Abstract
A novel enzyme which may be important in mucin degradation has been discovered in the mucin-utilizing anaerobe Prevotella strain RS2. This enzyme cleaves terminal 2-acetamido-2-deoxy-β-d-glucopyranoside 6-sulfate (6-SO3-GlcNAc) residues from sulfomucin and from the model substrate 4-nitrophenyl 2-acetamido-2-deoxy-β-d-glucopyranoside 6-sodium sulfate. The existence of this mucin-desulfating glycosidase (sulfoglycosidase) suggests an alternative mechanism by which this bacterium may desulfate sulfomucins, by glycosidic removal of a sulfated sugar from mucin oligosaccharide chains. Previously, mucin desulfation was thought to take place by the action of a specific desulfating enzyme, which then allowed glycosidases to remove desulfated sugar. Sulfate removal from sulfomucins is thought to be a rate-limiting step in mucin degradation by bacteria in the regions of the digestive tract with a significant bacterial flora. The sulfoglycosidase was induced by growth of the Prevotella strain on mucin and was purified 284-fold from periplasmic extracts. Tryptic digestion and sequencing of peptides from the 100-kDa protein enabled the sulfoglycosidase gene to be cloned and sequenced. Active recombinant enzyme was made in an Escherichia coli expression system. The sulfoglycosidase shows sequence similarity to hexosaminidases. The only other enzyme that has been shown to remove 6-SO3-GlcNAc from glycoside substrates is the human lysosomal enzyme β-N-acetylhexosaminidase A, point mutations in which cause the inheritable, lysosomal storage disorder Tay-Sachs disease. The human enzyme removes GlcNAc from glycoside substrates also, in contrast to the Prevotella enzyme, which acts on a nonsulfated substrate at a rate that is only 1% of the rate observed with a sulfated substrate.
A barrier of mucus covers the surface of the gastrointestinal tract. It is believed that this barrier protects the underlying cells from damage by endogenous bacterial enzymes, toxins, extremes of pH, and harmful molecules. The main structural components of the mucus layer are the secretory mucins. Mucins are polymerized, high-molecular-weight glycoproteins with many oligosaccharide side chains attached to a central peptide core by O-glycosidic linkages, and they form a viscous, hydrated mucus gel at the mucosal surface (13). The mucins can be lost from the mucus layer by sloughing, by the slow action of endogenous proteases, and by the action of enzymes from certain gut bacteria. A number of specific glycosidases and proteinases, probably originating from several different bacteria, are required for the complete degradation of mucins (16).
In the colon the secreted mucins have oligosaccharide side chains that are more heavily sulfated (14) than the side chains of the secreted mucins in regions of the digestive tract with lower bacterial numbers. There is evidence that this sulfation of mucins makes them less susceptible to degradation by bacterial glycosidases, and thus the protective barrier is thought to remain more intact (7, 14, 16, 25). However, some bacteria possess mucin-desulfating sulfatases which desulfate sulfomucin, allowing glycosidases to access and act on the mucins. The mucin-desulfating sulfatases that have been described include sulfatases specific for the β-d-galactopyranosyl 3-sulfate, β-d-galactopyranosyl 6-sulfate, and 2-acetamido-2-deoxy-β-d-glucopyranosyl 6-sulfate (6-SO3-β-GlcNAc) building blocks of the oligosaccharide chains (26, 28, 29).
We recently discovered a new enzyme in the anaerobic colon bacterium Prevotella strain RS2 which desulfates mucin by a different mechanism. This enzyme removes the sulfated sugar 6-SO3-β-GlcNAc by glycosidic bond cleavage, suggesting that during the degradation of sulfated mucin oligosaccharide side chains, the removal of a 6-SO3-β-GlcNAc residue by a sulfatase followed by an N-acetylglucosaminidase could be bypassed by the action of a single enzyme. We termed this enzyme a sulfoglycosidase (SGL). No enzyme with this specificity has been described previously from a bacterial source. Currently, only one other enzyme, the human lysosomal enzyme subunit β-N-acetylhexosaminidase A, is known to remove 6-SO3-β-GlcNAc from glycoside substrates (10). This protein has been extensively studied, as point mutations in it cause the inheritable, autosomal, recessive, lysosomal storage disorder Tay-Sachs disease (10, 24).
In this paper we describe the discovery and purification of SGL from Prevotella strain RS2, cloning of the gene, and expression of a recombinant enzyme. The specificity of the recombinant SGL was also characterized, and other anaerobic gut bacteria were screened for the presence of SGL activity.
MATERIALS AND METHODS
Bacterial culture.
Prevotella strain RS2 was originally isolated from pig colonic mucosa and was selected for its ability to grow on colonic mucin as an energy source (22). It was grown at 37°C in broth culture anaerobically under CO2 by using a basal medium modified from medium 10 (4, 17) and supplemented with either 0.2% (wt/vol) galactose or 0.3% pig gastric mucin (PGM) (described below) as an energy source. After four subcultures on PGM-supplemented medium, the bacterium was grown overnight in 500 ml of medium.
Pig gastric mucin preparation.
Stomach mucus gel was gently scraped from the body region of washed pig stomachs and stored at −20°C until it was needed. PGM was prepared by solubilizing mucus from the harvested gel and precipitating the crude mucin with ethanol. Each batch of mucus (100 ml) was mixed with 200 ml of KCl (0.1 M) and 7.5 ml of potassium phosphate buffer (1 M, pH 7.0), and the pH was readjusted to 7.0. The preparation was heated at 80°C for 40 min under N2; then cysteine-HCl (0.05%, wt/vol) was added, and the preparation was incubated for a further 15 min to reduce the disulfide bonds between mucin subunits. After cooling, the mixture was centrifuged (160 × g, 5 min), and the supernatant containing solubilized mucin was precipitated by addition of ethanol to a concentration of 70% (vol/vol). The precipitate was centrifuged, suspended in distilled water, and freeze-dried. The crude PGM was used as a bacterial growth substrate.
A purer preparation of pig gastric mucin, PGMt, was prepared for studies of cleavage of sulfated sugar from mucin. This mucin was prepared as previously described (19, 21) by 2-mercaptoethanol reduction to reduce disulfide linkages, DNase treatment, and trypsin treatment to cleave the mucin chain at regions with low glycosylation. The pig stomach mucus used was harvested from the fundus region, as fundal mucin is more heavily sulfated than the mucin in the body region. PGMt is a preparation of soluble mucin subunits, which is suitable for enzyme studies and free from major contaminants.
Enzyme assays.
SGL was assayed by measuring p-nitrophenol (pNP) release from 4-nitrophenyl 2-acetamido-2-deoxy-β-d-glucopyranoside 6-sodium sulfate (6-SO3-β-GlcNAc-1-pNP). This substrate was synthesized at Industrial Research Ltd., Lower Hutt, New Zealand (5). The substrate (1 mM 6-SO3-β-GlcNAc-1-pNP) and buffer A (20 mM l-histidine buffer [pH 6.0] containing 10 mM 2-mercaptoethanol) were incubated at 37°C with the SGL preparation in a 0.075-ml (final volume) mixture. Reactions were terminated after 15 to 45 min by addition of Na glycine (0.925 ml, 0.5 M, pH 9.6). Negative controls, in which SGL was added only after incubation and glycine addition, were included. The absorbance at 410 nm, due to free pNP, was determined, and the control value was subtracted. One unit of SGL activity was defined as production of 1 μmol of pNP min−1. Kinetic measurements were obtained by using four preparations of recombinant SGL. The Km and Vmax values calculated from the most extensive data set are presented below. Ten data points obtained by using substrate concentrations between 0.5 and 10 mM were computer fitted to the Michaelis-Menten equation, and 90% confidence ranges for Km and Vmax were determined.
During column chromatography, SGL fractions were assayed in 96-well Nunc microwell plates. Substrate (0.005 ml), buffer A (0.04 ml), and 0.001 to 0.01 ml of a column fraction were added to a well and incubated for 30 min. Na glycine (pH 9.6, 0.17 ml) was added to stop the reaction by pH change, and the absorbance at 410 nm was determined. The data were used to identify active fractions.
Sulfatase (MdsA) activity was assayed with 6-SO3-β-GlcNAc-1-pNP plus the auxiliary enzyme hexosaminidase at pH 7.4 (18) or by using the coupled glucose oxidase assay (17). Hexosaminidase activity was measured as described above for SGL, except that p-nitrophenyl 2-acetamido-2-deoxy-β-d-glucopyranoside (β-GlcNAc-1-pNP) was used as the substrate. Protein was measured by the dye-binding method of Bradford (3), using bovine serum albumin as the standard.
Sulfoglycosidase purification methods.
Prevotella strain RS2 cells were collected by centrifugation and washed, and periplasmic extracts in 50 mM Tris-HCl buffer (pH 7.0) containing 10 mM 2-mercaptoethanol were made by using a method based on the method of Witholt et al. (27). A protease inhibitor cocktail tablet (one Complete Mini tablet per 10 ml of extract; Roche Diagnostics Corp., Indianapolis, Ind.) was added. The periplasmic extract was fractionated by ammonium sulfate precipitation, and the protein which precipitated between 50 and 75% ammonium sulfate saturation was redissolved in buffer A and dialyzed against three changes of 500 ml of buffer A. The extract was chromatographed on a Q Sepharose Fast Flow anion-exchange column (bed volume, 20 ml; Amersham Pharmacia Biotech, Uppsala, Sweden) in buffer A, using an NaCl gradient (0 to 0.25 M, 350 ml). By using hydroxyapatite prepared by the procedure of Bernadi (2), the major SGL activity peak was fractionated on a hydroxyapatite column (bed volume, 5 ml; containing 50% [vol/vol] Sephadex G-25 to improve the flow rate) preequilibrated with buffer A. After the SGL sample was loaded, the column was washed with buffer A (30 ml) and eluted with a linear gradient of sodium phosphate buffer (0 to 0.3 M, pH 6.0) in buffer A (300 ml).
Electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of proteins was performed on a 10% polyacrylamide gel (12), and this was followed by staining with Coomassie brilliant blue.
Sequence analysis of peptides.
The larger SGL peak eluting from the hydroxyapatite column was subjected to SDS-PAGE, and the major band (100 kDa) was excised. N-terminal sequencing of selected tryptic peptides was performed as previously described (28).
Molecular techniques.
PCR (20) and inverse PCR (15) amplifications were conducted by standard methods, using genomic DNA from Prevotella strain RS2 as the template. The genomic DNA was isolated under anaerobic conditions until the DNA extraction step, as this minimized DNA damage due to an oxygen-activated endogenous DNase. PCR products were cloned into the pGEM-T Easy vector (Promega Corp., Madison, Wis.). Sequencing of inserts was performed by automated sequencing. Full-length sgl DNA sequences were constructed from overlapping fragments of DNA sequenced in both directions.
Molecular cloning of sulfoglycosidase gene and expression in Escherichia coli.
The putative sgl gene, including its leader sequence, was amplified by PCR by using primers JR18F (5′ GGG AAT TCC ATA TGA AAA AAC TGT GTT TTG CAC 3′; bases −11 to 22 of the sgl gene modified to contain an NdeI site) and JR19R (5′ CCG GCT CGA GCT TCA TAA ACA CCT TCT GGG C 3′; bases 2703 to 2683 of the sgl gene modified to contain a XhoI site). The product was digested with NdeI and XhoI and ligated into pET-22b(+) (Novagen Inc., Madison, Wis.), and the plasmid containing the insert was used to transform E. coli DH5α (Invitrogen, Carlsbad, Calif.). E. coli BL21(λDE3), which had been pretransformed with pRI592 (8) to provide rare tRNAs, was further transformed with isolated pET-22b(+) containing the sgl insert, and transformants containing both plasmids were selected on Luria-Bertani agar containing ampicillin and chloramphenicol. Colonies were grown for 4 h in 1.5 ml of Luria-Bertani broth (20) containing ampicillin (50 mg liter−1) and chloramphenicol (34 mg liter−1). Enzyme expression was induced for 3 h by adding 0.4 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). Pelleted cells were suspended in buffer A, and SGL was assayed in the cell suspension.
A second DNA construct containing the putative sgl gene, without its signal sequence, was amplified from genomic DNA by PCR. The primers used were JR26F (5′ GTA TGC TAC ATG TCG CAG ACG ATA ACA ACC TAC AGT C 3′; bases 58 to 79; modified to contain a BspLU11I site) and JR19R. The PCR product was digested with BspLU11I and XhoI. Vector pET-22b(+) was digested with XhoI, NcoI (giving a cohesive end with BspLU11I), and EcoRI (to cut the excised 128-bp plasmid fragment). The PCR product was ligated into the plasmid and used to transform E. coli DH5α (Invitrogen). This construct excluded the leader sequence of Prevotella SGL and replaced it with the E. coli pelB leader sequence for periplasmic translocation.
A hexahistidine tag was present at the C terminus of both sgl constructs. E. coli BL21(λDE3), pretransformed with pRI592 as described above, was transformed with isolated pET-22b(+) containing one of the putative sgl insert constructs, and transformants containing both plasmids were selected. Cells were then grown and the recombinant protein was expressed as described above.
Purification of recombinant sulfoglycosidase.
BL21(λDE3) recombinants were grown in 1 ml of Luria-Bertani broth containing ampicillin and chloramphenicol for 12 h at 28°C. An inoculum (0.5 ml) was added to 500 ml of fresh medium and grown to an absorbance at 600 nm of 0.7. Recombinant SGL was induced by addition of IPTG (0.5 mM), and incubation was continued for 16 h. The harvested cells were sonically disrupted, and the debris was removed by centrifugation. Recombinant SGL was purified by Ni-nitrilotriacetic acid (NTA) adsorption and elution with imidazole buffer, hydroxyapatite column chromatography, and Q Sepharose column chromatography at 4°C. The purified enzyme was concentrated by using a 10-kDa-cutoff membrane filter (Vivaspin 6) at 4°C.
Sulfomucin reaction with sulfoglycosidase and paper chromatography of products.
PGMt (5 mg) was incubated with recombinant SGL (0.5 U) in 0.5 ml of buffer A for 4 h at 37°C. High-molecular-weight mucin and the enzyme were removed by filtration (10-kDa cutoff). The filtrate was freeze-dried, resuspended in 0.02 ml of water, and then spotted on Whatman no. 1 filter paper. Suitable controls (no enzyme, no substrate, and buffer alone) were included. Sugar standards were also spotted. Ascending chromatography was carried out by using one of the following solvent systems to develop each chromatogram, as described below: system A (1-butanol-acetic acid-1 M NH4OH [2:3:1, vol/vol/vol]), system B (1-propanol—nitromethane-H2O [7:2:2, vol/vol/vol]), or system C (pyridine-ethyl acetate-acetic acid-H2O [5:5:1:3, vol/vol/vol/vol]). The paper was removed after 18 h and dried. Reducing sugars, including sulfated sugars, were visualized by using the dip method of Trevelyan et al. (23), as follows: 0.5% silver nitrate in acetone, followed by 0.5 N NaOH in ethanol, 20 min in 0.63 M sodium thiosulfate, and then a water wash.
A sulfated sugar product from an identical incubation was located by cutting the chromatography paper after development and staining the lane at its edges. It was eluted from the paper by water capillary action. After concentration, it was incubated with recombinant mucin-desulfating sulfatase (MdsA) (28) under suitable conditions. The new product was filtered, concentrated, and run on a paper chromatogram, and its identity was determined.
Synthesis of methyl 2-acetamido-2-deoxy-6-O-sulfamoyl-β-d-glucopyranoside.
Calcium carbonate (77 mg, 0.77 mmol) was added to a stirred solution of methyl 2-acetamido-3,4-di-O-acetyl-2-deoxy-β-d-glucopyranoside (1) (53 mg, 0.17 mmol) in anhydrous N,N-dimethylformamide (1 ml) under argon. Sulfamoyl chloride (11) (65 mg, 0.56 mmol) was added, and the mixture was stirred at the ambient temperature overnight. The solids were filtered off and discarded, and the filtrate was evaporated to a yellow oil, which was dissolved in a 2 M methanolic ammonia solution and left at the ambient temperature overnight. The solvent was evaporated, and the residue was purified by flash chromatography on silica (CH2Cl2-methanol-H2O, 14:5:1) to obtain the title compound as a colorless slightly hygroscopic solid (31 mg, 59%). The 1H nuclear magnetic resonance (1H-NMR) (CD3OD) data were as follows: δ 4.45 (dd, 1 H, J 10.8, 1.7 Hz), 4.34 (d, 1 H, J 8.4 Hz), 4.26 (dd, 1 H, J 10.8, 5.8 Hz), 3.65 (dd, 1 H, J 10.1, 8.6 Hz), 3.58-3.41 (m 2 H), 3.45 (s, 3 H), 3.33 (br. t, 1 H, J 9.2 Hz), 1.98 (s, 3 H). 13C-NMR (CD3OD) data were as follows: δ 174.0 (s), 103.6 (d), 76.1 (d), 75.5 (d), 71.9 (d), 70.0 (t), 57.5 (q), 57.2 (d), 23.1 (q); [α] D21 −21° (c 1.4, methanol). Fast atom bombardment mass spectrometry: m/z calculated for C9H19N2O8S (M+H)+, 315.0862; found, 315.0872. The optical rotation was determined with a Perkin-Elmer 241 polarimeter and was expressed in 10−1 degrees per square centimeter per gram (conventionally degrees). NMR spectra were recorded with a Bruker AC300E spectrometer at 300 MHz (1H) in CD3OD [internal reference, (CH3)4Si, δ 0] or at 75.5 MHz (13C) in CD3OD (center line at δ 49.2). The 13C spectra gave unambiguous data for the numbers of protons bound to each carbon atom; these data were expressed as s, d, t, and q, which were the multiplicities expected in C, H undecoupled spectra. The high-resolution +ve fast atom bombardment mass spectrometry determination was performed with a VG70-250S double-focusing, magnetic sector mass spectrometer (VG Analytical) in a nitrobenzyl alcohol matrix.
SGL activity screening in closely related anaerobic gut bacteria.
Bacterial strains from anaerobic stabs were subcultured twice for 16 h in 1 ml of modified medium 10 (as described above) supplemented with 0.2% (wt/vol) PGM plus 0.07% galactose as energy sources under a CO2 atmosphere. Cells from the second subculture were harvested by centrifugation (5,000 × g, 5 min), suspended in 0.2 ml of 50 mM histidine buffer (pH 5.5) containing 10 mM 2-mercaptoethanol (buffer B), and broken by five 10-s sonication treatments with a microprobe, with intervals of cooling on ice between treatments. The broken cells were separated by centrifugation (10,000 × g, 5 min) into a soluble extract and a pellet of damaged cells and membranes (which was resuspended in 0.2 ml of buffer B). One-half of each fraction was incubated (0°C for 16 h) with 0.5 mM methyl 2-acetamido-2-deoxy-6-O-sulfamoyl-β-d-glucopyranoside (6-SO2NH2-β-GlcNAc-1-Me). Studies of Prevotella strain RS2 MdsA sulfatase, performed by using the glucose-6-sulfatase assay (17), which is not interfered with by SGL activity, and studies of pure recombinant MdsA sulfatase expressed from Bacteroides thetaiotaomicron (28) indicated that 0.5 mM 6-SO2NH2-β-GlcNAc-1-Me inhibits MdsA more than 99% after overnight preincubation. In contrast, recombinant SGL incubated for a similar time with this concentration of inhibitor showed only 25 to 40% inhibition (unpublished data).
SGL and sulfatase activities were measured in a 96-well microwell plate by incubating 0.003 ml of uninhibited extract (or suspended particulate fraction) with 6-SO3-β-GlcNAc-1-pNP (1 mM), buffer B, and added auxiliary hexosaminidase (0.001 U of an Aspergillus oryzae extract; Sigma G7138) in a 0.19-ml (final volume) mixture for 60 min at 37°C. SGL activity alone was measured in a similar incubation experiment by using inhibitor-treated extract. The incubations were stopped by adding 0.23 ml of 0.5 M sodium glycine (pH 9.6). The difference between the measurements gave an indication of the sulfatase activity at pH 5.5, after partial inhibition of the SGL was taken into account.
Nucleotide sequence accession number.
The nucleotide sequence of the sgl gene plus upstream and downstream sequences has been deposited in the GenBank database under accession number AY158021.
RESULTS
Discovery of sulfoglycosidase activity.
The SGL activity was discovered in periplasmic extracts of Prevotella strain RS2 when the rather cumbersome assay initially used to purify the mucin-desulfating sulfatase (MdsA) (17, 28), involving glucose-6-sulfate desulfation followed by estimation of the free glucose released by using glucose oxidase, was replaced with a simpler coupled assay involving 6-SO3-β-GlcNAc-1-pNP desulfation and subsequent pNP release from the β-GlcNAc-1-pNP by added hexosaminidase (18). It became apparent that a previously undetected enzyme, acting on 6-SO3-β-GlcNAc-1-pNP, was present in fractions purified from periplasm. Preliminary experiments showed that this enzyme did not require added hexosaminidase to release pNP from 6-SO3-β-GlcNAc-1-pNP, had no sulfatase activity as determined by the glucose-6-sulfate desulfation assay above, had a different molecular size than MdsA, was not very sensitive to MdsA inhibitors, and had a different pH optimum than MdsA (data not shown). Clearly, it was not the known mucin-desulfating sulfatase, MdsA.
A comparison of the SGL activities in cytoplasmic and periplasmic fractions prepared from mucin-grown cells revealed 44-fold-higher activity in the latter fraction, in contrast to the cytoplasmic enzyme malate dehydrogenase activity, which was 6-fold higher in the former fraction (data not shown).
Purification of sulfoglycosidase.
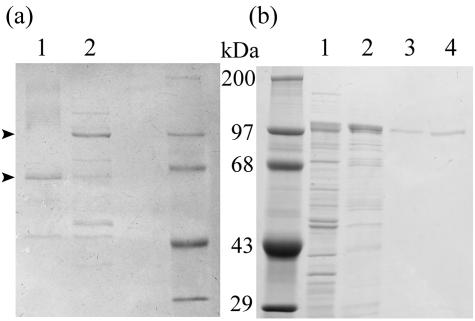
Periplasmic extract was prepared from Prevotella strain RS2 cultured in 500 ml of basic medium supplemented with 0.3% (wt/vol) pig gastric mucin. The extract was fractionated with ammonium sulfate (pH 6), and the proteins precipitating between 50 and 75% saturation were collected and dialyzed to remove salts (Table 1). The concentrated proteins were separated by anion-exchange chromatography on Q Sepharose (pH 6). This step separated SGL into several peaks with activity, and the major peak was selected. The activity in this fraction showed 98-fold purification and contained 40% of the SGL applied to the column. Q Sepharose fractions were also assayed for hexosaminidase and sulfatase activities in parallel, and these activities separated well from the major SGL peak, indicating that sequential actions of these enzymes did not result in apparent SGL activity. Next, the major SGL fraction was chromatographed on a hydroxyapatite column. A small peak of SGL activity eluted at 60 mM phosphate (peak A, 5.7 U mg of protein−1) and was well separated from a large peak of SGL activity at 150 mM phosphate (peak B, 19.4 U mg of protein−1). The latter fraction represented 284-fold purification compared with the periplasmic extract (Table 1). Pooled fractions from peaks A and B were electrophoresed on an SDS-PAGE gel and stained with Coomassie brilliant blue, which resulted in major bands at 65 and 100 kDa, respectively (Fig. 1a). Several minor bands were detected in each fraction. Since more activity was present in peak B, this peak was selected for further work. In-gel digestion of the 100-kDa band with trypsin was performed, followed by high-performance liquid chromatography separation of peptides and sequencing of likely peptides. Only two of the five peaks analyzed gave clear peptide sequences longer than seven amino acids (the rest of the sequences were short sequences or mixed peptide sequences that were later found in the complete sequence). Peak T27 had the sequence T(X)GFNT(E,A)GITNP, and peak T29 had the sequence (T)VQGTF(X)TEHV (where the amino acids in parentheses are uncertain). Neither of these peptide sequences was found in a Swiss-Prot protein database search.
TABLE 1.
Purification of sulfoglycosidase from Prevotella strain RS2 periplasmic extracta
| Purification step | Protein (mg) | Activity (u) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Periplasmic extract (pH 7.0)b | 91.01 | 9.0 | 0.099 | 1 | 100 |
| (NH4)2SO4 (50-75%) fractionb | 23.02 | 4.5 | 0.195 | 2 | 50 |
| Q Sepharose (pH 6.0) | 0.098 | 1.9 | 19.39 | 196 | 21.1 |
| Hydroxyapatite fraction A (pH 6.0) | 0.035 | 0.2 | 5.71 | 58 | 2.1 |
| Hydroxyapatite fraction B (pH 6.0) | 0.032 | 0.9 | 28.1 | 284 | 10.0 |
Enzymic activity was determined as described in the text.
This fraction may contain some MdsA sulfatase and N-acetylhexosaminidase activities.
FIG. 1.
SDS-PAGE of sulfoglycosidase. (a) SGL purified from Prevotella strain RS2. Lane 1, hydroxyapatite column fraction A (2.9 μg of protein) (see Table 1); lane 2, hydroxyapatite column fraction B (2.0 μg); right lane, protein molecular weight standard (0.5 μg). (b) Recombinant SGL. Left lane, protein molecular weight standard (5 μg); lane 1, supernatant from sonicated E. coli, containing recombinant SGL; lane 2, enzyme eluted from Ni-NTA column; lane 3, enzyme eluted from hydroxyapatite chromatography column; lane 4, enzyme eluted by Q Sepharose chromatography. The gels were stained with Coomassie brilliant blue.
PCR and inverse PCR amplification.
Degenerate forward and reverse primers were designed from the sequences described above. Primers JR8F (5′-CA[A/G]GGIACITT[C/T]IIIACIGA[A/G]CA[C/T]G-3′) and JR7R (5′-GT[A/G/T]ATICCI[G/T]CIGT[A/G]TT[A/G]AAICC-3′) gave a 600-bp product after PCR when anaerobically isolated Prevotella strain RS2 genomic DNA was used as the template. The translated open reading frame showed homology to hexosaminidases in protein databases, as anticipated from the SGL's specificity for sulfate-substituted N-acetylglucosamine, linked by a glycosidic bond.
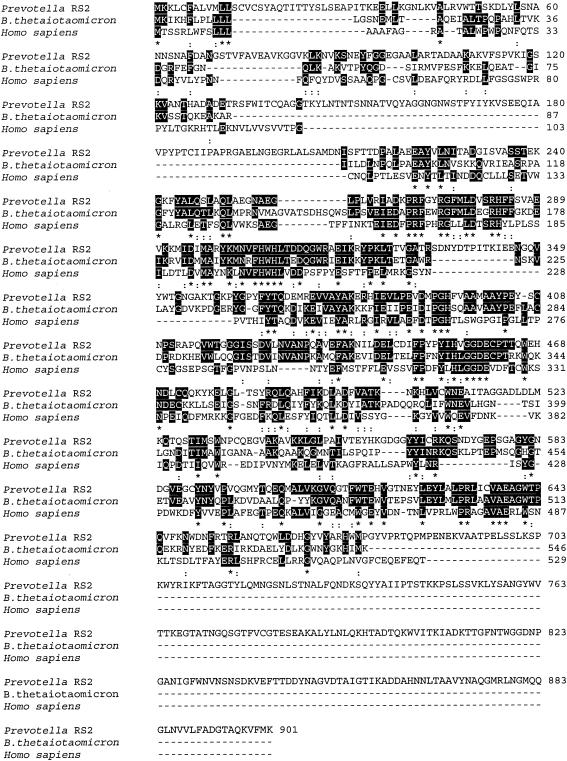
The genomic DNA sequences flanking the 600-bp product were amplified by inverse PCR by using the following primers based on nucleotides in the 600-bp product: JR10F (5′-ATGGGTCATCACCAAGATCGC-3′; nucleotides 2406 to 2426 in the final gene sequence) and JR11R (5′-GCCAGGTACTCCAAGTATTCG-3′; nucleotides 1866 to 1886). When these primers were used for inverse PCR with MseI-digested genomic DNA fragments circularized by ligation as the template, a 2.2-kb product was obtained. This product plus the original 600-bp sequence revealed a 2,703-bp open reading frame encoding a 901-amino-acid protein that included a leader sequence (amino acids 1 to 19) for translocation (Fig. 2). A DNA sequence 598 bp upstream of the putative sgl gene was obtained by inverse PCR by using a circularized Sau3AI-generated template and primers designed from the 5′ end of the putative gene. All six reading frames in this region contained frequent stop codons. A 167-bp sequence downstream from the putative sgl gene was generated by inverse PCR by using a circularized PstI-generated template and primers designed from the 3′ end of the putative gene. This sequence also contained frequent stop codons in all frames. The putative sgl gene was sequenced in both directions.
FIG. 2.
Alignment of the deduced amino acid sequence of sulfoglycosidase from Prevotella strain RS2 with homologous sequences. Prevotella RS2, sulfoglycosidase from Prevotella strain RS2; B. thetaiotaomicron, β-hexosaminidase precursor from B. thetaiotaomicron; Homo sapiens, β-N-acetylhexosaminidase subunit A from H. sapiens. Alignment was carried out by using Clustal W (1.82) multiple-sequence alignment. Fully conserved residues (asterisks) and residues with one conservative replacement (colons) are highlighted.
CAZy family homology of the SGL sequence.
When the National Center for Biotechnology Information BLAST protein search engine was used to examine the Swiss-Prot database, the 901-amino-acid sequence of the Prevotella putative SGL showed high levels of homology to many β-N-acetylglucosaminidases. The sequences were aligned by ClustalW analysis of the EMBL-EBI database. The highest levels of homology were with three gene sequences from B. thetaiotaomicron (the sequence for database accession number NP_813305 is shown in Fig. 2) and a previously characterized β-hexosaminidase from Porphyromonas gingivalis (accession number PO6865) (data not shown). An InterProScan sequence search of the EMBL-EBI database showed that the Prevotella putative SGL belongs to CAZy glycoside hydrolase family 20 (GH20 family) (EC 3.2.1.-.) on the basis of sequence homology. The members of this family that have been characterized are β-hexosaminidases (EC 3.2.1.52) or lacto-N-biosidases (EC 3.2.1.140).
The Prevotella putative SGL also showed 29% identity with residues 104 to 519 of the Homo sapiens lysosomal enzyme β-N-acetylhexosaminidase subunit A (Fig. 2), and the sequence indicated that this enzyme is also in the GH20 family. This enzyme can cleave the glycosidic bonds of both 2-acetamido-2-deoxy-β-d-glucopyranosyl and 6-SO3-β-GlcNAc residues (10).
Cloning and expression of the sulfoglycosidase gene in E. coli.
The putative sgl gene, including its own leader sequence, was amplified by PCR by using primers JR18F and JR19R and was inserted into pET-22b(+). Recombinant protein with a C-terminal hexahistidine tag was expressed in E. coli strain BL21(λDE3) by IPTG induction. Cell lysate showed an inducible band at 100 kDa but only low levels of SGL activity. Most of the induced protein had a lower molecular weight, probably because of the action of endogenous proteases in E. coli BL21(λDE3) and possibly because the Prevotella leader sequence, which was needed for translocation, was not recognized in this host (data not shown).
Another construct was made, in which the E. coli pelB leader sequence in pET-22b(+) was used to replace the natural sgl leader sequence. Genomic DNA was amplified by using primers JR26F and JR19R, and the product was ligated into pET-22b(+). Recombinant protein with a C-terminal hexahistidine tag was expressed in E. coli strain BL21(λDE3) by IPTG induction. Two additional amino acids were present at the N terminus of this recombinant protein after signal sequence cleavage, and the new sequence began with MTQTI. The induced recombinant protein, expressed as a 100-kDa protein in the periplasm, was stable and had good SGL activity (Fig. 1b and Table 2).
TABLE 2.
Substrate specificity of recombinant sulfoglycosidase
| Substrate | Sp act (U/mg) |
|---|---|
| β-GlcNAc-1-pNP | 1.1 |
| 6-SO3-β-GlcNAc-1-pNP | 94.1 |
| β-GalNAc-1-pNP | <0.3 |
| 6-SO3-β-Gal-1-pNP | <0.3 |
| 3-SO3-β-Gal-1-pNP | <0.3 |
| 4-(βGal)-6-SO3-β-GlcNAc-1-pNP | <0.3 |
| PGMt | +a |
See Fig 3. The product was 6-SO3-GlcNAc.
Properties of the recombinant sulfoglycosidase.
The recombinant protein was expressed only during IPTG induction. Cell extract obtained after sonic disruption was purified by Ni-NTA adsorption, hydroxyapatite chromatography, and ion-exchange (Q Sepharose) column chromatography. The pure recombinant SGL migrated as a single 100-kDa protein band on an SDS-PAGE gel (Fig. 1b). The amount of recombinant SGL obtained after purification from a 500-ml batch culture was 0.98 mg of protein with a specific activity of 80 U mg of protein−1, as determined by the standard assay for SGL. The pH optimum of SGL was 5.5 to 6.0. Kinetics measurements, obtained by using the purified recombinant SGL, 6-SO3-β-GlcNAc-1-pNP as the substrate, and pH 6.0, resulted in a Vmax of 122 U mg of protein−1 (90% confidence range, 110 to 135 U mg of protein−1) and a Km of 0.18 mM (90% confidence range, 0.11 to 0.44 mM).
The substrate specificity of SGL was studied by using (sulfated) mono- and disaccharide model substrates with structures analogous to structures found in mucin oligosaccharide chains (Table 2). The disaccharide substrate p-nitrophenyl 2-acetamido-2-deoxy-4-O-(β-d-galactopyranosyl)-β-d-glucopyranoside 6-sodium sulfate [4-(β-Gal)-6-SO3-β-GlcNAc-1-pNP], which was tested to determine whether the SGL can act as an endoglycosidase, did not exhibit glycosidic cleavage of the pNP. p-Nitrophenyl-β-d-galactopyranoside 3-sulfate (3-SO3-β-Gal-1-pNP) and p-nitrophenyl-β-d-galactopyranoside 6-sulfate (6-SO3-β-Gal-1-pNP) showed no glycosidic cleavage. β-GlcNAc-1-pNP (1 mM) was cleaved at a rate that was only 1% of the rate observed for 6-SO3-β-GlcNAc-1-pNP (1 mM). Further investigation of the SGL kinetics by using β-GlcNAc-1-pNP as the substrate gave a Vmax of 15 U mg of protein−1 (90% confidence range, 11 to 19 U mg of protein−1) and a Km of 4.3 mM (90% confidence range, 2.6 to 12.1 mM), indicating that a decreased Vmax and an increased Km were responsible for the lower activity.
To establish whether 6-SO3-β-GlcNAc residues present in sulfomucin oligosaccharide chains are substrates for the SGL, the purified recombinant enzyme was incubated with purified subunit pig gastric mucin (PGMt) originating from the fundus region. After incubation, the high-molecular-weight molecules, including the enzyme, were removed by passage through a 10-kDa-cutoff filter, and the filtrate was concentrated by freeze-drying. The product was chromatographed on Whatman paper, using solvent system A, and the reducing sugars were visualized with the stain of Trevelyan et al. (23). A single spot that comigrated with the 6-SO3-GlcNAc standard was found (Fig. 3). Similar migration results were obtained when solvent systems B and C were used. An unstained band of this product was eluted from the paper with water, freeze-dried, incubated with recombinant mucin-desulfating sulfatase (MdsA) (28), filtered to remove protein, concentrated, and examined again by paper chromatography. In this analysis the product comigrated with the GlcNAc sugar standard (data not shown). The results show that SGL removes terminally bound 6-SO3-β-GlcNAc residues from mucin and thus has a mucin-desulfating action.
FIG. 3.
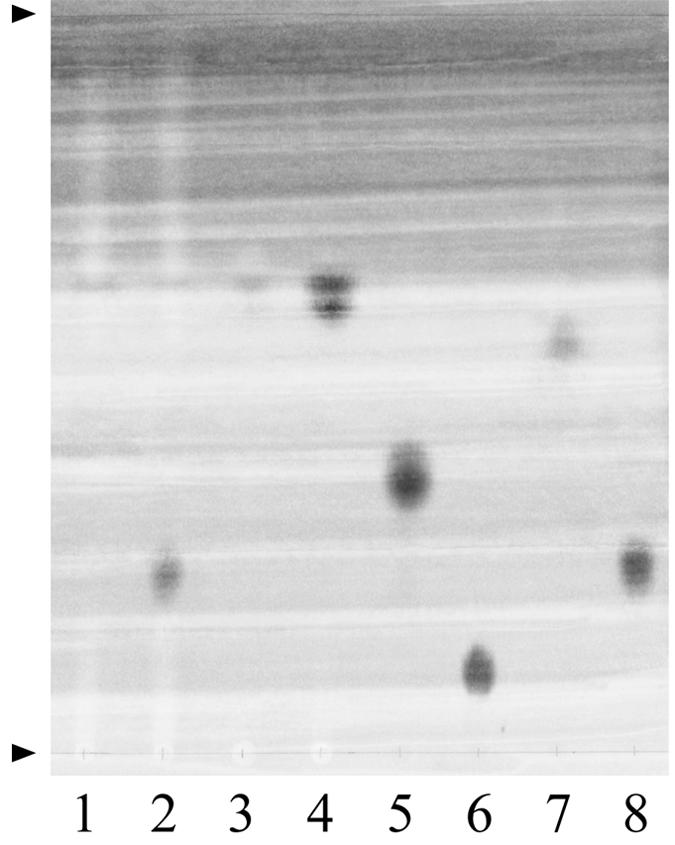
Paper chromatography of product from digestion of pig gastric mucin by recombinant sulfoglycosidase. PGMt was incubated with recombinant SGL as described in the text. The product was subjected to ascending paper chromatography performed with solvent system A. Lane 1, PGMt plus buffer (negative control, no SGL); lane 2, PGMt incubated with SGL and buffer; lane 3, buffer (negative control, no PGMt, no SGL); lane 4, SGL plus buffer (negative control, no PGMt); lane 5, galactose standard (0.1 μmol); lane 6, 6-SO3-Gal standard (0.1 μmol); lane 7, GlcNAc standard (0.1 μmol); lane 8, 6-SO3-GlcNAc standard (0.1 μmol). The arrowheads indicate the sample loading line and the solvent front. The chromatogram was stained by using the dip method of Trevelyan et al (23). The spot at an Rf of 0.62 occurs in all lanes containing material filtered through a dialysis membrane (cutoff, 10 kDa) (lanes 1 to 4) and is likely to be glycerin or a sulfur compound eluted from the membrane.
Assay of sulfoglycosidase activity in closely related anaerobic gut bacteria.
The list of bacteria belonging to the family Bacteroidaceae in which genes encoding members of the CAZy GH20 family (EC 3.2.1.-) have been identified is large, but the proteins encoded by many of these genes have not been isolated. For example, Bacteroides fragilis YCH46 and B. thetaiotaomicron VPI-5482 contain 3 and 15 open reading frames encoding proteins belonging to the GH20 family, although the encoded enzymes have not been characterized. To determine whether some of these putative β-hexosaminidases could in fact be SGLs, cell extracts from 13 selected gut anaerobic bacterial isolates were examined for enzyme activity using 6-SO3-β-GlcNAc-1-pNP as the substrate.
The sulfatase inhibitor 6-SO2NH2-β-GlcNAc-1-Me (0.5 mM) completely inhibited Prevotella strain RS2 MdsA sulfatase activity after overnight incubation with the enzyme (unpublished results) when we used periplasmic extract, semipurified enzyme from the periplasm, and recombinant pure enzyme. Recombinant SGL incubated overnight with 0.5 mM 6-SO2NH2-β-GlcNAc-1-Me showed less effect (20 to 40% inhibition) (unpublished results). The pNP released from 6-SO3-β-GlcNAc-1-pNP was therefore measured in selected bacterial extracts after overnight incubation (0°C, pH 5.5) in the presence and absence of the sulfatase inhibitor described above at a concentration of 0.5 mM. After this treatment, the inhibitor was expected to completely inhibit Prevotella strain RS2 MdsA sulfatase activity, and the sulfatase would be well below its pH optimum (pH 7.4), and there would be only a minor effect on SGL activity. Prevotella strain RS2 extracts showed the expected release of pNP in the absence and in the presence of 6-SO2NH2-β-GlcNAc-1-Me; under the latter conditions the level of inhibition was 20 to 40%, suggesting that most of the activity measured at pH 5.5 was due to SGL (Table 3). B. fragilis fractions showed approximately one-half the activity after preincubation with the inhibitor (this represented the SGL-like activity) compared to Prevotella strain RS2. The localization of the former activity was mainly in the membrane fraction, and the level of 6-SO2NH2-β-GlcNAc-1-Me inhibition was only 10% or less. Prevotella melaninogenica extracts showed measurable pNP release activity, and the levels were 5 to 10% of the levels seen with Prevotella strain RS2. This bacterium has a mucin-desulfating sulfatase (unpublished data), which is consistent with the finding that 54 to 77% of the pNP release was inhibited by 0.5 mM 6-SO2NH2-β-GlcNAc-1-Me. B. thetaiotaomicron extracts also showed measurable but low pNP release (less that 10% of the activity seen in Prevotella strain RS2 extracts), but the incomplete inhibition by 6-SO2NH2-β-GlcNAc-1-Me suggested that this release was probably due to an SGL-like activity (Table 3). These data should be considered qualitative or indicative at present, since the action of 6-SO2NH2-β-GlcNAc-1-Me needs to be tested with purified sulfatases from bacteria other than Prevotella strain RS2 before it can be considered a reliable tool for detecting inhibition. The results indicate that SGL may be present in some other colon anaerobes belonging to the Bacteroidaceae.
TABLE 3.
Sulfatase and sulfoglycosidase activities in cell extracts of selected anaerobes able to react with 6-SO3-β-GlcNAc-1-pNP
| Bacteriuma | Cell fraction | Activity at pH 5.5
|
||
|---|---|---|---|---|
| Uninhibited activity (nmol/min/mg) | 6-SO2NH2-β-GlcNAc-1-Me-inhibited activity (nmol/min/mg)b | % Inhibition | ||
| Prevotella strain RS2 | Extract | 253.3 | 191.5 | 24.4 |
| Membrane | 353.1 | 224.5 | 36.4 | |
| P. melaninogenica ATCC 25845 | Extract | 10.5 | 4.8 | 54.3 |
| Membrane | 16.8 | 3.9 | 76.8 | |
| B. fragilis ATCC 25285 | Extract | 22.0 | 21.5 | 2.3 |
| Membrane | 121.6 | 108.7 | 10.6 | |
| B. thetaiotaomicron ATCC 29148 | Extract | 6.9 | 5.5 | 20.3 |
| Membrane | 19.5 | 10.0 | 48.7 | |
Soluble extracts and membrane preparations from the following isolates were unable to release pNP from 6-SO3-β-GlcNAc-1-pNP: Prevotella buccae ATCC 33577, Prevotella corporis ATCC 33547, Prevotella intermedia ATCC 25611 and WR 2122, Prevotella oralis ATCC 33269 and UI 0279, Prevotella bivia US 4025, Peptostreptococcus anaerobius ATCC 27337, and Peptostreptococcus asaccharolyticus ATCC 14963.
Incubation of 0.5 mM 6-SO2-β-GlcNAc-1-Me with enzyme (16 h at 0°C) completely inhibited N-acetylglucosamine-6-sulfatase (MdsA) activity but resulted in only 20 to 40% inhibition of SGL from Prevotella strain RS2.
Nine other isolates were examined for SGL-like activity. No measurable pNP release from 6-SO3-β-GlcNAc-1-pNP was found when we used soluble extracts and membrane preparations from the following bacteria: Prevotella buccae ATCC 33577, Prevotella corporis ATCC 33547, Prevotella intermedia ATCC 25611 and WR 2122, Prevotella oralis ATCC 33269 and UI 0279, Prevotella bivia US 4025, Peptostreptococcus anaerobius ATCC 27337, and Peptostreptococcus asaccharolyticus ATCC 14963. The preparations were not preincubated with 0.5 mM 6-SO2NH2-β-GlcNAc-1-Me since there was no activity to inhibit.
DISCUSSION
We report here the identification, characterization, purification, cloning, and expression of a novel enzyme that is able to remove sulfate from mucin oligosaccharide chains by glycosidic cleavage of terminal 6-SO3-β-GlcNAc residues. The enzyme was found in Prevotella strain RS2, an anaerobic bacterium originally isolated from pig colon mucosa. We termed this enzyme a sulfoglycosidase and showed that it differs from the mucin-desulfating sulfatase, MdsA, that has been described previously. The SGL activity is not due to sequential action of MdsA and hexosaminidase. SGL is translocated to the periplasm, as shown by cell fractionation studies and by the presence of a periplasmic translocation motif at the N terminus of the translated product. At least one other fraction catalyzing SGL activity was detected during purification, and we have not investigated yet whether this fraction is a degradation product of the 100-kDa protein or is an isoenzyme.
SGL was tested for activity against a number of model substrates which have structures analogous to the structures of sugars in mucin oligosaccharide chains (Table 2). Activity was found only with 6-SO3-β-GlcNAc-1-pNP, in agreement with data showing that SGL released free 6-SO3-GlcNAc from mucin. The inability of SGL to cleave pNP from the disaccharide substrate 4-(β-Gal)-6-SO3-β-GlcNAc-1-pNP strongly suggests that the enzyme is an exosulfoglycosidase.
When the translated 901-amino-acid sequence (before translocation of the SGL) was compared with BLAST protein database sequences, the closest matches were with β-hexosaminidases from B. thetaiotaomicron and Porphyromonas gingivalis. To the best of our knowledge, these enzymes have not been isolated and/or have not been tested for activity against sulfated substrates, and SGL activity has not been described previously in bacteria. When crude cell extracts from several common species of gut anaerobes were assayed for SGL-like activity, B. fragilis and B. thetaiotaomicron appeared to have some activity, although the levels were lower than the levels for Prevotella strain RS2. The possible activity detected in P. melaninogenica was most likely due to sulfatase plus hexosaminidase. The nine other isolates tested had no SGL-like activity. It seems likely that some of the uncharacterized members of the CAZy GH20 family may well catalyze SGL-type reactions rather than conventional β-N-acetylhexosaminidase reactions.
Only one other enzyme, human β-d-N-acetylhexosaminidase A, has been shown to remove intact 6-SO3-GlcNAc from 6-SO3-β-GlcNAc-1-pNP. Consistent with this specificity, the human enzyme can liberate 6-SO3-GlcNAc from the nonreducing end of keratan sulfate-derived oligosaccharides. The enzyme can also remove GlcNAc from β-GlcNAc-1-pNP and shows 20-fold-higher affinity for the nonsulfated substrate (10). In contrast, the Prevotella SGL cleaves the nonsulfated substrate at a rate that is only 1% of the rate seen with the sulfated substrate (with substrate at a concentration of 1 mM), and the difference is due to the fact that the nonsulfated substrate has a higher Km and a lower Vmax. The Km and Vmax values are for model substrates, however, and may not necessarily be correct for native substrates. The Prevotella enzyme has a sevenfold-higher affinity (Km = 0.18 mM) for 6-SO3-β-GlcNAc-1-pNP than the human enzyme has. We have not tested the Prevotella SGL with keratan sulfate-derived oligosaccharides.
The human β-d-N-acetylhexosaminidase A is better known for its alternative specificity in cleaving N-acetylgalactosamine from the oligosaccharide chain of GM2 ganglioside. The human recessive lysozomal storage disease (GM2 gangliosidosis) called Tay-Sachs disease (9) results from β-d-N-acetylhexosaminidase A deficiency due to inheritance of mutant forms.
The finding that both a mucin-desulfating sulfatase (MdsA) (28) and SGL are present in the periplasm of Prevotella strain RS2 (and perhaps other bacteria) leads us to propose that sulfate group metabolism in sulfomucins, sulfoglycoproteins, and/or sulfoglycolipids must have important functions in the colonic habitat, which the bacterium is attempting to modify. These enzymes may enhance the ability of the bacterium to survive in its environment. Both enzymes are partially induced by growth on mucin, and both can desulfate mucin. The product of SGL action is a substrate for MdsA sulfatase, but not vice versa. The proposed roles of SGL in mucin metabolism may be hypothesized to be (i) removal of inhibitory 6-SO3-β-GlcNAc groups from mucin chains that limit degradation of the chain by exoglycosidases and neuraminidases, (ii) removal of 6-SO3-β-GlcNAc groups from mucin chains (usually exposing a galactose as the new terminal residue), thus creating or removing sites for different adhesins, and (iii) provision of 6-SO3-GlcNAc as a readily hydrolysable substrate for MdsA sulfatase. The first two roles could have substantial effects on the colon mucosal habitat.
It has been well established that there is a lower sulfate content in secreted colonic mucin in ulcerative colitis patients (6) and that mucin is more susceptible to fecal-bacterial glycosidase action in the presence of added sulfatase (26). The role of the SGL, as well as mucin-specific sulfatase activities, needs to be taken into account when the significance of mucin sulfate levels and the rate of desulfation are considered in relation to mucus barrier effectiveness and susceptibility to degradation. The presence of an enzyme with this selective specificity also suggests that its targeted sulfated sugar may have had an important biological action in its own right before its removal, such as being part of a ligand. Such an action of negatively charged groups on carbohydrate-presenting molecules has been documented in the case of 9-O-acetylsialic acids or sulfate groups in siglec and selectin ligands, the loss of which diminishes or prevents their function.
Acknowledgments
This work was supported by the Marsden Fund of the Royal Society of New Zealand.
pNP model substrates were synthesized by the carbohydrate chemistry team led by R. H. Furneaux at Industrial Research Limited, Lower Hutt, New Zealand. The syntheses were carried out by G. B. Evans (3- and 6-SO3-Gal-1-pNP), P. M. Rendle and D. Lenz (6-SO3-β-GlcNAc-1-pNP), and P. C. Tyler (6-SO3-β-Gal-1-pNP). Protein sequencing was performed by C. G. Knight and I. Anthony at the Protein Sequencing Unit of the School of Biological Sciences, The University of Auckland, Auckland, New Zealand.
REFERENCES
- 1.Berkin, A., W. A. Szarek, and R. Kisilevsky. 2002. Synthesis and biological evaluation of a radiolabeled analog of methyl 2-acetamido-2,4-dideoxy-β-d-xylo-hexopyranoside directed towards influencing cellular glycosaminoglycan biosynthesis. Carbohydr. Res. 337:37-44. [DOI] [PubMed] [Google Scholar]
- 2.Bernadi, G. 1971. Chromatography of proteins on hydroxyapatite. Methods Enzymol. 22:325-339. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell, D. R., and M. P. Bryant. 1966. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl. Microbiol. 14:794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinch, K., G. B. Evans, R. H. Furneaux, P. M. Rendle, P. L. Rhodes, A. M. Roberton, D. I. Rosendale, P. C. Tyler, and D. P. Wright. 2002. Synthesis and utility of sulfated chromogenic carbohydrate model substrates for measuring activities of mucin-desulfating enzymes. Carbohydr. Res. 337:1095-1111. [DOI] [PubMed] [Google Scholar]
- 6.Corfield, A. P., N. Myerscough, N. Bradfield, C. D. O. Amaral-Corfield, M. Gough, J. R. Clamp, P. Durdey, B. F. Warren, D. C. Bartolo, K. R. King, and J. M. Williams. 1996. Colonic mucins in ulcerative colitis: evidence for loss of sulfation. Glycoconj. J. 13:809-822. [DOI] [PubMed] [Google Scholar]
- 7.Corfield, A. P., S. A. Wagner, L. J. O'Donnell, P. Durdey, R. A. Mountford, and J. R. Clamp. 1993. The roles of enteric bacterial sialidase, sialate O-acetyl esterase and glycosulfatase in the degradation of human colonic mucin. Glycoconj. J. 10:72-81. [DOI] [PubMed] [Google Scholar]
- 8.Del Tito, B. J., Jr., J. M. Ward, J. Hodgson, C. J. Gershater, H. Edwards, L. A. Wysocki, F. A. Watson, G. Sathe, and J. F. Kane. 1995. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J. Bacteriol. 177:7086-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gravel, R. A., J. T. R. Clarke, M. M. Kaback, D. Mahuran, K. Sandhoff, and K. Suzuki. 1995. The GM2 gangliosidoses, p. 2839-2882. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease, 7th ed., vol. II. McGraw-Hill, Inc. Health Professional Division, New York, N.Y.
- 10.Kresse, H., W. Fuchs, J. Glossl, D. Holtfrerich, and W. Gilberg. 1981. Liberation of N-acetylglucosamine-6-sulfate by human beta-N-acetylhexosaminidase A. J. Biol. Chem. 256:12926-12932. [PubMed] [Google Scholar]
- 11.Kristinsson, H., K. Neber, A. C. O'Sullivan, F. Struber, T. Winkler, and Y. Yamaguchi. 1994. A novel synthesis of sulfamoyl nucleotides. Tetrahedron 50:6825-6838. [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Neutra, M. R., and J. F. Forstner. 1987. Gastrointestinal mucus: synthesis, secretion and function, p. 975-1009. In L. R. Johnson (ed.), Physiology of the gastrointestinal tract, 2nd ed. Raven Press, New York, N.Y.
- 14.Nieuw Amerongen, A. V., J. G. Bolscher, E. Bloemena, and E. C. Veerman. 1998. Sulfomucins in the human body. Biol. Chem. 379:1-18. [DOI] [PubMed] [Google Scholar]
- 15.Ochman, H., F. J. Ayala, and D. L. Hartl. 1993. Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol. 218:309-321. [DOI] [PubMed] [Google Scholar]
- 16.Roberton, A. M., and A. P. Corfield. 1999. Mucin degradation and its significance in inflammatory conditions of the gastrointestinal tract, p. 222-261. In G. W. Tannock (ed.), Medical importance of normal microflora. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 17.Roberton, A. M., C. G. McKenzie, N. Sharfe, and L. B. Stubbs. 1993. A glycosulphatase that removes sulphate from mucus glycoprotein. Biochem. J. 293:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberton, A. M., D. I. Rosendale, and D. P. Wright. 1999. Assays for bacterial mucin-desulfating sulfatase, p. 417-426. In A. P. Corfield (ed.), Glycoprotein methods and protocols: the mucins. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 19.Roberton, A. M., and R. A. Stanley. 1982. In vitro utilization of mucin by Bacteroides fragilis. Appl. Environ. Microbiol. 43:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Stanley, R. A., S. P. Lee, and A. M. Roberton. 1983. Heterogeneity in gastrointestinal mucins. Biochim. Biophys. Acta 760:262-269. [DOI] [PubMed] [Google Scholar]
- 22.Stanley, R. A., S. P. Ram, R. K. Wilkinson, and A. M. Roberton. 1986. Degradation of pig gastric and colonic mucins by bacteria isolated from the pig colon. Appl. Environ. Microbiol. 51:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trevelyan, W. E., D. P. Procter, and J. S. Harrison. 1950. Detection of sugars on paper chromatograms. Nature 166:444-445. [DOI] [PubMed] [Google Scholar]
- 24.Triggs-Raine, B. L., B. R. Akerman, J. T. Clarke, and R. A. Gravel. 1991. Sequence of DNA flanking the exons of the HEXA gene, and identification of mutations in Tay-Sachs disease. Am. J. Hum. Genet. 49:1041-1054. [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai, H. H., A. D. Dwarakanath, C. A. Hart, J. D. Milton, and J. M. Rhodes. 1995. Increased faecal mucin sulphatase activity in ulcerative colitis: a potential target for treatment. Gut 36:570-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai, H. H., D. Sunderland, G. R. Gibson, C. A. Hart, and J. M. Rhodes. 1992. A novel mucin sulphatase from human faeces: its identification, purification and characterization. Clin. Sci. (London) 82:447-454. [DOI] [PubMed] [Google Scholar]
- 27.Witholt, B., M. Boekhout, M. Brock, J. Kingma, H. V. Heerikhuizen, and L. D. Leij. 1976. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal. Biochem. 74:160-170. [DOI] [PubMed] [Google Scholar]
- 28.Wright, D. P., C. G. Knight, S. G. Parkar, D. L. Christie, and A. M. Roberton. 2000. Cloning of a mucin-desulfating sulfatase gene from Prevotella strain RS2 and its expression using a Bacteroides recombinant system. J. Bacteriol. 182:3002-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright, D. P., D. I. Rosendale, and A. M. Roberton. 2000. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 190:73-79. [DOI] [PubMed] [Google Scholar]