additional regulatory factors, just as the vegetative RNA polymerase containing σ70 does.
The identification of a clearly defined σS consensus promoter sequence and therefore the prediction of σS-controlled promoters in upstream regions of genes in the E. coli genome have been notoriously difficult. σS is highly related to σ70, and genes that are dependent on EσS in vivo can often be transcribed in vitro by Eσ70, and vice versa. The current view of this “sigma selectivity paradox” is that EσS and Eσ70 in principle use very similar promoters but that minor differences, e.g., in the extended −10 region (6), can shift the preference towards one or the other holoenzyme. Also, transcription initiation by EσS is less affected by various deviations from the classical promoter consensus sequence (e.g., by degeneration of the −35 sequence) (20), which gives EσS an advantage at nonoptimal promoters (summarized in reference 25).
The present study was undertaken with a number of questions and goals in mind. How many and which genes in the E. coli genome are under σS control? Does a more or less complete set of these genes provide us with novel insights into the physiological function of the σS regulon? Can we use such a database of σS-dependent genes for unequivocal in silico identification of a σS consensus promoter sequence? Does expression of the majority of these genes just follow σS levels, or is differential regulation common among σS-dependent genes? In view of the similarity between σ70- and σS-controlled promoters, can EσS selectivity be conditional and can histone-like proteins globally affect EσS and/or Eσ70 promoter selectivity (as suggested for Lrp in a few cases so far)? The present study provides answers to these questions from a genome-wide perspective.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are derivatives of E. coli K-12 strain MC4100 (11). Mutant alleles previously described are rpoS359::Tn10 (35), rpoS::kan (7), and lrp-201::Tn10 (17). The gadX::cat insertion was isolated by one-step inactivation (15) with the following primers: 5′-GCGTGCTACATTAATAAACAGTAATATGTTTATGTAATATTAAGTCAACTGTGTAGGCTGGAGCTGCTTC-3′ and 5′-ATGTCTGAGTAAAACTCTATAATCTTATTCCTTCCGCAGAACGGTCAGTGCATATGAATATCCTCCTTAG-3′ (nucleotides shown in boldface type deviate from the gadX sequence). These mutations were introduced into MC4100 by P1 transduction (50).
Standard batch cultures were grown at 37°C under aeration in a rotary shaker in Luria-Bertani (LB) medium. Ampicillin (100 μg/ml) was added for plasmid-carrying strains. For hyperosmotic shift experiments, cultures were grown in M9 (50) with glycerol (0.4%) for three or more generations before 0.3 M NaCl was added. For pH downshift experiments, cultures were grown in LB medium for four or more generations before 170 mM 4-morpholine-methanesulfonic acid (MES) was added (this procedure acidifies the medium to pH 5). Growth was monitored by measuring the optical density at 578 nm (OD578).
Origin of E. coli DNA microarrays.
Genomic E. coli K-12 DNA microarrays were made by robotically spotting PCR products. The PCR products were generated with an ORFmer primer set (Genosys Biotechnologies, Cambridge, United Kingdom) giving full-length open reading frames as double-stranded DNA. Details of the spotting procedure and quality control of the microarrays were described previously (39, 54, 71).
RNA preparation and cDNA labeling.
For total RNA preparation, cultures were grown and harvested under three different conditions known to produce high σS levels and σS-dependent target gene expression in wild-type strains: (i) during transition into stationary phase in LB medium at an OD578 of 4.0, (ii) 20 min after the addition of 0.3 M NaCl (added at an OD578 of 0.3) in M9-0.4% glycerol, and (iii) 40 min after a shift to pH 5 in LB medium (MES was added at an OD578 of 0.4).
One volume of cell suspension was harvested on 0.5 volume of ice (−20°C) and centrifuged immediately for 2 min at 4,500 × g at 4°C. The pellet was resuspended in 700 μl of RLT buffer (RNeasy; QIAGEN, Hilden, Germany) and transfered to a vial with 1 g of 0.1-mm Zirconia/Silica beads (Roth, Karlsruhe, Germany). Cells were disrupted with a Mini-BeadBeater (Biospec Products, Inc.) at 5,000 rpm for three intervals of 30 s each. After centrifugation, the lysate was supplemented with 500 μl of ethanol and split in two portions, and total RNA was extracted with two RNeasy mini-columns according to the manufacturers instructions (QIAGEN). The isolated RNA was treated with 30 U of DNase I (RNase free; Roche, Mannheim, Germany) in DNase I buffer (1 M sodium acetate, 50 mM MgSO4 [pH 5.0]) for 20 min at 37°C, incubated for 10 min at 70°C for inactivation of DNase I, and extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1), followed by ethanol precipitation (39, 54, 58). RNA concentration and quality were checked photometrically and on formaldehyde gels according to standard procedures (58). Equal amounts of total RNA (each, 20 to 25 μg) were used for random hexamer-primed synthesis of fluorescence-labeled cDNA with the fluorescent nucleotide analogues Cy3-dUTP and Cy5-dUTP (Amersham Pharmacia), as described previously (39, 54, 71).
DNA microarray analysis.
Hybridization of the fluorescence-labeled cDNA to the microarrays and the washing protocol were described previously (39, 54, 71). Fluorescence at 532 nm (Cy3-dUTP) and 635 nm (Cy5-dUTP) was determined at a 10-μm resolution with a GenePix 4000 (Axon, Inc.) laser scanner. TIFF images were analyzed with the software GenePix Pro 3.0 (Axon). The normalized Cy5/Cy3 ratio for the median was taken to reflect relative RNA level changes (39, 54).
Data analysis.
Each microarray experiment was repeated independently at least three times (biological replicates). Genes were considered differentially expressed according to the following criteria. (i) Reliable detection was based on signal-to-noise ratios exceeding a factor of 3. (ii) Reliable detection was confirmed in at least two out of three repetitions. (iii) In a paired Student's t test, relative RNA levels were significantly different from the levels of the genomic DNA controls (P < 0.05) (39, 54). (iv) Average relative RNA level changes were at least twofold (in all three replicate experiments).
Functional grouping of genes was made according to the data from GenProtEC (http://genprotec.mbl.edu/) (60). MEME (http://meme.sdsc.edu/) (3) and BioProspector (http://robotics.stanford.edu/∼xsliu/BioProspector/) (40) were used for sequence analysis of upstream regions of coregulated open reading frames. Both algorithms seek conserved motifs in sets of unaligned sequences. The sequence logo was created with WebLogo (http://weblogo.berkeley.edu/) (14).
RNA preparation and primer extension.
For gadE mRNA detection by primer extension experiments, cells were grown under the same conditions as for the microarray experiments (see above). Total RNA was prepared and subject to reverse transcription as previously described (49). The primer used for reverse transcription was 5′-ATCTTTCAACTGCCAAAAGCCCTGT-3′.
Construction of lacZ reporter fusions and their transfer into the chromosome.
Chromosomal lacZ fusions to the gadA and gadB genes (as well as to the regulatory gene gadE) were isolated with the fusion vectors pJL28 and pJL30, respectively (42), which are pMLB1034 (62) derivatives carrying the polylinkers of pNM480 and pNM482 (52). The inserts were generated by PCR with MC4100 chromosomal DNA as a template, digested with EcoRI and SalI, and cloned into the fusion vector, which was digested with the same enzymes. The following primers were used for PCR: (i) 5′-GTGGATGAATTCGTAGCTTTCCTGC-3′ (upstream of gadA), (ii) 5′-GTGAGAATTCAGGAGACACAGAATGC-3′ (upstream of gadB), (iii) 5′-GATAATCTGAAAGTCGACATCATCGC-3′ (used for gadA and gadB, since the coding regions for the isoenzymes GadA and GadB are nearly identical), (iv) 5′-TTGAATTCCGCATAAATATCCGTGTCTCCAGACG-3′ (upstream of gadE), and (v) 5′-ATCTATAAGCTTTATCTTTCAACTGCCAAAAGCCCTG-3′ (reverse primer complementary to the coding sequence of gadE). These constructs result in translational fusions inserted after nucleotide 138 of the coding regions of gadA and gadB and after nucleotide 61 of gadE. Translational fusions were converted to transcriptional fusions, as previously described (56) (with a HindIII-ClaI fragment to replace a corresponding fragment carrying the fusion joint and thereby introducing stop codons in all three reading frames, a Shine-Dalgarno sequence, and an initiation codon for lacZ). PCR-derived parts of the resulting plasmids carrying all fusions were sequenced. All constructs were crossed onto λRS45, followed by lysogenization into MC4100 according to the method described in reference 63. Single lysogeny was tested by a PCR method (55).
β-Galactosidase assay.
β-Galactosidase activity was assayed with o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate and is reported as the number of micromoles of o-nitrophenol per minute per milligram of cellular protein (50).
RESULTS
Genome-wide identification of σS-dependent genes under three different growth and stress conditions.
We used genome-wide transcription profiling of isogenic rpoS+ and rpoS::Tn10 strains to identify σS-dependent genes of E. coli. To find as many such genes as possible, we used not only one but three different culture growth conditions, which are known to result in high cellular σS levels. These were (i) growth in LB medium to an OD578 of 4.0 (this corresponds to transition into stationary phase), (ii) growth in minimal medium to which 0.3 M NaCl was added at an OD578 of 0.3 (cells were harvested 20 min after this hyperosmotic shift), and (iii) growth in LB medium which was acidified by the addition of MES at an OD578 of 0.4 (cells were harvested 40 min after this shift to pH 5). Total RNA and labeled cDNA were prepared and hybridized to genomic E. coli microarrays (for details, see Materials and Methods).
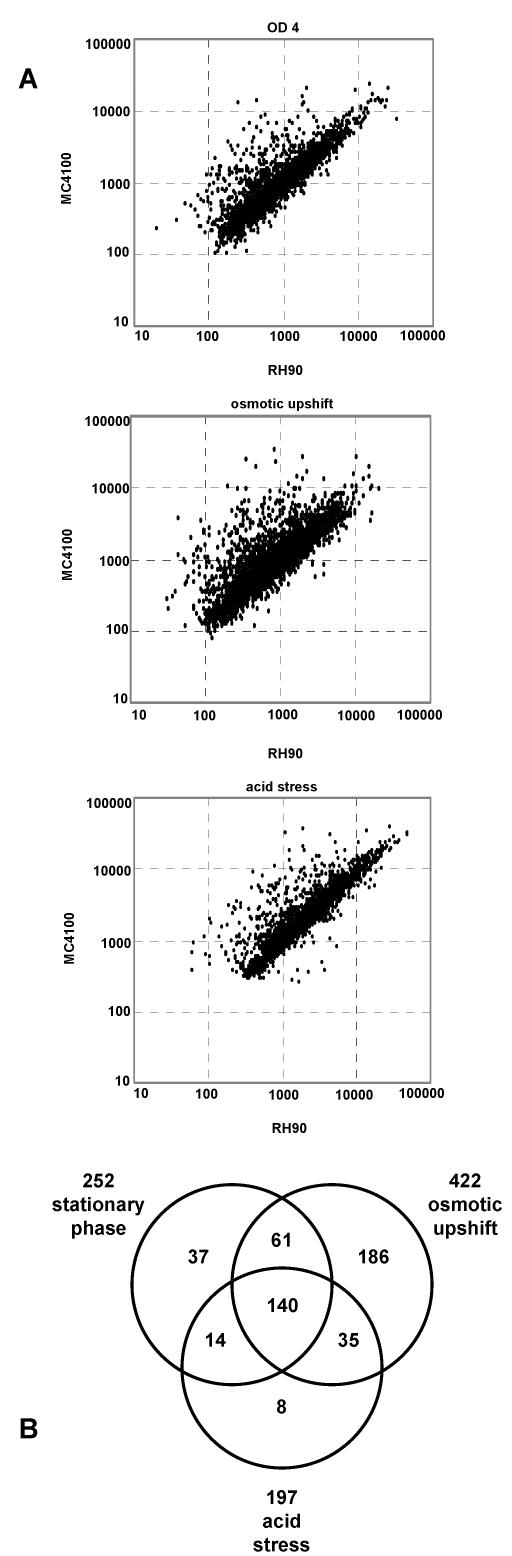
We identified a total of 481 genes, which exhibited >2-fold-higher expression in the rpoS+ strain than in the rpoS mutant (Fig. 1). In addition, 95 genes showed >2-fold-higher expression in the rpoS mutant (at least under one condition tested); i.e., they appeared to be negatively controlled by σS (Fig. 1A). Most likely, these latter genes are expressed by σ70-containing RNA polymerase (Eσ70) from promoters that are sensitive to the increase in the cellular concentration of Eσ70 that results from a lack of competition between σS and σ70 for core polymerase in the rpoS mutant. Alternatively, some of the negatively σS-controlled genes may be subject to repression by σS-dependent regulatory proteins.
FIG. 1.
(A) Comparison of genome-wide gene expression in rpoS+ and rpoS::Tn10 strains (MC4100 and RH90, respectively) under three different growth and stress conditions. RNA was prepared during entry into stationary phase in LB medium (OD578 = 4), 20 min after the addition of 0.3 M NaCl in minimal medium and 40 min after a shift to pH 5 in LB medium. Cy3- and Cy5-labeled cDNAs obtained from these RNA preparations were analyzed by whole-genome microarray analysis, and normalized intensities are visualized as scatter plots (MC4100 versus RH90). All data are the average of three independent identical experiments. (B) The numbers of σS-controlled genes (i.e., genes with an at least twofold difference in expression in rpoS+ and rpoS::Tn10 strains) identified under one, two, or all three conditions tested are shown as a Venn diagram.
Interestingly, only 140 of the positively σS-controlled genes were found under all three growth and stress conditions (Fig. 1B); expression ratios for rpoS+ and rpoS-negative strains as well as assigned functions of these genes are listed in Table 1. The expression of these genes may just change in parallel with σS levels, and we henceforth refer to them as the core set of σS-controlled genes. The other 341 genes revealed their σS dependence under only one or two of the growth and stress conditions used (with genes in all possible combinatorial groups) (Fig. 1B). Especially noteworthy was a large set of 186 genes, which was observed as σS controlled only under conditions of osmotic upshift (Fig. 1B). Thus, the majority of σS-controlled genes require not only the presence of σS but also specific environmental conditions for expression. Alternatively, certain subsets of genes may switch from σS to σ70 dependence under certain conditions. Taken together, these data indicate that the σS regulon (i) is much larger (up to 10% of all E. coli K-12 genes) and (ii) displays a higher degree of internal differential regulation than previously suspected.
TABLE 1.
Expression ratios of σS-controlled genes identified under three different σS-inducing conditions
| Function (reference) and gene | b no. | Gene product | Ratio
|
||
|---|---|---|---|---|---|
| OD 4a | NaClb | pH 5c | |||
| Metabolism (26) | |||||
| adhP | b1478 | Alcohol dehydrogenase, propanol preferring | 5.38 | 9.19 | 3.43 |
| amyA | b1927 | Cytoplasmic alpha-amylase | 12.21 | 11.02 | 10.57 |
| dkgV | b3012 | 2,5-Diketo-D-gluconate reductase A | 4.37 | 6.33 | 2.37 |
| fbaB | b2097 | Fructose-bisphosphate aldolase class I | 8.00 | 9.61 | 9.33 |
| gabD | b2661 | Succinate-semialdehyde dehydrogenase NADP dependent | 4.81 | 4.08 | 2.89 |
| hdhA | b1619 | alpha-hydroxysteroid dehydrogenase, NAD dependent | 2.71 | 6.97 | 2.43 |
| hycF | b2720 | Hydrogenase 3, putative quinone oxidoreductase, FeS related | 10.67 | 24.06 | 9.11 |
| IdcC | b0186 | Lysine decarboxylase 2, constitutive | 3.76 | 3.72 | 3.88 |
| narU | b1469 | Nitrate extrusion protein | 2.75 | 3.62 | 2.07 |
| narY | b1467 | Nitrate reductase 2, beta-subunit | 5.24 | 5.70 | 4.57 |
| poxB | b0871 | Pyruvate dehydrogenase/oxidase:FAD and thiamine PPi-binding | 17.67 | 45.89 | 29.60 |
| qor | b4051 | Quinone oxidoreductase, NADPH dependent | 2.39 | 2.73 | 2.64 |
| talA | b2464 | Transaldolase A | 6.79 | 13.08 | 13.83 |
| tam | b1519 | Trans-aconitate methyltransferase | 2.76 | 8.68 | 3.53 |
| tktB | b2465 | Transketolase 2, thiamin binding, isozyme | 14.08 | 35.87 | 10.76 |
| ugp | b3449 | Glycerophosphodiester phosphodiesterase, cytosolic | 2.30 | 3.69 | 2.01 |
| ybaS | b0485 | Putative glutaminase/carboxypeptidase with beta-lactamase/d-Ala-carboxypeptidase domain | 14.05 | 9.46 | 2.70 |
| ybaY | b0453 | Glycoprotein/polysaccharide metabolism | 6.15 | 8.67 | 6.09 |
| ycaC | b0897 | Putative enzyme with cysteine hydrolase domain | 13.34 | 5.92 | 5.88 |
| yfcF | b2301 | Putative glutathione S-transferase enzyme with thioredoxin-like domain | 2.55 | 2.44 | 2.40 |
| ygjG | b3073 | Putrescine:2-oxoglutaric acid aminotransferase | 8.93 | 16.90 | 7.45 |
| yhfW | b3380 | Putative phosphopentomutase, alkaline phosphatase-like domain | 4.00 | 6.67 | 4.46 |
| yjcS | b4083 | Putative enzyme with 2 metallohydrolase/oxidoreductase domains | 10.73 | 10.38 | 4.71 |
| yjgB | b4269 | Putative alcohol dehydrogenase with NAD(P)-binding and GroES domains | 6.98 | 10.80 | 7.40 |
| yncG | b1454 | Putative glutathione S-transferase with thioredoxin-like and glutathione S-transferases, C-terminal domain | 3.42 | 13.77 | 3.64 |
| ysgA | b3830 | Putative dienelactone hydrolase | 2.69 | 2.27 | 3.68 |
| Regulation (11) | |||||
| bolA | b0435 | Transcriptional activator of morphogenic pathway (BolA family), important in general stress response | 4.74 | 8.47 | 4.77 |
| chaB | b1217 | Cation transport regulator | 3.36 | 5.54 | 3.38 |
| csiR | b2664 | Putative transcriptional repressor with DNA-binding winged helix domain (GntR family) | 4.84 | 10.09 | 4.30 |
| gadW | b3515 | Transcriptional regulator for GadX (regulatory protein), glutamic acid decarboxylase (GadA, -B), and glutamate transport protein (GadC) (AraC-XylS family) | 6.12 | 5.98 | 3.09 |
| gadX | b3516 | Transcriptional regulator for glutamic acid decarboylase and transporter (GadA, GadBC) (AraC/XylS family) | 6.09 | 7.45 | 3.14 |
| gem | b1285 | RNase II modulator with PYP-like sensor domain | 6.78 | 3.83 | 6.94 |
| pdhR | b0113 | Transcriptional repressor for pyruvate dehydrogenase complex (GntR family) | 5.01 | 3.42 | 2.86 |
| rssB | b1235 | Response regulator involved in protein turnover, controls stability of RpoS | 4.68 | 7.19 | 4.08 |
| wrbA | b1004 | Flavodoxin-like protein, trp repressor-binding protein | 9.38 | 6.83 | 14.77 |
| yiaG | b3555 | Putative transcriptional regulator with DNA-binding domain | 8.40 | 9.20 | 4.08 |
| yjdC | b4135 | Putative regulator with homeodomain-like DNA-binding domain (TetR/AcrR family) | 2.49 | 2.34 | 2.46 |
| Transport and membrane (20) | |||||
| artM | b0861 | Arginine transport protein (ABC superfamily, membrane) | 2.20 | 2.79 | 2.61 |
| artP | b0864 | Arginine transport protein (ABC superfamily, ATP-binding subunit) | 3.50 | 4.56 | 4.71 |
| blc | b4149 | Outer membrane lipoprotein (lipocalin) | 4.77 | 8.99 | 8.10 |
| eutH | b2452 | Putative transport protein, ethanolamine utilization | 3.11 | 3.06 | 2.59 |
| gabP | b2663 | Gamma-aminobutyrate transport protein | 4.06 | 4.70 | 2.83 |
| mscL | b3291 | Mechanosensitive channel | 2.94 | 2.31 | 2.59 |
| potF | b0854 | Putrescine transport protein (ABC superfamily, periplasmic binding protein) | 2.76 | 4.25 | 2.24 |
| rssA | b1234 | Putative transmembrane protein | 2.50 | 3.19 | 2.26 |
| ugpB | b3453 | Sn-Glycerol 3-phosphate transport protein (ABC superfamily, periplasmic binding protein) | 3.29 | 5.55 | 3.29 |
| ugpC | b3450 | Sn-Glycerol 3-phosphate transport protein (ABC superfamily, ATP-binding subunit) | 2.39 | 2.50 | 2.44 |
| ybiO | b0808 | Putative transport protein, integral membrane location | 8.49 | 4.51 | 2.02 |
| ybiP | b0815 | Putative transmembrane protein (N terminal); putative phosphatase (C terminal) | 7.73 | 5.89 | 2.43 |
| ydcS | b1440 | Putative transport protein (ABC superfamily, periplasmic binding protein) | 21.66 | 4.83 | 4.36 |
| ydhJ | b1644 | Putative multidrug resistance membrane protein | 2.21 | 4.29 | 2.22 |
| yeaY | b1806 | Putative membrane protein | 2.43 | 3.21 | 4.06 |
| yebS | b1833 | Putative membrane protein | 2.08 | 2.50 | 2.48 |
| yhhT | b3474 | Putative permease (PerM family) | 2.30 | 3.89 | 2.92 |
| yhiD | b3522 | Putative membrane protein | 4.11 | 5.93 | 7.24 |
| yphA | b2543 | Putative transmembrane protein | 2.35 | 3.38 | 2.53 |
| yqaE | b2666 | Putative transport protein (YqaE family) | 2.42 | 4.78 | 3.16 |
| Adaptation to stress (16) | |||||
| aidB | b4187 | Putative acyl-CoA dehydrogenase (flavoprotein), adaptive response (transcription activated by Ada) | 4.11 | 2.08 | 3.47 |
| bfr | b3336 | Bacterioferritin, an iron storage homoprotein | 8.84 | 3.28 | 4.72 |
| dps | b0812 | Stress response DNA-binding protein with ferritin-like domain | 8.92 | 24.19 | 4.32 |
| gadA | b3517 | Glutamate decarboxylase, isozyme A | 32.38 | 5.80 | 2.33 |
| gadB | b1493 | Glutamate decarboxylase, isozyme B | 16.44 | 3.96 | 2.22 |
| katE | b1732 | Catalase; hydroperoxidase HPII (III), RpoS dependent | 11.14 | 9.00 | 7.37 |
| osmB | b1283 | Lipoprotein, osmotically inducible | 3.34 | 2.36 | 2.34 |
| osmC | b1482 | Resistance protein, osmotically inducible | 4.93 | 6.19 | 3.51 |
| osmY | b4376 | Hyperosmotically inducible periplasmic protein, RpoS-dependent stationary-phase gene | 50.66 | 68.76 | 19.08 |
| otsA | b1896 | Trehalose-6-phosphate synthase | 10.27 | 52.44 | 13.79 |
| otsB | b1897 | Trehalose-6-phosphate phosphatase, biosynthetic | 5.56 | 38.22 | 9.09 |
| psiF | b0384 | Induced by phosphate starvation | 9.48 | 10.53 | 7.42 |
| treA | b1197 | Trehalase, periplasmic | 7.98 | 12.71 | 2.50 |
| treF | b3519 | Trehalase, cytoplasmic | 2.07 | 9.25 | 5.20 |
| uspB | b3494 | Universal stress protein B | 4.52 | 5.91 | 3.61 |
| gadC | b1492 | Putative glutamate:gamma-aminobutyric acid antiporter (APC family) | 10.28 | 3.48 | 2.22 |
| Protein processing (7) | |||||
| cbpA | b1000 | Curved DNA-binding protein, cochaperone of DnaK (Hsp40 family) | 4.97 | 4.16 | 2.05 |
| hchA | b1967 | Molecular chaperone independent of ATP/ADP cycle, with class I glutamine amidotransferase-like domain | 6.02 | 15.64 | 4.89 |
| hycI | b2717 | Protease involved in processing C-terminal end of HycE | 4.00 | 4.16 | 2.79 |
| rpsV | b1480 | 30S ribosomal subunit protein S22 | 5.20 | 6.07 | 4.60 |
| ycfF | b1103 | Putative inhibitor of protein kinase C; contains a transferase domain | 2.68 | 2.99 | 2.10 |
| ylgH | b4248 | Putative translation factor | 6.04 | 4.70 | 3.06 |
| yhbO | b3153 | Putative intracellular proteinase with class I glutamine amidotransferase-like domain | 4.94 | 23.00 | 12.10 |
| Other or undefined function (60) | |||||
| csiD | b2659 | Conserved protein with clavaminate synthase-like domain | 6.90 | 7.82 | 3.68 |
| elaB | b2266 | Unknown CDSd | 8.11 | 17.82 | 11.80 |
| erfK | b1990 | Conserved hypothetical protein with NAD(P)-binding Rossmann-fold domain | 2.24 | 4.55 | 2.18 |
| fic | b3361 | Possible cell filamentation protein, induced in stationary phase | 12.94 | 17.76 | 10.82 |
| msyB | b1051 | Acidic protein suppresses mutants lacking function of protein export | 17.37 | 8.60 | 8.89 |
| phnB | b4107 | Conserved protein with glyoxalase and dihydroxybiphenyl dioxygenase domain | 4.81 | 5.37 | 3.38 |
| yaiA | b0389 | Unknown CDS | 3.92 | 4.72 | 2.41 |
| yajO | b0419 | Putative oxidoreductase, NAD(P) dependent | 2.21 | 6.76 | 3.42 |
| ybdK | b0581 | Conserved hypothetical protein | 8.78 | 14.70 | 12.71 |
| ybeL | b0643 | Conserved hypothetical protein | 2.42 | 9.80 | 4.27 |
| ybgA | b0707 | Conserved protein | 6.53 | 16.97 | 13.24 |
| ybgS | b0753 | Conserved protein | 8.43 | 38.08 | 16.12 |
| ybhE | b0767 | Putative isomerase | 2.66 | 5.57 | 2.38 |
| yhjP | b0865 | Conserved hypothetical protein | 3.66 | 6.53 | 6.24 |
| yccJ | b1003 | Unknown CDS | 10.42 | 17.09 | 16.33 |
| yceK | b1050 | Unknown CDS | 14.93 | 14.50 | 10.14 |
| ycgB | b1188 | Conserved hypothetical protein | 10.36 | 35.43 | 11.19 |
| ycgZ | b1164 | Unknown CDS | 5.11 | 5.53 | 4.00 |
| yciF | b1258 | Conserved protein | 2.98 | 20.43 | 2.04 |
| yciG | b1259 | Conserved hypothetical protein | 18.16 | 84.92 | 19.19 |
| ydaM | b1341 | Conserved protein with PYP-like sensor domain | 6.74 | 9.54 | 3.15 |
| ydfN | b1547 | Qin prophage; putative tail fiber protein | 2.25 | 4.71 | 2.35 |
| ydgA | b1614 | Conserved protein | 2.99 | 5.89 | 2.63 |
| yeaG | b1783 | Conserved protein, nucleotide triphosphate hydrolase domain | 19.44 | 26.61 | 21.48 |
| yeaH | b1784 | Conserved hypothetical protein | 11.89 | 14.66 | 14.73 |
| yebF | b1847 | Conserved hypothetical protein | 4.04 | 2.79 | 2.17 |
| yeeP | b1999 | CP4-44 prophage; putative GTP binding factor | 2.80 | 4.87 | 3.08 |
| yegP | b2080 | Conserved hypothetical protein | 17.85 | 17.67 | 7.86 |
| yegS | b2086 | Conserved protein with PDZ-like domain | 8.87 | 6.52 | 13.93 |
| yfiL | b2602 | Conserved hypothetical protein | 3.22 | 3.63 | 3.75 |
| ygaF | b2660 | Putative enzyme | 5.28 | 6.57 | 5.18 |
| ygaM | b2672 | Conserved hypothetical protein | 9.39 | 14.13 | 13.02 |
| ygaU | b2665 | Conserved hypothetical protein with LysM domain | 7.54 | 11.75 | 8.51 |
| ygbA | b2732 | Conserved hypothetical protein | 2.67 | 2.80 | 2.08 |
| ygfS | b2886 | Putative 4Fe-4S ferredoxin-type protein | 3.06 | 5.92 | 3.66 |
| yggE | b2922 | Conserved protein | 3.35 | 2.57 | 2.52 |
| yghA | b3003 | Putative oxidoreductase; NAD(P)-binding domain | 4.55 | 26.14 | 6.54 |
| ygiW | b3024 | Conserved hypothetical protein | 2.86 | 2.67 | 2.16 |
| yhfG | b3362 | Conserved hypothetical protein | 6.03 | 8.40 | 5.95 |
| yhhA | b3448 | Conserved protein | 4.05 | 8.31 | 3.28 |
| yhjG | b3524 | Conserved protein | 3.89 | 7.11 | 4.58 |
| yjbJ | b4045 | Unknown CDS with YmbJ domain | 21.96 | 20.09 | 7.60 |
| yjdI | b4126 | Conserved hypothetical protein | 8.41 | 9.78 | 7.15 |
| yjdJ | b4127 | Putative acyl-CoA N-acyltransferase | 6.34 | 9.34 | 6.96 |
| yjeB | b4178 | Conserved protein, winged helix domain | 2.22 | 2.49 | 2.08 |
| yjgG | b4247 | Unknown CDS | 3.62 | 3.87 | 2.22 |
| yjgR | b4263 | Putative enzyme contains nucleoside triP hydrolase domain | 2.70 | 2.94 | 2.74 |
| yjhT | b4310 | Putative enzyme contains galactose oxidase-like central domain | 3.05 | 7.04 | 3.05 |
| ymgA | b1165 | Unknown CDS | 4.98 | 5.87 | 4.63 |
| yncB | b1449 | Putative dehydrogenase, with NAD(P)-binding and GroES-like domains | 2.08 | 9.30 | 2.70 |
| ynhG | b1678 | Putative ATP synthase subunit with LysM domain | 5.46 | 8.64 | 6.71 |
| ynjF | b1758 | Putative transferase | 2.95 | 2.87 | 2.69 |
| yodC | b1957 | Unknown CDS | 5.46 | 3.02 | 2.15 |
| yodD | b1953 | Unknown CDS | 8.49 | 18.20 | 15.16 |
| yohF | b2137 | Putative oxidoreductase with NAD(P)-binding domains | 3.97 | 11.26 | 5.21 |
| ygjC | b3097 | Conserved protein | 6.77 | 14.06 | 5.17 |
| ygjD | b3098 | Conserved hypothetical protein | 4.88 | 4.75 | 4.59 |
| ygjE | b3099 | Conserved protein | 5.26 | 5.36 | 4.65 |
| ygjG | b3102 | Putative glutathione S-transferase enzyme with thioredoxin-like domain | 4.77 | 9.03 | 5.65 |
| ygjK | b3100 | Conserved hypothetical protein | 3.43 | 5.60 | 3.80 |
Average relative mRNA levels (rpoS+/rpoS mutant ratio) determined at transition into stationary phase in LB medium. OD 4, OD578 value of 4.0.
Hyperosmotic shift in M9 medium.
pH downshift in LB medium.
CDS, coding sequence.
To our knowledge, previous publications have described 87 E. coli genes as being σS controlled. With our microarrays, we identified 54 of these genes as σS dependent (with 36 belonging to the core set of σS-controlled genes). Many of the other 33 genes exhibited ratios of expression of just below 2. Likely explanations are that these genes can be expressed from more than one promoter with not all promoters being σS dependent, as is known, for example, for glgS (26), proP (48), or ftsQAZ (19) or from promoters which can be activated by EσS as well as by Eσ70, as demonstrated for osmE (8) and csiE (45).
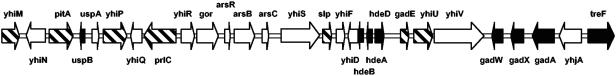
In principle, σS-dependent genes are scattered all over the E. coli chromosome. Nevertheless, there were a few clusters with a rather high local density of σS-controlled genes. One example was a region of approximately 91 kb around 79.3 min on the chromosome, which features 29 σS-controlled genes, including several regulatory and structural genes involved in acid resistance (further analyzed and discussed below) (Fig. 2). Another cluster of approximately 13 kb included the previously described csiD-ygaF-gabDTP operon (49), as well as the genes ygaU, yqaE, yqaM, and nrdE (around 60.2 min on the chromosome). Additional such regions included dps and poxB (among 27 σS-dependent genes clustered over approximately 89 kb, located at around 18.6 min of the chromosome) or sodC, cfa, ihfA, and katE (among 34 σS-dependent genes clustered over approximately 120 kb, located around 38.4 min of the chromosome). On the other hand, the only region which over a long distance (approximately 540 kb) was essentially free of σS-dependent genes (a single exception is the ysgG gene) included the replication initiation region oriC (data not shown). The biological significance of this absence of σS control in this large segment of the genome is currently unknown.
FIG. 2.
Cluster of σS-controlled genes at around 79 min on the E. coli chromosome that includes several acid resistance genes (gad and hde genes). Genes identified as σS controlled under all three growth and stress conditions (core genes) are shown by black arrows, and σS-controlled genes identified only under one or two conditions are indicated by hatched arrows.
In silico identification of a σS consensus promoter sequence.
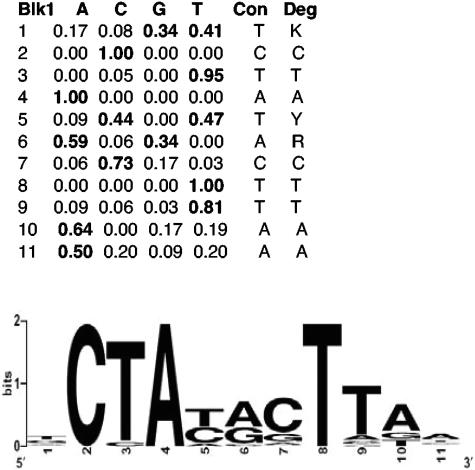
To identify putative promoter sequences recognized by σS-containing RNA polymerase, the upstream noncoding regions (200 bp) of the 140 σS-dependent core genes were searched for common motifs with the programs MEME (3, 4) and BioProspector (40). Both algorithms identified the sequence 5′-TCTATACTTAA-3′ with high statistical significance. This sequence is shown in Fig. 3 as a sequence logo with base frequencies represented by the height of a stack of letters at each position; note that even at positions where the sequence logo does not appear impressive, preference for one or two nucleotides is still highly significant, as can be seen in the accompanying table.
FIG. 3.
A common sequence pattern in the 200-bp regions upstream of σS-controlled core genes strongly resembles an extended −10 region previously discussed for σS-dependent promoters. Relative frequencies of nucleotides as identified by BioProspector (40) are shown in the table and correspond to positions −14 to −4 in putative σS-dependent promoters. A consensus sequence (Con) is given as well as a degenerate consensus (Deg), which also takes into account the second-most-frequent nucleotide if it occurs in more than 30% of the sequences identified (K stands for T or G, Y stands for T or C, and R stands for A or G). The consensus sequence is also shown as a sequence logo (14).
This sequence motif represents an extended version of the sequence previously discussed as a putative −10 region for σS-specific promoters (see discussion below and reference 25). In several cases where genes were already known to be σS dependent, this sequence coincided with experimentally demonstrated promoters (e.g., for dps, poxB, osmC, and otsB; for compilations of experimentally verified σS-dependent promoters, see references 6, 18, 37, and 41). The presence of such a common promoter motif suggests that these promoters are subject to the same regulatory mechanism, i.e., direct recognition by σS-containing RNA polymerase. This motif could be clearly identified from the core set of σS-dependent genes, but neither MEME nor BioProspector recognized this pattern upstream of noncore σS-controlled genes, i.e., genes which display σS dependence under only one or two of the three growth and stress conditions tested (Fig. 1). This indicates either that these noncore σS-controlled genes have more degenerate σS-dependent promoters, which require additional activation mechanisms, or that they are under the indirect control of σS.
Modules within the σS network: the case of acid resistance genes.
Our finding that many regulatory genes are under σS control (Table 1; Fig. 6) suggested that the σS regulon constitutes a large regulatory network with a hierarchical (cascade-like), modular internal architecture. Thus, secondary regulators may impose special regulatory patterns upon subsets of σS-dependent genes (modules). This includes a high potential for additional signal input into specific modules. From our data, it is directly seen that a group of known acid resistance genes constitutes such a module with an interesting regulatory pattern. These include gadA and the gadBC operon (encoding two glutamate decarboxylases and a glutamate-GABA exchange carrier involved in cytoplasmic proton scavenging) (57), the hde genes (which have also been implicated in acid resistance) (68), and the regulatory genes gadX (yhiX), gadW (yhiW), and gadE (yhiE). With the exception of the gadBC operon, all these genes are located in one of the chromosomal clusters of σS-dependent genes (Fig. 2), which has also been referred to as a “fitness island for acid adaptation” in E. coli (27). At least under certain conditions, the regulators GadX and GadW seem to play opposite roles in the expression of gadA, gadBC, and the hde genes, whereas GadE seems to be an essential activator for these genes (27, 43, 44, 47, 66). Our microarray data (Table 2) not only demonstrated that all these genes are under σS control (previously demonstrated for some genes) (43, 44, 65) but also revealed an interesting pattern of regulation: while these genes were strongly σS dependent during entry into stationary phase, their σS dependence was reduced or even abolished under acid stress conditions (Table 2).
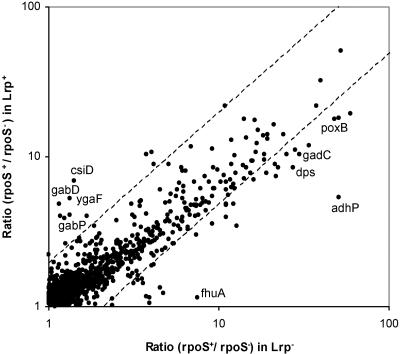
FIG. 6.
σS dependence of many genes is altered in the absence of the global regulator Lrp. Ratios of expression in rpoS+ and rpoS mutant strains were determined by microarray analysis of lrp+ and lrp::Tn10 backgrounds and are shown in a scatter plot. Genes with a >2-fold difference in this ratio (i.e., in σS dependence) in lrp+ and lrp mutant strains fall outside of the diagonal field marked by hatched lines.
TABLE 2.
Expression ratios of σS-controlled acid resistance genesa
| Gene | b no. | Gene product | Ratio
|
|||
|---|---|---|---|---|---|---|
| OD 4 | NaCl | pH 5 | OD 4 (Irp mutant)b | |||
| gadA | b3517 | Glutamate decarboxylase A, isozyme, PLP dependent | 32.38 | 5.80 | 2.33 | 39.58 |
| gadB | b1493 | Glutamate decarboxylase, PLP dependent, isozyme beta | 16.44 | 3.96 | 2.22 | 21.23 |
| gadC | b1492 | Putative glutamate gamma-aminobutyric acid antiporter (APC family) | 10.28 | 3.48 | 2.22 | 29.72 |
| hdeA | b3510 | Conserved protein with protein HNS-dependent expression | 11.26 | 9.16 | 1.65 | 9.17 |
| hdeB | b3509 | Conserved hypothetical protein | 6.61 | 4.46 | 1.28 | 5.87 |
| hdeD | b3511 | Putative membrane protein | 2.52 | 2.07 | 1.27 | 3.64 |
| slp | b3506 | Outer membrane protein, induced after carbon starvation | 3.68 | 2.79 | 1.08 | 5.19 |
| yhiU | b3513 | Multidrug resistance protein (lipoprotein) | 3.48 | 1.38 | 0.99 | 3.23 |
| gadE | b3512 | Transcriptional regulator for gadABC operon, activates glutamate decarhboxylase-dependent acid resistance | 13.06 | 8.68 | 1.14 | 19.38 |
| gadW | b3515 | Transcriptional regulator for GadX (regulatory protein), glutamic acid decarboxylase (GadA, -B), and glutamate transport protein (GadC) | 6.12 | 5.98 | 3.09 | 12.39 |
| gadX | b3516 | Transcriptional regulator for glutamic acid decarboylase and transporter (gadA, gadBC) | 6.09 | 7.45 | 3.14 | 9.39 |
For explanations of data, see the footnotes to Table 1.
Average relative mRNA levels (ratio of the Irp rpoS+ mutant to the Irp rpoS mutant) determined at transition into stationary phase in LB medium.
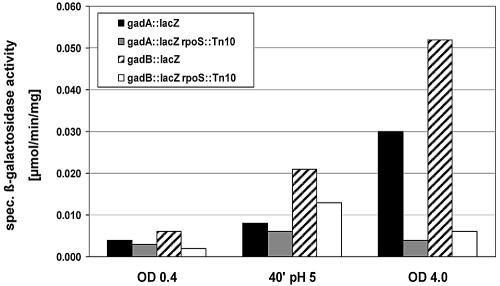
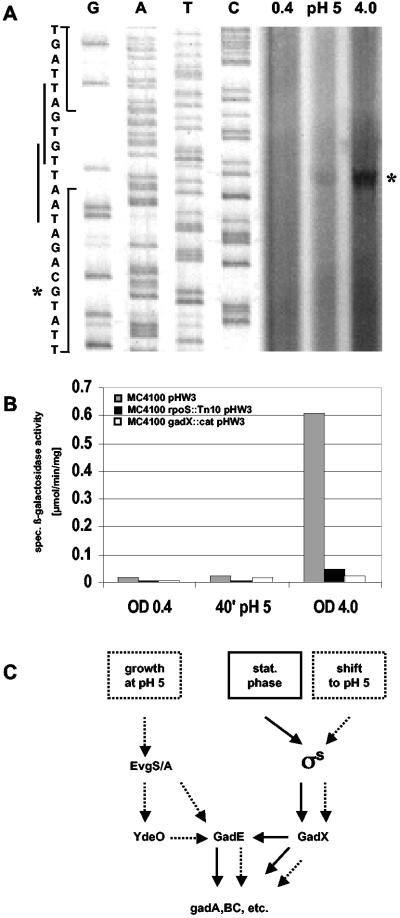
To confirm this apparent change in sigma factor dependence and to assay induction ratios in response to acid shift or entry into stationary phase, we constructed lacZ reporter fusions in the two target genes gadA and gadB and in the regulatory gene gadE. Expression of the single-copy transcriptional lacZ fusions in gadA and gadB was tested in wild-type and rpoS mutant backgrounds under the same conditions used for array analysis. The results (Fig. 4) demonstrated that expression of gadA and gadB was activated under stress conditions and that this expression required σS during entry into stationary phase but was nearly σS independent after acid shift. By contrast, transcription of gadE was not stimulated by shift to pH 5 but was strongly activated during entry into stationary phase, as demonstrated by primer extension experiments (Fig. 5A) and transcriptional gadE::lacZ fusion analysis (Fig. 5B). The transcriptional start site shown in Fig. 5A was the same as for a previously described gadE transcript (a second transcript, which was observed in an hns mutant, could not be detected in any of our experiments and probably indicates the presence of a second H-NS-silenced promoter) (27). Also, stationary-phase induction of gadE was strongly dependent on σS and on GadX (Fig. 5B). Thus, σS dependence of gadE during entry into stationary phase was probably indirect via GadX, consistent with the putative −10 region of the stationary-phase-inducible gadE promoter (Fig. 5A) not showing similarity to the EσS consensus promoter (Fig. 3). Taken together, these data suggest that this module of acid resistance genes can switch σS and/or σ70 dependence, depending on specific environmental or stress conditions, since σS is part of only one of the pathways that allow activation of these genes (Fig. 5C).
FIG. 4.
Expression and σS dependence of gadA::lacZ and gadB::lacZ fusions under different growth and stress conditions. Strains JK86 and JK87, which carry transcriptional single-copy lacZ fusions in gadA and gadB (black and hatched bars, respectively), as well as their rpoS::Tn10 derivatives (grey and white bars, respectively) were grown in LB medium. During log-phase growth, an aliquot of the culture was shifted to pH 5 (see Materials and Methods for details). Samples were taken during log phase (OD578 = 0.4; pH 7), 40 min after shift to pH 5, and during entry into stationary phase (OD578 = 4; pH 7). Specific β-galactosidase activities were measured (the data given represent the average of three independent experiments each).
FIG. 5.
The role of σS, GadX, and GadE in the expression of acid resistance genes under different stress conditions. (A) The gadE transcriptional start site was determined by primer extension experiments with RNA prepared from strain MC4100 carrying the translational gadE::lacZ fusion plasmid (see Materials and Methods for details). During log-phase growth in LB, an aliquot of the culture was shifted to pH 5. Samples for RNA preparation were taken during log phase (OD578 = 0.4; pH 7), 40 min after shift to pH 5, and during entry into stationary phase (OD578 = 4; pH 7). The reverse transcript and the transcriptional start site in the sequence are indicated by asterisks, and two putative −10 regions are indicated along the sequence. (B) Ex-pression of gadE was assayed by a transcriptional gadE::lacZ fusion present on a plasmid, because in single-copy constructs, measurable activities were extremely low. Translational single-copy fusions, which exhibit higher activities, yielded results similar to those obtained with the multicopy transcriptional fusions. Strain MC4100 and its rpoS and gadX mutant derivatives carrying these fusions were grown and sampled as described in the legend to Fig. 4, and specific β-galactosidase activities were measured. (C) Summarizing model. Solid arrows indicate regulatory influences relevant during entry into stationary phase, and dotted arrows indicate regulatory influences upon shift to or growth at acidic pH.
Relationship between σS and Lrp in global regulation.
A factor that appears to directly affect σS and/or σ70 selectivity at certain promoters is the global regulatory protein Lrp, as was previously reported for the σS-controlled genes osmY (13), osmC (9), and aidB (32). To find out whether Lrp affects sigma factor selectivity of promoters in a more global way, we again determined ratios of expression in rpoS+ and rpoS mutant strains, i.e., σS dependence on genomic microarrays but now for strains that were defective in the lrp gene. For these experiments, we chose one of the conditions previously tested, i.e., entry into stationary phase in LB medium (OD578), since many putative stationary-phase-induced σS-controlled genes are repressed by Lrp (64). However, a difference in the σS dependence in lrp+ and lrp mutant strains (in contrast to a mere difference in expression levels) would only be expected if Lrp differentially affected promoter access and/or activation by either σS-containing or σ70-containing RNA polymerase, in other words, affected sigma factor selectivity.
As is demonstrated in Fig. 6, where σS dependence (i.e., ratios of expression in rpoS+ and rpoS strains) is shown for lrp+ and lrp mutant backgrounds, there are indeed σS-controlled genes with such changes in sigma factor selectivity. All genes for which such changes were more than twofold (Fig. 6) are listed in Table S3 in the supplemental material. An example of reduction or nearly complete loss of σS dependence is the csiD-ygaF-gabDTP operon (49). These microarray data are consistent with a previous report of a modulatory role of Lrp in the control of the csiD promoter, observed with lacZ fusions (21). On the other hand, a number of genes exhibited a clear increase in σS dependence in the lrp mutant background (Fig. 6; Table S3 in the supplemental material). We identified 28 genes for which the ratio of σS dependence increased more than twofold in the lrp mutant background. Thirteen of these genes (adhP, cfa, dps, gadC, gadW, mlrA, poxB, otsB, otsA, yciG, yeaG, yjgB, and yohF) were also described as Lrp repressed by Tani et al. (64). Also, the acid resistance genes gadA, gadB, and gadC, as well as their regulatory genes gadE, gadW and gadX, exhibited increased σS dependence in the lrp mutant (although the increase in general was less than twofold) (Table 2).
Extensive overlap between σS and cAMP-CRP in global regulation.
CRP plays a complex role in the regulation of the expression of rpoS itself (34; F. Scheller and R. Hengge, unpublished results). In addition, several previously identified σS-dependent genes (e.g., csiD and osmY) are also under direct positive or negative control by cAMP-CRP (13, 46). To find out whether coregulation by σS-containing RNA polymerase and cAMP-CRP is a more general phenomenon, the upstream regions (200 bp) of all 481 positively σS-controlled genes identified here were screened for putative cAMP-CRP boxes, defined as TGTGA(N6)TCACA, with a maximum of three mismatches allowed.
We identified 263 σS-controlled genes (i.e., 55%) featuring putative cAMP-CRP binding sites (with one, two, or three mismatches from the consensus) upstream of their coding regions. Some of these genes have two or more cAMP-CRP boxes. Even though not all of these putative binding sites may actually play physiologically relevant regulatory roles, this is a striking number that indicates a strong overlap between the σS and cAMP-CRP regulons.
For 55 of 140 σS-dependent core genes, putative extended −10 promoter regions (Fig. 3) could be unequivocally identified or were known before (this corresponds to a total of 64 such regions, as 9 genes displayed 2 of these putative promoter regions). More than half of these genes (30 of 55) also contained putative cAMP-CRP-binding sites in their promoter regions. Nineteen genes exhibited one cAMP-CRP box, 9 had two, and 2 (osmC and talA) had three such sites. The maximum number of cAMP-CRP boxes, i.e., five, was found in the pdhR promoter region, where the first such site overlapped with the putative promoter and the additional ones followed further downstream. Among these 48 putative cAMP-CRP-binding sites, 5 sites were located at typical activator positions (class I or II) (10), 14 sites were found at typical repressor positions (i.e., overlapping with the promoter and/or the transcriptional start site), 3 sites were situated between typical activator and repressor sites (around −50), 12 sites were situated too far upstream to exert a direct activating effect (but may act indirectly, e.g., by bending DNA), and 14 sites were located downstream of the transcriptional start site (the latter groups could in principle also serve other non-σS-dependent promoters that may contribute to the expression of the respective genes).
Physiological functions of σS-dependent genes.
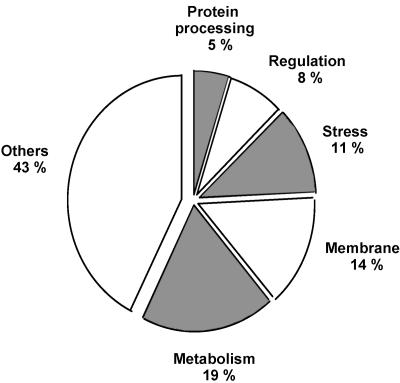
For approximately 57% of all σS-dependent genes identified here, functional annotations exist. In Table 1, the core set of σS-controlled genes is ordered in functional categories (according to a simplified version of the system used by Riley and coworkers) (60, 61), and the relative occurrence of genes belonging to each category is shown in Fig. 7. Besides genes with known functions in stress management (11%), nearly all σS-controlled core genes with known or probable functions fall into three groups. They encode either metabolic enzymes (19%), transport proteins and/or intrinsic membrane proteins of unclear function (which are likely to be transporters as well) (14%), or regulatory proteins (8%).
FIG. 7.
Functional annotations of σS-controlled genes. Numbers shown were obtained for core genes. For gene names and further functional details, see Table 1 and the text.
Upon closer inspection of the metabolic genes, an interesting pattern became apparent. A number of genes involved in central energy metabolism (glycolysis, fermentation, anaerobic respiration, and the pentose phosphate shunt) exhibited positive σS control at least under one condition tested (Table S4 in the supplemental material). In addition, interesting regulatory antagonisms were observed. Thus, not only pyruvate oxidase (poxB) was strongly σS activated, but also the repressor (encoded by pdhR) for the housekeeping pyruvate-oxidizing enzyme, i.e., pyruvate dehydrogenase, was under σS control. While fumarate reductase (frdA) was positively σS controlled, succinate dehydrogenase (sdhCDAB) was negatively affected by σS. In the pentose phosphate shunt, there was opposite σS control of the two genes for transketolase (tktA and tktB), indicating that in stationary phase or under other stress conditions, the tktA-encoded major enzyme may be replaced by TktB, for which only very low expression levels were previously reported (29). Overall, these data indicate that induction of σS in starving or otherwise stressed cells may contribute to decreasing aerobic respiration in favor of a more fermentative and/or anaerobic respiration-based energy metabolism.
DISCUSSION
Genome-wide identification of σS-dependent genes and in silico identification of a σS consensus promoter sequence.
Among the 481 positively σS-controlled genes identified here, there is a core group of 140 genes which were found to be σS controlled under all three growth and stress conditions used here, which known to result in high cellular σS levels (Fig. 1). Thus, the expression of these genes may just follow the cellular σS concentration. Expression of the remaining 341 genes seems to require some special conditions and therefore specific regulation in addition to high σS levels (see also below).
The large number of σS-dependent genes identified here provided an ideal database for an in silico search for common regulatory motifs upstream of the coding sequences. From the core group of σS-dependent genes, two pattern-searching programs, MEME and Bioprospector, identified a common motif with high statistical significance. The motif consensus sequence is TCTATACTTAA (or KCTAYRCTTAA, which takes into account the second-most-frequent nucleotides when present in more than 30% of the sequences analyzed; K stands for T or G, Y stands for T or C, and R stands for A or G) (Fig. 3). This sequence represents an extended version (nucleotide −14 to −4) of a −10 promoter region previously proposed to be recognized by EσS (6, 18, 25, 38). By contrast, it was recently suggested that a C instead of a T at the −12 position strongly contributes to EσS selectivity of a promoter (30). However, our compilation of putative −10 regions found upstream of σS-dependent core genes (as well as the previously published shorter lists of σS-controlled promoters mentioned above) indicates that a C (−12) is possible but actually quite rare (5%) (Fig. 3, with A and G being completely absent). Thus, a C (−12) is obviously not part of an EσS-selective consensus promoter but may be an occasionally occurring deviation from the consensus that is better tolerated by EσS than by Eσ70 and thereby contributes to Eσ70 selectivity.
The extended −10 consensus sequence identified here features all the nucleotides, which have been found experimentally to be important in promoter binding and activation by EσS. T (−14) and above all C (−13) were shown to directly interact with a specific amino acid (K173) in region 3.0 (2.5) of σS (6). Strong conservation of T (−12), A (−11), and T (−7) reflects the special importance of these nucleotides in EσS-mediated promoter melting (36-38). Finally, in the context of the σS-controlled rssAB promoter, the TAA motif (−6 to −4) has also been found to stimulate σS-dependent activation in stationary phase (56). Interestingly, both pattern identification algorithms used here could not identify a motif corresponding to a −35 region, which is consistent with suggestions that the −35 regions of naturally evolved σS-dependent promoters may be more degenerate (20, 36, 37). Taking the data together, we would like to suggest that the motif identified here, KCTAYRCTTAA, represents an extended −10 region of a directly EσS-recognized and -activated promoter. The length of this sequence motif should allow the identification of putative σS-controlled promoters in silico with high probability.
For many of the 140 core σS-dependent genes, this putative extended −10 region can easily be recognized with only one or two mismatches. In those cases where such a sequence is less apparent, several explanations are possible. Either the promoter is further upstream than the 200 upstream nucleotides screened in our study (e.g., the osmY promoter) (33), especially if genes are part of an operon. On the other hand, such a gene may be under the control of a σS-dependent activator (whose gene would belong to the core set of σS-controlled genes), which may then activate promoters in conjunction with Eσ70.
σS-controlled genes represent a large and complex network with differentially controlled modules and connections to other global regulons.
Two major observations reported here indicate that σS controls not only a regulon but rather a regulatory network with an intrinsic hierarchical and modular structure. (i) The majority (71%) of the positively σS-dependent genes were found only under one or two conditions characterized by high cellular σS levels, and even the core genes exhibited very different degrees of σS dependence under different conditions (Table 1). (ii) Quite a large number of σS-dependent genes identified here encode regulatory proteins (Table 1 and Fig. 6), which can be expected not only to affect the expression of subsets of σS-dependent genes but also to serve as additional signal integrators. If the target genes of these regulators are also directly dependent on σS-containing RNA polymerase, this would establish feed-forward regulatory circuits (51), which may fine-tune or boost the expression of subsets of σS-controlled genes (i.e., modules) under specific conditions. Knockout mutants of such secondary regulatory genes are currently isolated to identify their spectrum of target genes.
The internal architecture of the σS network is also dynamic, i.e., subject to environmental regulation. σS-controlled target genes may be flexibly allocated to additional global regulons, as suggested by the apparently strong overlap with cAMP-CRP control. More than half of all σS-controlled genes exhibit putative cAMP-CRP binding sites in their 200-bp upstream regions. The locations of these sites in cases where the promoters were either known or can be identified with high probability with our −10 region consensus (Fig. 3) indicate that cooperation between σS and cAMP-CRP can be positive, negative, or more complex (e.g., involving several cAMP-CRP boxes or additional promoters). These connections between the σS and the cAMP-CRP networks also extend to the level of the master regulators, as cAMP-CRP controls σS itself in a complex and as-yet-unclarified manner (reference 34; Scheller and Hengge, unpublished). In other words, σS and cAMP-CRP seem to tightly cooperate in integrating the responses to multiple (general) stresses and specific C starvation, respectively. Thus, cAMP-CRP may have a global regulatory role that goes beyond mediating catabolite control of gene expression. In a recent transcriptome study of CRP-dependent catabolite control, the strong overlap with the σS network did not become apparent, although many stress genes encoding chaperones and proteases, for example, exhibited glucose-CRP-dependent regulation (22). An obvious explanation is that this study used cells that grew rapidly in LB medium under conditions in which σS is hardly present in the cell. To further analyze the cooperation between σS and cAMP-CRP, future experiments will have to assay the effects of crp mutations under σS-inducing stress conditions (such as those used in the present study).
Depending on specific stress conditions, acid resistance genes belong to the σS network or not.
By our microarrays and reporter gene fusion analysis, we identified a subset of σS-controlled genes, which were clearly stationary-phase-induced in a σS-dependent manner but exhibited only minor or no σS dependence upon acidic shift (Table 2; Fig. 4). The expression of these genes probably becomes σ70 dependent, as no other E. coli sigma factor is known to be induced or activated under acidic conditions. These genes are crucial for acid resistance and include gadA, gadBC, and the hde genes (as well as other less-well-characterized genes), together with their regulatory genes gadE (yhiE), gadX (yhiX), and gadW (yhiW) (Table 2). Thus, whether these genes belong to the σS network or not depends on environmental conditions.
The GadE regulator was reported to be essential for the expression of gadA, gadBC, and the hde genes, whereas GadX and GadW play a complex and conditional modulatory role, which becomes dispensable if GadE is overproduced (27, 43, 47). Reduced σS dependence and even a lack of induction upon acid shift is especially pronounced for the regulator GadE (Fig. 5; Table S3 in the supplemental material), and the corresponding regulatory pattern of the downstream target genes may be a reflection of this change in gadE control. Moreover, we show here that strong gadE expression during entry into stationary phase also requires the GadX regulator. This generates a feed-forward loop in which the target genes gadA and gadB and probably others are under both direct and indirect control of GadX (the latter via GadE) (Fig. 5C).
These and the previously published data on the complex control of acid resistance genes can be summarized in a model in which the essential module activator GadE is under the control of two pathways (Fig. 5C): (i) the stationary-phase induction pathway using σS, which is required for GadX expression (this control includes the small σS-dependent gadY RNA which affects the stability of gadX mRNA) (53), which in turn activates gadE, and (ii) a regulatory cascade involved in gadE expression in cells growing at low pH, which comprises the EvgS-EvgA two-component system and YdeO (47). Thus, the switch in sigma factor dependence of the acid resistance genes would be due to σS being part of only one of the activating pathways for gadE. This model explains earlier reports of acid tolerance being σS dependent only in stationary phase (12) and is also reflected in the absence of σS in the regulatory network of these acid resistance genes as presented by Masuda and Church (47). The previously reported strong derepression of acid resistance genes in hns mutants involves gadX (28) and most likely reflects the strong negative regulation of σS by H-NS (5, 69). Interestingly, σS itself is strongly induced by a shift to pH 5, so why is σS not sufficient and apparently not even relevant for activation of these acid resistance genes at low pH? We have recently observed that acid induction of σS is transient, i.e., σS levels do not remain high in cells growing continuously at pH 5 (M. Metzner and R. Hengge, unpublished results) and this transient σS induction upon sudden acid shift does not result in the activation of gadE (Fig. 5A and B). However, continuous expression of gadA, gadBC, and the hde genes is likely necessary to cope with permanent acid stress; this requires continuously high expression of GadE, probably mediated by the EvgSA/YdeO pathway.
EσS-Eσ70 promoter selectivity can be conditional and influenced by the abundant nucleoid protein Lrp.
As outlined above, a switch between σS and σ70 dependence can be a consequence of using different activating pathways that converge on a module regulator (such as GadE). On the other hand, the similarity of EσS- and Eσ70-recognized promoters allows a switch between σS and σ70 dependence, even at the same promoter, i.e., the sigma factor selectivity of a promoter can be conditional. An example of such regulation is provided by the dps gene, which is strongly activated by EσS in stationary phase or under other stress conditions (dps is a core gene) (Table 1) but which can also be activated by Eσ70 cooperating with the H2O2-activated regulator OxyR (1). In fact, of the 22 genes that belong to the OxyR regulon (70), 10 genes were observed here to be under σS control, including the core genes dps and yaiA, the noncore genes dsbG and hemH, and the sufABCDSE operon (data not shown). The general pattern that seems to emerge here is that certain modules of genes within the σS network can be recruited as subsets of other regulons under special growth or stress conditions. This also indicates that the internal architecture of stress-responsive regulatory networks is not static but is itself subject to regulation.
Another module within the σS network comprises genes that are also under the control of the global regulator Lrp. Approximately half of the 140 core σS-dependent genes identified here are also under repression by Lrp (64). Previous studies with the osmY, osmC, and aidB promoters suggested that Lrp not only acts as an ordinary repressor but can also affect sigma factor selectivity (9, 13, 32). Our data presented here indicate that this is a more general phenomenon. We identified 28 genes for which σS dependence was >2-fold higher in the lrp mutant background (Fig. 6; Table S3 in the supplemental material). For almost half of these genes, this correlates with repression by Lrp (64). On the other hand, 13 genes showed a reduction or even loss of σS dependence in the lrp mutant background. The most striking example is the csiD-ygaF-gabDTP operon (Fig. 5), which lost σS dependence in the lrp mutant background and for which a role of Lrp as a positive modulator at the csiD promoter was suggested previously (21).
How can Lrp affect the preference for either σS- or σ70-containing RNA polymerase holoenzyme? Lrp is known as an abundant regulatory and chromosome-organizing protein that is further induced during entry into stationary phase (2, 31). Lrp can bend DNA and often assembles at multiple adjacent sites along one side of the DNA helix (67). This means that Lrp can induce complex DNA superstructures, which in general would have an inhibitory effect on gene expression but at some promoters may less affect EσS than Eσ70. At other promoters, Lrp may also somewhat optimize the positioning of the −35 and −10 regions and perhaps also of operator sites required for expression by Eσ70. In all these cases, the absence of Lrp would shift relative EσS-Eσ70 dependence, as EσS is less demanding with respect to using a nonoptimal −35 region (20, 25) or to nonoptimal spacing of the −35 and −10 regions (for a summary, see reference 25; A. Typas and R. Hengge, unpublished data). Taken together, our data suggest that the abundant Lrp protein not only acts as a global repressor (or activator in some cases) but also affects EσS-Eσ70 selectivity at many stationary-phase-induced promoters, probably by modulating local DNA topology.
Physiological functions of the σS network.
Besides genes directly involved in coping with the detrimental effects of stress (e.g., dps, katE, the otsBA operon, or the gad genes), the other annotated σS-dependent genes (core and noncore genes) in principle encode three major functional groups of proteins (Fig. 7): (i) regulatory factors (about 8%) (implications are discussed above), (ii) known transport systems or other intrinsic membrane proteins (14%), and (iii) metabolic enzymes, many of which belong to central energy metabolism (19%) (Table S4 in the supplemental material). This suggests that overall membrane traffic is significantly altered in stressed or stationary-phase cells. Thus, σS control may contribute to scavenging of various nutrients under nutrient-limiting conditions, as well as to increased resistance against various toxic compounds, by inducing putative efflux pumps. Moreover, σS seems to have a more pronounced influence on energy metabolism than previously suspected and may be crucial in the transition from growth to maintenance metabolism in stressed or stationary-phase cells. Important genes involved in glycolysis, fermentation, anaerobic respiration, electron transport, and the pentose phosphate shunt turned out to be under positive σS control (Table S4 in the supplemental material). These data suggest that induction of σS may prepare the cells for a shift away from oxidative respiration towards a more fermentative or anaerobic respiratory energy metabolism. This may serve to counteract the increased production of reactive oxygen species in aerobic respiration during entry into a starvation situation (16), but detailed implications will have to be studied in the future.
Finally, it will certainly be interesting to compare the σS networks of various bacterial species. So far, genome-wide profiling of σS-controlled genes has only been reported for Pseudomonas aeruginosa (59). There, the σS network is at least as large as in E. coli (14% of the genes in the genome are affected). A large group of σS-dependent genes specifically found in Pseudomonas is involved in quorum sensing. As E. coli does not possess the corresponding quorum-sensing systems, this demonstrates that even in relatively closely related species, the same global regulators and their regulatory networks have been recruited to serve different functions in ways that may reflect differences in natural environments and lifestyles.
Supplementary Material
Acknowledgments
Financial support for this study was provided by the Deutsche Forschungsgemeinschaft (Gottfried-Wilhelm-Leibniz program and He1556/11-1), the state of Baden-Württemberg (Landesforschungspreis), and the Fonds der Chemischen Industrie.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altuvia, S., M. Almirón, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and σS in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 2.Azam, T. A., A. Iwata, A. Nishimura, S. Ueda, and A. Ishihama. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, T., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 4.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. [DOI] [PubMed] [Google Scholar]
- 5.Barth, M., C. Marschall, A. Muffler, D. Fischer, and R. Hengge-Aronis. 1995. A role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σS and many σS-dependent genes in Escherichia coli. J. Bacteriol. 177:3455-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, G., and R. Hengge-Aronis. 2001. What makes an Escherichia coli promoter σS-dependent? Role of the −13/−14 nucleotide promoter positions and region 2.5 of σS. Mol. Microbiol. 39:1153-1165. [DOI] [PubMed] [Google Scholar]
- 7.Bohannon, D. E., N. Connell, J. Keener, A. Tormo, M. Espinosa-Urgel, M. M. Zambrano, and R. Kolter. 1991. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of σ70. J. Bacteriol. 173:4482-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordes, P., R. Repoila, A. Kolb, and C. Gutierrez. 2000. Involvement of differential efficiency of transcription by EσS and Eσ70 RNA polymerase holenzymes in growth phase regulation of the Escherichia coli osmE promoter. Mol. Microbiol. 35:845-853. [DOI] [PubMed] [Google Scholar]
- 9.Bouvier, J., S. Gordia, G. Kampmann, R. Lange, R. Hengge-Aronis, and C. Gutierrez. 1998. Interplay between global regulators of Escherichia coli: effect of RpoS, H-NS and Lrp on transcription of the gene osmC. Mol. Microbiol. 28:971-980. [DOI] [PubMed] [Google Scholar]
- 10.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 11.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 12.Castanie-Cornet, M.-P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colland, F., M. Barth, R. Hengge-Aronis, and A. Kolb. 2000. Sigma factor selectivity of Escherichia coli RNA polymerase: a role for CRP, IHF and Lrp transcription factors. EMBO J. 19:3028-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dukan, S., and T. Nyström. 1998. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 12:3431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernsting, B. R., M. R. Atkinson, A. J. Ninfa, and R. G. Matthews. 1992. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J. Bacteriol. 174:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinosa-Urgel, M., C. Chamizo, and A. Tormo. 1996. A consensus structure for σS-dependent promoters. Mol. Microbiol. 21:657-659. [DOI] [PubMed] [Google Scholar]
- 19.Flärdh, K., T. Garrido, and M. Vicente. 1997. Contribution of individual promoters in the ddlB-ftsZ region to the transcription of the essential cell-division gene ftsZ in Escherichia coli. Mol. Microbiol. 24:927-936. [DOI] [PubMed] [Google Scholar]
- 20.Gaal, T., W. Ross, S. T. Estrem, L. H. Nguyen, R. R. Burgess, and R. L. Gourse. 2001. Promoter recognition and discrimination by EσS RNA polymerase. Mol. Microbiol. 42:939-954. [DOI] [PubMed] [Google Scholar]
- 21.Germer, J., G. Becker, M. Metzner, and R. Hengge-Aronis. 2001. Role of activator site position and a distal UP-element half-site for sigma factor selectivity at a CRP/H-NS activated σS-dependent promoter in Escherichia coli. Mol. Microbiol. 41:705-716. [DOI] [PubMed] [Google Scholar]
- 22.Gosset, G., Z. Zhang, S. Nayyar, W. A. Cuevas, and M. H. Saier Jr. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 186:3516-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 24.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS subunit of RNA polymerase in Escherichia coli. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hengge-Aronis, R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-dependent? Curr. Opin. Microbiol. 5:591-595. [DOI] [PubMed] [Google Scholar]
- 26.Hengge-Aronis, R., and D. Fischer. 1992. Identification and molecular analysis of glgS, a novel growth phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol. Microbiol. 6:1877-1886. [DOI] [PubMed] [Google Scholar]
- 27.Hommais, F., E. Krin, J.-Y. Coppée, C. Lacroix, E. Yeramian, A. Danchin, and P. Bertin. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150:61-72. [DOI] [PubMed] [Google Scholar]
- 28.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J.-P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 29.Iida, A., S. Teshiba, and K. Mizobuchi. 1993. Identification and chracterization of the tktB gene encoding a second transketolase in Escherichia coli K-12. J. Bacteriol. 175:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacour, S., A. Kolb, and P. Landini. 2003. Nucleotides from −16 to −12 determine specific promoter recognition by bacterial σS-RNA polymerase. J. Biol. Chem. 278:37160-37168. [DOI] [PubMed] [Google Scholar]
- 31.Landgraf, J. R., J. Wu, and J. M. Calvo. 1996. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 178:6930-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landini, P., L. I. Hajec, L. H. Nguyen, R. R. Burgess, and M. R. Volkert. 1996. The leucine-responsive regulatory protein (Lrp) acts as a specific repressor for σS-dependent transcription of the Escherichia coli aidB gene. Mol. Microbiol. 20:947-955. [DOI] [PubMed] [Google Scholar]
- 33.Lange, R., M. Barth, and R. Hengge-Aronis. 1993. Complex transcriptional control of the σS-dependent stationary phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary phase response of Escherichia coli. J. Bacteriol. 175:7910-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 35.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. J., and J. D. Gralla. 2003. Open complex formation in vitro by σ38 (rpoS) RNA polymerase: roles for region 2 amino acids. J. Mol. Biol. 329:941-948. [DOI] [PubMed] [Google Scholar]
- 37.Lee, S. J., and J. D. Gralla. 2002. Promoter use by σ38 (rpoS) RNA polymerase. J. Biol. Chem. 49:47420-47427. [DOI] [PubMed] [Google Scholar]
- 38.Lee, S. J., and J. D. Gralla. 2001. σ38 (rpoS) RNA polymerase promoter engagement via −10 region nucleotides. J. Biol. Chem. 276:30064-30071. [DOI] [PubMed] [Google Scholar]
- 39.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 40.Liu, X., D. L. Brutlag, and J. S. Liu. 2001. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 127-138. [PubMed]
- 41.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 42.Lucht, J. M., P. Dersch, B. Kempf, and E. Bremer. 1994. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon on Escherichia coli. J. Biol. Chem. 269:6578-6586. [PubMed] [Google Scholar]
- 43.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309-1320. [DOI] [PubMed] [Google Scholar]
- 44.Ma, Z., H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184:7001-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marschall, C., and R. Hengge-Aronis. 1995. Regulatory characteristics and promoter analysis of csiE, a stationary phase-inducible σS-dependent gene under positive control of cAMP-CRP in Escherichia coli. Mol. Microbiol. 18:175-184. [DOI] [PubMed] [Google Scholar]
- 46.Marschall, C., V. Labrousse, M. Kreimer, D. Weichart, A. Kolb, and R. Hengge-Aronis. 1998. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on σS and requires activation by cAMP-CRP. J. Mol. Biol. 276:339-353. [DOI] [PubMed] [Google Scholar]
- 47.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 48.McLeod, S. M., J. Xu, and R. C. Johnson. 2000. Coactivation of the RpoS-dependent proP P2 promoter by Fis and cyclic AMP receptor protein. J. Bacteriol. 182:4180-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzner, M., J. Germer, and R. Hengge. 2004. Multiple stress signal integration in the regulation of the complex σS-dependent csiD-ygaF-gabDTP operon in Escherichia coli. Mol. Microbiol. 51:799-811. [DOI] [PubMed] [Google Scholar]
- 50.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 51.Milo, R., S. Shen-Orr, S. Itzkovitz, N. Kashtan, D. Chklovskii, and U. Alon. 2002. Network motifs: simple building blocks of complex networks. Science 298:824-827. [DOI] [PubMed] [Google Scholar]
- 52.Minton, N. P. 1984. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene 31:269-273. [DOI] [PubMed] [Google Scholar]
- 53.Opdyke, J. A., J.-G. Kang, and G. Storz. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polen, T., D. Rittmann, V. F. Wendisch, and H. Sahm. 2003. DNA microarray analyses of the long-term adaptive response of Escherichia coli to acetate and propionate. Appl. Environ. Microbiol. 69:1759-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Powell, B. S., D. L. Court, Y. Nakamura, M. P. Rivas, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruteanu, M., and R. Hengge-Aronis. 2002. The cellular level of the recognition factor RssB is rate-limiting for σS proteolysis: implications for RssB regulation and signal transduction in σS turnover in Escherichia coli. Mol. Microbiol. 45:1701-1714. [DOI] [PubMed] [Google Scholar]
- 57.Richard, H. T., and J. W. Foster. 2003. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52:167-186. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 60.Serres, M. H., S. Goswami, and M. Riley. 2004. GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res. 32:D300-D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serres, M. H., and M. Riley. 2000. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics 5:205-222. [DOI] [PubMed] [Google Scholar]
- 62.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 63.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 64.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tucker, D. L., N. Tucker, Z. Ma, J. W. Foster, R. L. Miranda, P. S. Cohen, and T. Conway. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185:3190-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, Q., and J. M. Calvo. 1993. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of higher-order nucleoprotein structure. EMBO J. 12:2495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waterman, S. R., and P. L. C. Small. 1996. Identification of σS-dependent genes associated with the stationary-phase acid resistance phenotype of Shigella flexneri. Mol. Microbiol. 21:925-940. [DOI] [PubMed] [Google Scholar]
- 69.Yamashino, T., C. Ueguchi, and T. Mizuno. 1995. Quantitative control of the stationary phase-specific sigma factor, σS, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 14:594-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.