Abstract
The recent availability of the whole genome of Synechococcus sp. strain WH8102 allows us to have a global view of the complex structure of the phycobilisomes of this marine picocyanobacterium. Genomic analyses revealed several new characteristics of these phycobilisomes, consisting of an allophycocyanin core and rods made of one type of phycocyanin and two types of phycoerythrins (I and II). Although the allophycocyanin appears to be similar to that found commonly in freshwater cyanobacteria, the phycocyanin is simpler since it possesses only one complete set of α and β subunits and two rod-core linkers (CpcG1 and CpcG2). It is therefore probably made of a single hexameric disk per rod. In contrast, we have found two novel putative phycoerythrin-associated linker polypeptides that appear to be specific for marine Synechococcus spp. The first one (SYNW2000) is unusually long (548 residues) and apparently results from the fusion of a paralog of MpeC, a phycoerythrin II linker, and of CpeD, a phycoerythrin-I linker. The second one (SYNW1989) has a more classical size (300 residues) and is also an MpeC paralog. A biochemical analysis revealed that, like MpeC, these two novel linkers were both chromophorylated with phycourobilin. Our data suggest that they are both associated (partly or totally) with phycoerythrin II, and we propose to name SYNW2000 and SYNW1989 MpeD and MpeE, respectively. We further show that acclimation of phycobilisomes to high light leads to a dramatic reduction of MpeC, whereas the two novel linkers are not significantly affected. Models for the organization of the rods are proposed.
Phycobilisomes (PBSs) are the major light-harvesting systems used by cyanobacteria and red algae. This type of photosynthetic antenna consists of a macrocomplex composed of chromophorylated proteins designated phycobiliproteins (PBPs). These are organized as a core connected with photosystems and generally surrounded by six peripheral rods (for a review, see e.g., reference 37). PBSs are very variable in structure and pigment composition, and this has allowed cyanobacteria to colonize environments showing a great diversity in terms of light quantity and quality. The PBSs of open-ocean Synechococcus cyanobacteria are among the most complex ones described to date since they possess four types of constitutive PBPs: allophycocyanin (APC), phycocyanin (PC), and two forms of phycoerythrin, PEI and PEII, the latter being specific for these organisms. Each PBP covalently binds one or two types of tetrapyrrole chromophores (designated phycobilins) to cysteinyl residues, via a thioether bond. PEI and PEII bind both phycourobilin (PUB; absorption maximum [Amax], 495 nm) and phycoerythrobilin (PEB; Amax, 550 nm) in different proportions, PC may bind both PEB and phycocyanobilin (PCB; Amax, 620 nm), whereas APC binds only PCB (14, 29).
All PBPs are made up of two types of chains, α and β, carrying most of the phycobilin molecules. These chains are organized as trimers (αβ)3, themselves coupled to form hexameric disks (αβ)6. These disks are stabilized by linker polypeptides which also ensure the correct structural assembly of the different PBPs within the rods (14, 16, 25). Although cyanobacterial rod linkers are generally nonpigmented, PEII has been shown to possess a specific type of γ linker (called MpeC) which binds one PUB molecule (49). MpeC is the sole PEII-associated linker described to date, although other PBP types generally possess several. For instance, the sole PE type of the freshwater cyanobacterium Fremyella diplosiphon possesses three linker proteins: CpeC, -D, and -E (12).
Light quality and quantity are among the major factors affecting the composition of PBSs. In F. diplosiphon, the relative proportion of PC and PE can vary within the PBS rods in response to a change in the light climate (1, 18), but such complementary chromatic adaptation is rare among marine Synechococcus spp. (32, 46). Changes in photon fluxes also have an effect on the structure of PBSs. Marine cyanobacteria mainly respond to high light stress by decreasing the content of PBSs per cell (20, 38). This decrease is mainly attributable to a reduction of the surface of thylakoid membranes (19). In freshwater cyanobacteria and rhodophytes, it has been shown that the PBS rods may also shorten after cells have been transferred from low to high light conditions (4, 23, 26, 27, 36). However, whether this phenomenon also occurs in marine Synechococcus and to what extent have been little studied so far (38).
In this study, we used a combination of genomic and biochemical analyses to study the composition of PBSs of the marine Synechococcus strain WH8102 and structural changes occurring in PBSs as a result of photoacclimation. This strain is characterized by its very high PUB/PEB ratio, typical of open-ocean populations of Synechococcus, and its genome has recently been published (33). Although the PBSs of several marine Synechococcus strains have been extensively analyzed through an excellent series of papers by A. N. Glazer and coworkers (see, e.g., references 29-31 and 47-49), the availability of the whole genome of Synechococcus sp. strain WH8102 has allowed us to have a global view of the organization of PBS genes. This analysis revealed a number of novelties, including the presence of two novel genes encoding putative PE-associated linker polypeptides. Using a biochemical approach, we have been able to confirm the presence of these linkers in the PBSs and to characterize some of their properties, including their probable localization within the PBS rods.
MATERIALS AND METHODS
Genomic analysis.
The methods used for sequencing and modeling the genome of Synechococcus sp. strain WH8102 (GenBank/EMBL accession number NC_005070) have been described elsewhere (33). Results from the automated annotation were checked manually and completed by BlastP analyses (2) of the genome data bank with individual PBS proteins known from other marine Synechococcus spp. or freshwater cyanobacteria. Alignments of linker polypeptides were made using ClustalW (43) and refined by hand.
Culture conditions.
The axenic marine Synechococcus sp. clone WH8102 was grown at 23 ± 1°C in 8 liters of polycarbonate culture flasks (Nalgene) containing 0.2 μm filtered PCR S-11 medium (34). Cultures were progressively acclimated for at least 1 month to a range of continuous white irradiances (from 15 to 250 μmol of photons · m−2 · s−1) provided by 10 daylight 865/36W fluorescent tubes (Mazdafluor Prestiflux) and various combinations of density and/or diffusion filters (LEE Filters; Panavision, Paris, France). For comparison of PBS structure, Synechococcus sp. clone WH7803 was grown at 15 μmol of photons · m−2 · s−1. The origins of both strains have been described elsewhere (13).
Biochemical analysis of nondissociated phycobilisomes. (i) Extraction of intact PBS.
Frozen cells were thawed, washed in 0.75 M K-Na phosphate buffer (pH 7), and resuspended in 10 ml of the same buffer complemented with a 1 mM concentration of each of the following protease inhibitors: Pefabloc (Interchim), EDTA, and phenylmethylsulfonyl fluoride (Sigma Aldrich). The solution was homogenized with a Thoma potter and passed twice through a French press at 20,000 lb/in2 at 4°C. A final concentration of 5% (wt/vol) Triton X-100 was added, and the samples were allowed to incubate with stirring at room temperature for 45 min. The solution of broken cells was centrifuged for 15 min at 7,000 × g to allow phase separation. The lower red layer containing nondissociated PBS was collected, loaded onto a discontinuous sucrose gradient in phosphate buffer (0.75, 0.62, 0.50, 0.37, and 0.25 M), and ultracentrifuged for 15 h at 40,000 × g at 12°C in a Beckman L8 M-55 ultracentrifuge (SW28B rotor). The heaviest PBS fraction (see Results) was carefully collected and characterized by its absorption spectra, using a double-wavelength spectrophotometer (Aminco Chance DW2), and 77 K emission fluorescence spectra with excitation at 495 nm on filters immersed in liquid nitrogen. The latter spectra were recorded with a spectrofluorimeter (Hitachi F4500) with 5 nm/2.5 nm (excitation/emission) slit widths.
(ii) LiDS-PAGE analysis.
For lithium dodecyl sulfate (LiDS)-polyacrylamide gel electrophoresis (PAGE) analysis, aliquots of PBS fractions were precipitated with 10% trichloroacetic acid on an equal PCB basis (optical density at 640 nm) and denatured for 3 min at 70°C in 3% LiDS denaturation buffer. The denatured samples were then loaded on an LiDS polyacrylamide gradient gel (10 to 20%). Electrophoresis was run at 4°C and a constant 20 mA for 6 h in Laemmli buffer (24) containing 0.1% LiDS. In addition, aliquots of the PBP-enriched fractions (see below) were loaded on a similar LiDS-polyacrylamide gel. For fluorescence visualization under UV light, gels were preincubated for 15 min in 20 mM zinc acetate to enhance PBP fluorescence (3).
Analysis of isolated phycobiliproteins. (i) Phycobilisome dissociation.
Cells were harvested by centrifugation at 7,000 × g at room temperature, washed, and resuspended in a low-strength ionic buffer (10 mM TES buffer [pH 7]; Sigma Aldrich) supplemented with protease inhibitors (see above). Cells were broken twice by using a cooled French press at 20,000 lb/in2. All subsequent steps were carried out in darkness at 4°C. The solution of broken cells was ultracentrifuged for 3 h at 130,000 × g in a Beckman XL-70 ultracentrifuge (SW41Ti rotor) in order to remove remnants of thylakoid membranes. Aliquots of the reddish supernatant (containing different PBPs resulting from the dissociation of PBSs) were snap-frozen in liquid nitrogen and stored at −80°C until isoelectric focusing (IEF) analysis (see below).
For separating the PBS dissociation products, the supernatant was loaded on a continuous sucrose gradient (0 to 30%) in 10 mM TES buffer (pH 7) in the presence of protease inhibitors and ultracentrifuged for 12 h at 130,000 × g and 12°C. The different PBP-enriched fractions were carefully collected, dialyzed against TES buffer, and concentrated by using 5-kDa-cutoff membrane columns (Vivascience, Hannover, Germany). The fractions were then characterized by their absorption spectra with a double-beam spectrophotometer (Uvikon UV/Vis 943) as previously described (38). Samples were frozen in liquid nitrogen and stored at −80°C until PBP separation on an IEF gel.
(ii) IEF analysis.
Aliquots of the concentrated PBP-enriched fractions or total dissociated PBS-enriched extracts were mixed with glycerol (15% final concentration) and loaded onto a 7% acrylamide minigel containing ampholyte carriers pH 4 to 6.5 (Amersham Pharmacia). The electrophoresis system was a mini-Protean II system (Bio-Rad), where internal glass plates were replaced with alumina plates (Hoefer). The cathode tank was filled with 20 mM NaOH, and the anode one contained 20 mM orthophosphoric acid. The PBPs were allowed to focus during about 2 h by increasing steps of 50 V every 10 min up to 350 V. Voltages higher than 400 V led to overfocusing, resulting in strong band distortion. The run was interrupted before complete focusing when the band separation looked optimal. Longer runs led all PBPs to focus towards an apparent isoelectric point of ca. 5.
The different-colored bands were carefully cut out and then were crushed in 10 mM Tricine buffer (pH 7.8) by using an electric grinder. Acrylamide remnants were removed by a 5-min centrifugation at 20,000 × g. Absorption spectra were recorded and corrected for the influence of dissolved acrylamide by fitting and subtracting a two-parameter exponential decay curve: y = a · e(−b[r] · x[r]).
Emission fluorescence spectra were determined at room temperature with a Perkin-Elmer L50B spectrofluorimeter with 7-nm (excitation/emission) slit widths, and the maxima were normalized to 1 to ease comparisons.
For comparative IEF analyses, PBPs were loaded after adjusting their optical densities at 650 nm to get similar APC concentrations. Prior to this step, the samples were concentrated on 5-kDa-cutoff membranes (Nalgene) to limit the disorganization of APC molecules.
Mass spectrometry. (i) In-gel digestion.
The identification of the major linker polypeptides from LiDS-PAGE analysis was performed by mass spectrometry, and some of them were confirmed by N-terminal sequencing (see Results). Reduction was achieved by a 1-h treatment of the gel spots excised from the gel in 10 mM dithiothreitol at 57°C. An alkylation reaction was performed using 25 mM iodoacetamide for 45 min at room temperature with protection from light. Tryptic digestion was performed in 12.5 ng of trypsin (Promega, Madison, Wis.) μl−1 in 25 mM NH4HCO3 (freshly diluted) at 37°C overnight. Gel pieces were centrifuged, and 5 μl of an H2O-CH3CN-HCOOH (25/70/5, vol/vol/vol) solution was added to extract peptides. The supernatant solvent was completely evaporated in order to remove all acetonitrile from the sample. Then 10 μl of H2O-5% HCOOH was added and injected in the high-performance liquid chromatography system.
(ii) Nano LC-MS/MS.
Nanoscale capillary liquid chromatography-tandem mass spectrometric (LC-MS/MS) analysis of the digested proteins were performed using a capillary LC system (CapLC; Micromass, Manchester, United Kingdom) coupled to a hybrid quadripole orthogonal acceleration time-of-flight tandem mass spectrometer (Q-TOF II). The LC-MS union was made with a PicoTip (New Objective, Woburn, Mass.) fitted on a Zspray (Micromass) interface. Chromatographic separations were conducted on a reverse-phase capillary column (Pepmap; C18, 75-μm internal diameter, 15-cm length, LC packing) with a 200-nl min−1 flow. The gradient profile used consisted of a linear gradient from 95% A (H2O, 0.1% HCOOH) to 45% B (CH3CN, 0.1%HCOOH) over 35 min followed by a linear gradient to 95% B for 1 min. Mass data acquisitions were piloted by MassLynx software using automatic switching between MS and MS-MS modes. Mass data collected during an LC-MS/MS analysis were processed to be submitted to the search software Protein Lynx Global Server 2.0 (both softwares are from Micromass). Searches were done with a tolerance on mass measurement of 0.2 Da in MS mode and 0.5 Da in MS/MS mode.
For three of the major linker polypeptides, N-terminal sequences were also determined after blotting proteins onto a polyvinylidene difluoride membrane and staining with 0.1% amido black in 50% methanol. The sequences were determined on an ABI 494A system (Applied Biosystems) as described previously (42).
RESULTS
Genomic analysis of phycobilisome structure.
The Synechococcus sp. strain WH8102 genome contains at least 29 open reading frames (ORFs) encoding polypeptides with similarity to proteins involved in PBS assembly, phycobilin biosynthesis or ligation, and PBS regulation. The presence in the genome of apcABCDEF homologs (with apcEABC making a cluster and the other two genes being isolated in the genome) indicates that APC cores likely have a classical structure made up of two types of α chains (α and α-Β) and β chains (β and β-18), as well as LC and LCM linker polypeptides.
Synechococcus sp. strain WH8102 possesses a single rpcBA operon, encoding the α and β subunits of PC, so-called R-PCII in marine Synechococcus spp. (8). An additional ORF (SYNW1996) also shows some weak similarity to a β subunit of PC, but the function of its putative product is unclear. Two cpcG homologs (rpcG1 and rpcG2), encoding rod-core linker polypeptides (LRC), are also identifiable in the Synechococcus sp. strain WH8102 genome (compared to, e.g., four in Anabaena sp. strain PCC 7120) but are not part of the rpcBA cluster. Absence of PC rod linker (LR) genes such as cpcC or cpcD (5) suggests that under all light conditions there is only one hexameric PC disk per PBS rod. This may explain why this strain is unable to perform chromatic adaptation (CA), a process involving light quality-induced variations of the relative rod content in either the sole PE (CA group II) or both PC and PE (CA group III) (41). Expectedly, this strain also lacks a number of genes which in other cyanobacteria have been shown to be involved in the regulation of this phenomenon (rcaA to rcaG [6, 21, 28, 39]).
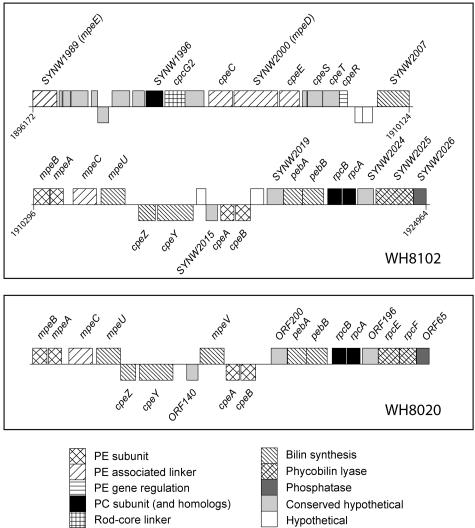
All PE genes, as well as most genes involved in the biosynthesis or binding of phycobilins (PEB, PUB, and PCB), are gathered into a large cluster, also including the PC genes cpcG2 and SYNW1996, as well as the rpcBA operon (Fig. 1). Besides the cpeBA and mpeBA operons, encoding the β and α subunits of PEI and PEII, respectively, there are five genes encoding proteins with similarity to the PE linkers: cpeC, cpeE, mpeC, and two novel ones, SYNW1989 and SYNW2000. Three of them are included in a possible operon cpeC-SYNW2000-cpeE. The region including these genes and the three genes downstream shows a strong similarity to the cpeCDESTR gene region from F. diplosiphon (7). However, SYNW2000 is much longer than F. diplosiphon cpeD (548 vs. 249 residues) and appears to be a fusion gene. The protein alignment shown in Fig. 2 clearly shows that the C terminus of the SYNW2000 gene product has a strong homology to the cpeD gene products from F. diplosiphon (11, 12) and Gloeobacter violaceus whereas its N terminus is homologous to Synechococcus sp. strain WH8102 and WH8020 mpeC gene products, PEII γ linkers (49). SYNW1989 encodes a product of more classical length for a PBP linker (300 residues). The closest homolog of SYNW1989 is also mpeC, although it differs from it by encoding a longer N terminus and a fairly divergent C terminus (Fig. 2).
FIG. 1.
Complete phycobilisome rod gene cluster from Synechococcus sp. strain WH8102 and partial homologous cluster from Synechococcus sp. strain WH8020 (8). These sequences were retrieved from GenBank (accession numbers NC_005070 and M95288, respectively). For WH8102, numbers indicate the positions of the genes in the genome.
FIG. 2.
Alignment of the novel putative phycoerythrin II linkers deduced from the sequences of the SYNW2000 and SYNW1989 genes from Synechococcus sp. strain WH8102 with MpeC homologs from the same organism (deduced from SYNW2010) and from the marine Synechococcus sp. strain WH8020 (Q02181) and with CpeD homologs from F. diplosiphon (P18543) (Fdi) and G. violaceus (Glr1264, retrieved from Cyanobase) (Gvi). Identical residues in at least two aligned sequences are indicated in white type on a black background. Gray blocks are conservative substitutions within amino acid groups with similar properties: ILV, FWY, AGS, ST, DE, DN, KR, and EQ. Cysteinyl residues conserved in at least three sequences are indicated with an asterisk.
To check whether these two novel genes were expressed and the resulting linkers were functional, we performed a biochemical analysis of the linkers in intact and dissociated PBSs.
Biochemical analysis of linkers from phycobilisome fractions.
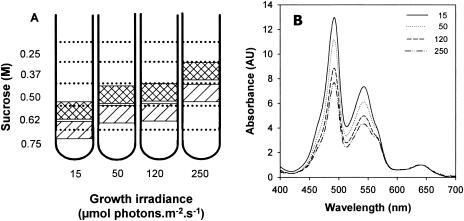
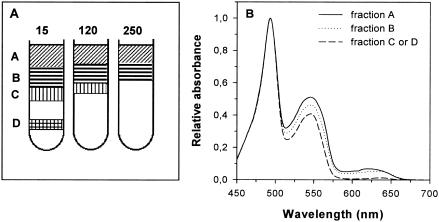
Sucrose gradient centrifugation was used to obtain purified fractions of nondissociated PBSs. The gradient exhibited two pink bands enriched in PBSs having different apparent densities depending on growth irradiance (Fig. 3A). At 15 μmol of photons · m−2 · s−1, the two PBS-enriched bands were retrieved in the range of 0.62 to 0.75 M sucrose, while at 250 μmol of photons · m−2 · s−1, they were in the range 0.37 to 0.5 M sucrose, indicating that the size of PBS complexes decreased at high light. The upper fractions (Fig. 3A, hatched) contain several non-PBS contaminants, and therefore we used the lower fractions for further analysis.
FIG. 3.
Schematic representations of discontinuous sucrose gradients of PBS particles from Synechococcus sp. strain WH8102 grown at four growth irradiances (A) and absorption spectra of the heaviest (hatched) PBS bands (B).
The 77 K fluorescence emission spectra of these fractions showed a strong contribution of the PBS terminal acceptor (679 nm) compared to PE (570 nm) and PC (648 nm), indicating efficient energy transfer between the PBPs (data not shown). The absorption spectra of these fractions from different light levels normalized at the PCB maximum (640 nm) showed a strong (54%) relative decrease of the PUB peak (at 492 nm) between 15 and 250 μmol of photons · m−2 · s−1 (Fig. 3B). In oceanic Synechococcus spp., PCB is a chromophore associated with PC and APC, whereas PUB is found only in PE. Thus, this decrease indicates that the level of PE is specifically reduced relative to PC and APC in PBSs at high light. The peak at 543 nm (A543) in absorption spectra is due to PEII-associated PEB, whereas that at 565 nm (A565) is attributable to PEI- plus PC-associated PEB (38). Thus, the decrease of the A543/A565 absorbance ratio further suggests that PEII varies much more than PEI during acclimation of the PBSs to high light.
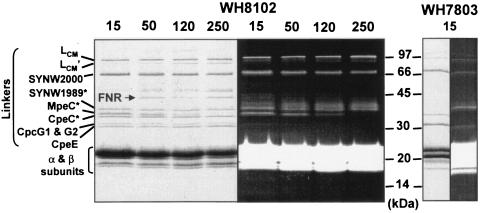
LiDS-PAGE analysis of these PBS fractions allowed the clear separation of seven bands corresponding to PBS linkers, including five with an apparent molecular mass between 30 and 40 kDa (Fig. 4). It also revealed the presence of ferredoxin-NADP reductase (FNR) which, as in other cyanobacteria (17), seems to be associated with PBSs. The identities of the FNR and of most linker polypeptides were determined by mass spectrometric analysis. The only exceptions were the core-membrane linker LCM and its degradation product LCM′, which can be unambiguously identified by their large apparent molecular masses, and ApcC, which was not detected with the electrophoretic conditions used. Data were totally consistent with N-terminal sequencing also performed for three of these proteins (Fig. 4). Visualization of the gel under UV light in the presence of zinc acetate indicated that four of these linkers were chromophorylated: LCM (and LCM′), SYNW2000, SYNW1989, and MpeC, a PEII-associated γ linker. The green fluorescence of the last three proteins under UV light (Fig. 4, right panel) suggests that their chromophore is PUB, as confirmed by the 77 K fluorescence emission of the corresponding bands cut out from the gel (see spectrum of SYNW2000 in Fig. 7A). In contrast, the PEI-associated rod linkers CpeC and CpeE, as well as the phycocyanin rod-core linkers CpcG1 and CpcG2 (which made a single band on SDS-PAGE), were not chromophorylated. A prominent red fluorescent smear was also observed above SYNW1989 under UV light, but its origin remains unclear since there was no corresponding band on the Coomassie blue-stained gel. The LiDS-PAGE analysis of PBSs from Synechococcus sp. strain WH8102 grown at different irradiances unambiguously showed that the level of MpeC dramatically decreased with increasing growth irradiances, until it became undetectable at 250 μmol of photons · m−2 · s−1. Another surprising outcome of this photoacclimation study was the apparent increase of FNR at high light relative to PBS proteins, but the reason for that is unclear.
FIG. 4.
LiDS-PAGE gel (polyacrylamide gradient, 10 to 20%) of PBSs from Synechococcus sp. strain WH8102 cells acclimated at four irradiances (in micromoles of photons per square meter per second) and WH7803 cells grown at 15 μmol of photons · m−2 · s−1. (Right) Coomassie staining. (Left) Same gel under UV light in the presence of 20 mM zinc acetate. The identity of star-labeled polypeptides was determined by N-terminal sequencing (in addition to mass spectrometry).
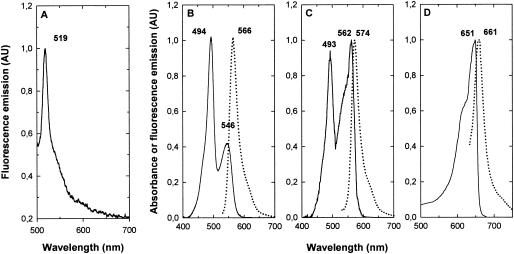
FIG. 7.
(A) Emission fluorescence spectrum at 77 K (with excitation at 440 nm) of the LiDS-PAGE band of the PUB-chromophorylated linker SYNW2000. Room temperature absorption (continuous line) and fluorescence emission spectra (dotted line) of PEII (B), PEI (C), and APC (D) from Synechococcus sp. strainWH8102. Numbers indicate the positions of maxima in nanometers.
Comparison of the PBS linker composition of the high-PUB Synechococcus sp. strainWH8102 and the low-PUB Synechococcus sp. strain WH7803 (Fig. 4) at low light showed that they share the presence of a chromophorylated linker with an apparent molecular mass around 68 kDa. This strongly suggests that WH7803 possesses a homolog of the WH8102 SYNW2000 linker (note that the latter has a predicted mass of only 59,409 Da without the chromophore).
Isolation and characterization of phycobiliproteins.
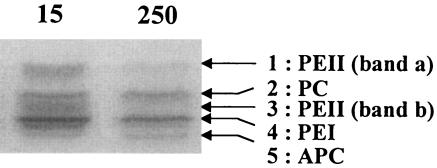
In order to better characterize the different PBPs of Synechococcus sp. strain WH8102, the PBSs were dissociated in a low ionic buffer and then the PBP complexes were isolated by IEF. When PBP-enriched fractions were directly loaded onto IEF gels, five bands were observed (Fig. 5). Their colors provided a visual indication of their nature: the orange bands 1 and 3 were PEII (likely in two different organization states), the pink band 4 was PEI, the violet band 2 was PC, and the blue band 5 was APC. PBS extracts from samples grown at 15 and 250 μmol of photons · m−2 · s−1 were loaded on an IEF gel on the basis of an equal APC amount (Fig. 5). This clearly showed that at high light the two PEII bands dramatically decreased whereas the PEI and PC remained at a similar level.
FIG. 5.
Comparative IEF gel of relative cellular PBP content in Synechococcus sp. strain WH8102 acclimated at 15 and 250 μmol of photons · m−2 · s−1. PBPs were loaded on the basis of an equal APC content (A650).
Before characterizing spectrometrically these PBPs, we purified them further by adding one more step of centrifugation on a continuous sucrose gradient prior to the IEF. The resolution of the PBP separation was then increased, likely because these samples contained fewer contaminant soluble proteins than the initial preparations of dissociated PBS. The band pattern on the sucrose gradient varied depending on growth irradiance (Fig. 6A). Under all light conditions, samples showed two upper pinkish bands (A and B). Their absorption characteristics (Fig. 6B) show that they contain all three phycobilin types (PUB, PEB, and PCB), the upper band (A) being richer in PCB than the second one (band B). Two additional orange bands (C and D) were seen at the lower part of the gradient under low light conditions but progressively disappeared as growth irradiance increased (Fig. 6A). The absorption spectra of bands C and D were identical and revealed that these bands contained only traces of PCB (Fig. 6B).
FIG. 6.
PBS dissociation products separated on continuous sucrose gradients (0 to 30%) from Synechococcus sp. strain WH8102 acclimated at several irradiances (A) (irradiances in micromoles of photons per square meter per second) and absorption spectra normalized at 495 nm of dissociated PBS fractions from Synechococcus sp. strain WH8102 acclimated at 15 μmol of photons · m−2 · s−1 (B).
On IEF gels, the orange fractions C and D presented identical patterns and got separated into two orange bands, corresponding to bands 1 and 3 in Fig. 5. These two bands exhibited identical spectral properties (Fig. 7B). Room temperature fluorescence emission spectra with excitation at 495 nm showed a single maximum at 566 nm, whereas absorption spectra showed two maxima at 494 nm (PUB) and 546 nm (PEB). These results confirm that these orange fractions contained nearly pure PEII, with a PUB/PEB ratio of 2.5 ± 0.06, very similar to that previously described for the PEII from Synechococcus sp. strain WH8103 (29).
Fractions A and B from the sucrose gradient displayed patterns of bands similar to those in Fig. 5 but with a reduced contribution of bands 1 and 3 (data not shown). Identification of band 4 as PEI was confirmed by its absorption properties, with a PUB maximum at 493 nm and a shouldered PEB peak at 562 nm, and its fluorescence emission maximum at 574 nm (Fig. 7C). Due to the weak amount of PC (band 2 in Fig. 5) and its comigration with PEII (band 3b in Fig. 5), we could not get a pure PC fraction. Indeed, the fluorescence emission spectra with excitation at 495 nm (PUB) of band 2 showed a single maximum at 566 nm, typical of PEII (Fig. 7C). Nevertheless, excitation at 545 nm (PEB) and 620 nm (PCB) both yielded an emission maximum at 639 nm, typical of PC fluorescence (data not shown). This suggests that the PC of Synechococcus sp. strain WH8102 binds two types of chromophores, PEB and PCB, confirming its identification as R-PCII, as was expected from the close phylogenetic relatedness of this strain to the R-PCII-containing Synechococcus sp. strain WH8103 (30).
The optical properties of band 5 confirmed that it was APC (Fig. 7D). It is known that APC organization may change depending on the ionic environment and/or the APC concentration itself (45). When concentrated enough (Amax > 0.1), APC was mostly in the trimeric form (αβ)3 and its absorption and fluorescence emission maxima were 651 and 661 nm, respectively (Fig. 7D).
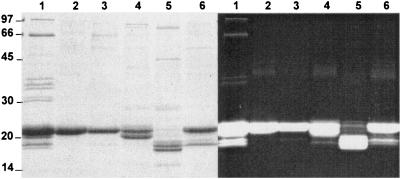
An LiDS-PAGE analysis of these PBP-enriched fractions revealed that most linker polypeptides were lost during the purification procedure (compare lane 1 and lanes 2 through 6 in Fig. 8). However, a thin band was found at 66 kDa in lane 3 and possibly lane 4, corresponding to PEII band b and PEI, respectively. This band is indicative of the presence of the SYNW2000 linker, though at too low a concentration to exhibit a clear green fluorescence under UV light. The presence of SYNW2000 in these two fractions is consistent with the hypothesis that this novel linker would be associated with both PEs, as discussed below.
FIG. 8.
LiDS-PAGE gel (acrylamide gradient, 10 to 20%) of PBP-enriched fractions from a Synechococcus sp. strain WH8102 culture grown at 15 μmol of photons · m−2 · s−1. Lanes: 1, whole PBSs; 2, PEII band a; 3, PEII band b; 4, PEI; 5, APC; 6, PC. (Left) Coomassie staining. (Right) Same gel under UV light in the presence of 20 mM zinc acetate.
The PEII α and β subunits comigrated at ca. 22 kDa (Fig. 8, lanes 2 and 3), although their theoretical molecular masses without chromophores are 17.7 and 18.1 kDa, respectively. In contrast, the PEI α and β subunits (theoretical molecular masses = 17.8 and 19.1 kDa, respectively) were separated as 20- and 22-kDa bands, respectively (Fig. 8, lane 4). The PC α and β subunits comigrated and could not be separated from the contaminating PEII α and β subunits (Fig. 8, lane 6). In this PC-enriched fraction, there was also an orange band (fluorescing green under UV light) at 19 kDa which was not present in the PEII fraction. The nature of this PUB-chromophorylated polypeptide apparently associated with PC remains unclear. The APC α and β subunits migrated as two well-separated bands (blue under visible light and red fluorescing under UV light) at about 18 and 19 kDa (Fig. 8, lane 5) despite similar theoretical molecular masses (17.4 and 17.5 kDa, respectively).
DISCUSSION
The genomic analysis of PBS genes revealed that most PE- and PC-associated genes, as well as those involved in the biosynthesis or binding of phycobilins, are gathered into a large cluster covering more than 28 kb (Fig. 1, top panel). This specialized region is significantly more extended than the 15-kb “phycobiliprotein rod gene cluster” studied in the marine Synechococcus sp. strain WH8020 by Wilbanks and Glazer (48). The latter corresponds to the region from mpeB to SYNW2026 in WH8102 (Fig. 1, bottom panel). The nature and order of genes within these homologous regions are fairly well conserved between the two marine Synechococcus strains, although they belong to distinct genetic clades (28) and WH8020 but not WH8102 is thought to be capable of a special type of chromatic adaptation (CA group IV [32, 38]). Homologs of three genes with unknown functions in WH8102 (SYNW2015, SYNW2019, and SYNW2024) are found at the same positions in WH8020 (ORF140, ORF200, and ORF196), pointing out their probable roles in phycobilisome biosynthesis, structure, or regulation. Both genomic regions end with a phosphatase gene (SYNW2026 in WH8102, ORF65 in WH8020), with the phosphatase perhaps having a role in the phosphorylation/dephosphorylation of PBS proteins under some yet-unknown conditions, such as, e.g., during photoacclimation (48). There are only two notable differences between the two homologous gene regions. First, the putative bilin attachment enzyme gene mpeV found upstream of cpeBA in WH8020 is absent in WH8102. Since it is found nowhere else in this genome, this gene, a paralog of mpeU, is likely not critical for PBS assembly. Second, the rpcEF operon, encoding a putative phycoerythrobilin lyase in WH8020 (8), is replaced in WH8102 by a single fusion gene (SYNW2025). Consequently, its product is an unusual phycobilin lyase consisting of a single unit, instead of two subunits as in all other lyases of the CpcE/F, RpcE/F, and PecE/F families (9, 10, 40, 44). Further characterization of this enzyme is needed in order to determine (i) its PBP specificity (PC, PEI, or PEII only or several of these) and (ii) if it is PEI and/or PEII specific, whether it is a mere PEB lyase or rather a PEB lyase/isomerase, i.e., an enzyme which would bind a PEB molecule and concomitantly isomerize it to PUB (40).
Genes encoding the two novel linkers that were studied in more detail are both found in the region upstream of mpeB (Fig. 1). While the first one (SYNW1989) is isolated from the rest of the region by a number of hypothetical or conserved hypothetical genes, the second one is included in a gene cluster which shows high homology to the cpeCDESTR cluster in F. diplosiphon (7). However, a major difference between these two clusters is that in WH8102, the cpeD gene is replaced by a much larger gene (SYNW2000). Sequence comparison suggests that this new linker gene resulted from the fusion of an mpeC paralog and a cpeD-like linker gene (no other cpeD-like gene is found elsewhere in the WH8102 genome). Indeed the N-terminal part of the protein sequence derived from the SYNW2000 gene shares several of the specific features previously reported for MpeC from Synechococcus sp. strain WH8020 (49). This includes an N-terminal extension (ca. 48 residues in MpeC and ca. 43 residues in SYNW2000) relative to other types of PE γ linkers, such as CpeC. Furthermore, SYNW2000 shares with MpeC several conserved cysteinyl residues not found in non-bilin-bearing γ linkers (Fig. 2). It has been shown experimentally in the MpeC of Synechococcus sp. strain WH8020 that only Cys-49 would be a PUB attachment site (49), but we lack direct experimental evidence to confirm if this is also the case in WH8102. A third conserved cysteine (i.e., a potential phycobilin attachment site) is found in WH8102 MpeC, SYNW2000, and SYNW1989 (at positions 99, 104, and 118, respectively) but is missing in WH8020 MpeC (Fig. 2). Further analyses, e.g., using reverse-phase high-performance liquid chromatography analysis of this linker after in-solution digestion, are needed to check whether this additional cysteinyl residue may also bind a chromophore.
Given the higher PUB/PEB ratio of WH8102 compared to that of WH8020 (1.9 ± 0.06 versus 0.8 ± 0.03) and the fact that the γ subunits of red algal R-PEs (22) and B-PEs (15) have been shown to carry multiple bilins, the hypothesis that in WH8102 one or several of the MpeC paralogs (including the SYNW2000 N terminus) possess more than one chromophore is plausible. In contrast, no conserved cysteinyl residues are found in the CpeD-like part of the SYNW2000 protein, consistent with the fact that in freshwater PE-containing cyanobacteria CpeD is nonchromophorylated.
SYNW1989 also possesses an N-terminal extension compared to other types of γ linkers, even longer than in MpeC (Fig. 2). In both proteins, this extension is compensated by a shorter C terminus (49 and data not shown). The special features of this novel linker (similarity to MpeC, chromophorylation with PUB) strongly suggest that it is associated with PEII. We therefore propose to designate it MpeE (Table 1). Similarly, the hybrid structure of SYNW2000, made up of a PEI linker polypeptide and of a PEII linker homolog, suggests that in the rods it is linked to both of these PBPs. It is reasonable to assume that its C terminus is located in the central channel of the most core-distal PEI trimer and its N terminus is located in the channel of the most core-proximal PEII trimer. To differentiate it from the CpeD linker protein found in other cyanobacteria such as F. diplosiphon (7), but also to acknowledge its many similarities with it, including the fact that its gene is located in a region between the two other PE linker genes cpeC and cpeE, we propose to designate this novel linker MpeD (Table 1). It is worth noting that a chromophorylated linker with a similar apparent molecular mass on LiDS-PAGE gels (ca. 68 kDa) is found in several other marine Synechococcus clones, including WH7803 (Fig. 4), as well as in WH8020 and WH8103 (data not shown). Since WH8020, WH7803, and WH8102/WH8103 belong to three distinct clades (28), this previously unknown linker appears to be widespread among marine, PEII-containing Synechococcus spp.
TABLE 1.
Phycobilisome linker polypeptides of Synechococcus sp. strain WH8102 as derived from genomic analysis and classified according to their predicted molecular masses (without phycobilins)a
| ORF no. | Predicted molecular mass (Da) | Chromo- phorylated | Gene | Proposed function of product |
|---|---|---|---|---|
| SYNW0486 | 104,989 | Yes | apcE | AP-LCM |
| SYNW2000 | 59,409 | Yes | mpeD | PEI/II-LR59 |
| SYNW1999 | 33,147 | No | cpeC | PEI-LR33.1 |
| SYNW1989 | 32,805 | Yes | mpeE | PEII-LR32.8 |
| SYNW2010 | 31,953 | Yes | mpeC | PEII-LR32 |
| SYNW0314 | 28,340 | No | cpcG1 | PC-LRC28.3 |
| SYNW1997 | 28,102 | No | cpcG2 | PC-LRC28.1 |
| SYNW2001 | 27,055 | No | cpeE | PEI-LR27 |
| SYNW0483 | 7,714 | No | apcC | AP-LC7.7 |
The two novel linkers are shown in bold letters. A function is proposed for each linker, following the international nomenclature for phycobiliproteins. The abbreviations AP, PEI, PEII, and PC indicate allophycocyanin, phycoerythrin I, phycoerythrin II, and phycocyanin, respectively. LLCM, LR, and LRC correspond to core-membrane linker, rod linker, and rod-core linker, respectively. The superscript numbers correspond to the molecular masses in kilodaltons.
Data derived from photoacclimation experiments (Fig. 4) allowed us to get some further insights into the localization of the different PE linkers within the rods. They show that the relative MpeC content rapidly decreases with growth irradiance to become undetectable at 250 μmol of photon · m−2 · s−1. This strongly suggests a more distal position compared to MpeD and MpeE.
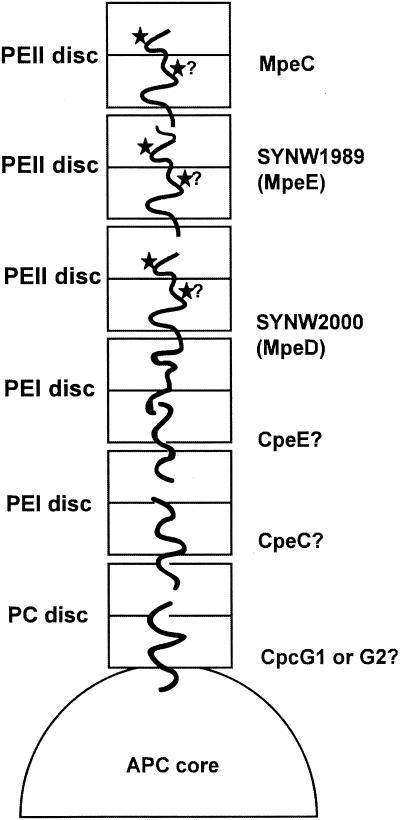
Figure 9 shows a possible model of the arrangement of linkers within the PBS rods of Synechococcus sp. strain WH8102. This model, which extends that proposed for the sole MpeC by Wilbanks and Glazer (49), rests on several assumptions: (i) that the morphology of WH8102 PBSs resembles that documented for freshwater cyanobacterial PBSs (see, e.g., reference 37), (ii) that all rods have homogeneous PBP and linker compositions, and (iii) that linkers do not overlap themselves within the rods. In this model, each rod is suggested to be composed of six disks at low light, namely three PEII disks, two PEI disks, and one PC disk. This proportion is compatible with our estimates of the relative content of PBPs within PBSs at 15 μmol of photons · m−2 · s−1 after separating them by IEF (data not shown). The presence of a single PC disk per rod can reasonably be assumed from the fact that no PC rod linkers (only rod-core linkers) are found in the genome (33 and this study). Three PEII disks are needed to fit the three Mpe linkers. The intermediate position of MpeE in between MpeC and MpeD was deduced from its structural characteristics and its much lower relative decrease at high light than that of MpeC. Unfortunately, although the three-dimensional structures of a number of PBPs have been resolved, including the PUB-containing R-PE from the red alga Griffithsia monilis (35), the precise arrangement of γ linkers is still unclear since they cannot be seen in electron density maps. Furthermore, there is no direct visualization by transmission electron microscopy of whole PBS particles in marine Synechococcus spp. published to date and our own attempts to get such pictures for WH8102 and WH7803 have failed so far, possibly due to the instability of these particular PBSs. So whether rods can be as long as six disks at low light, as we assume in our model, cannot be ascertained yet. Our model also suggests that the most distal PEII linker polypeptide is MpeC since it is the most rapidly lost during acclimation to high light. It remains to be tested whether MpeE would not also be released at higher light levels than the ones tested here. We hypothesize that SYNW2000 (or MpeD) would firmly anchor the proximal PEII disk to the rest of the rod whereas the short C terminus of MpeC (and possibly MpeE as well) make it more susceptible to release/breakage during photoacclimation processes. This hypothesis could explain that while the PEII content per PBS is much lower at very high light compared to low light, some PEII is still found under the former conditions (Fig. 5).
FIG. 9.
Putative model of the localization of linker proteins in a phycobilisome rod of Synechococcus sp. strain WH8102, as deduced from sequence data and biochemical observations. The exact positions of CpeC and CpeE are not known and can be the reverse of that shown. Stars indicate the putative positions of PUB chromophores.
Given the uncertainties relating to the rod morphology mentioned above, other models for the arrangement of rods and linkers cannot be excluded. Indeed, it is possible that the rods are in fact more compact than we assumed, with short rods having a heterogeneous linker composition. Some rods might have one combination of three PE linkers (e.g., CpeC, MpeD, and MpeC), while others would have another combination (e.g., CpeE, MpeD, and MpeE). The presence of two rod-core linker types (CpcG1 and CpcG2) also might indicate a heterogeneous rod linker composition.
Acknowledgments
This work was supported by the EC program MARGENES (QLRT 2001-01226) and the French programs PROOF-BIOSOPE and PROOF-UVECO. C.S. is supported by a grant from the Ministère de la Recherche et des Nouvelles Technologies (France).
We thank Brian Palenik for useful comments on the manuscript.
REFERENCES
- 1.Allen, J. F., and H. C. P. Matthijs. 1997. Complementary adaptations, photosynthesis and phytochrome. Trends Plant Sci. 2:41-43. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkelman, T. R., and J. C. Lagarias. 1986. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal. Biochem. 156:194-201. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, C., A. L. Etienne, and J. C. Thomas. 1996. Synthesis and binding of phycoerythrin and its associated linkers to the phycobilisome in Rhodella violacea (Rhodophyta): compared effects of high light and translation inhibitors. J. Phycol. 32:265-271. [Google Scholar]
- 5.Bryant, D. A., V. L. Stirewalt, M. Glauser, G. Frank, W. Sidler, and H. Zuber. 1991. A small multigene family encodes the rod-core linker polypeptides of Anabaena sp. PCC7120 phycobilisomes. Gene 107:91-99. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, G. G., M. R. Schaefer, and A. R. Grossman. 1992. Complementation of a red-light-indifferent cyanobacterial mutant. Proc. Natl. Acad. Sci. USA 89:9415-9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobley, J. G., A. C. Clark, S. Weerasurya, F. A. Queseda, J. Y. Xiao, N. Bandrapali, I. D'Silva, M. Thounaojam, J. F. Oda, T. Sumiyoshi, and M. H. Chu. 2002. CpeR is an activator required for expression of the phycoerythrin operon (cpeBA) in the cyanobacterium Fremyella diplosiphon and is encoded in the phycoerythrin linker-polypeptide operon (cpeCDESTR). Mol. Microbiol. 44:1517-1531. [DOI] [PubMed] [Google Scholar]
- 8.de Lorimier, R., S. M. Wilbanks, and A. N. Glazer. 1993. Genes of the R-phycocyanin II locus of marine Synechococcus spp., and comparison of protein-chromophore interactions in phycocyanins differing in bilin composition. Plant Mol. Biol. 21:225-237. [DOI] [PubMed] [Google Scholar]
- 9.Fairchild, C. D., and A. N. Glazer. 1994. Oligomeric structure, enzyme kinetics, and substrate specificity of the phycocyanin alpha subunit phycocyanobilin lyase. J. Biol. Chem. 269:8686-8694. [PubMed] [Google Scholar]
- 10.Fairchild, C. D., J. Zhao, J. Zhou, S. E. Colson, D. A. Bryant, and A. N. Glazer. 1992. Phycocyanin alpha-subunit phycocyanobilin lyase. Proc. Natl. Acad. Sci. USA 89:7017-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Federspiel, N. A., and A. R. Grossman. 1990. Characterization of the light-regulated operon encoding the phycoerythrin-associated linker proteins from the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 172:4072-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Federspiel, N. A., and L. Scott. 1992. Characterization of a light-regulated gene encoding a new phycoerythrin-associated linker protein from the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 174:5994-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller, N. J., D. Marie, F. Partensky, D. Vaulot, A. F. Post, and D. J. Scanlan. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69:2430-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glazer, A. N. 1989. Light guides—directional energy transfer in a photosynthetic antenna. J. Biol. Chem. 264:1-4. [PubMed] [Google Scholar]
- 15.Glazer, A. N. 1977. Structure and molecular organization of the photosynthetic accessory pigments of cyanobacteria and red algae. Mol. Cell. Biochem. 18:125-140. [DOI] [PubMed] [Google Scholar]
- 16.Glazer, A. N., R. C. Williams, S. W. Yeh, and J. H. Clark. 1985. Kinetics of energy flow in the phycobilisome core. Science 230:1051-1053. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Lojero, C., B. Perez-Gomez, G. Shen, W. M. Schluchter, and D. A. Bryant. 2003. Interaction of ferredoxin:NADP(+) oxidoreductase with phycobilisomes and phycobilisome substructures of the cyanobacterium Synechococcus sp. strain PCC 7002. Biochemistry 42:13800-13811. [DOI] [PubMed] [Google Scholar]
- 18.Grossman, A. R., M. R. Schaefer, G. G. Chiang, and J. L. Collier. 1993. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 57:725-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kana, T. M., and P. M. Glibert. 1987. Effect of irradiances up to 2000 μE m−2 s−1 on marine Synechococcus WH7803-I. Growth, pigmentation, and cell composition. Deep Sea Res. 34:479-485. [Google Scholar]
- 20.Kana, T. M., and P. M. Glibert. 1987. Effect of irradiances up to 2000 μE m−2 s−1 on marine Synechococcus WH7803 - II. Photosynthetic responses mechanisms. Deep-Sea Res. 34:497-516. [Google Scholar]
- 21.Kehoe, D. M., and A. R. Grossman. 1996. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273:1409-1412. [DOI] [PubMed] [Google Scholar]
- 22.Klotz, A., V., and A. N. Glazer. 1985. Characterization of the bilin attachment sites in R-phycoerythrin. J. Biol. Chem. 260:4856-4863. [PubMed] [Google Scholar]
- 23.Koller, K. P., and W. Wehrmeyer. 1975. B-phycoerythrin from Rhodella violacea. Archiv. Microbiol. 104:255-261. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.MacColl, R. 1998. Cyanobacterial phycobilisomes. J. Struct. Biol. 124:311-334. [DOI] [PubMed] [Google Scholar]
- 26.Miskiewicz, E., A. G. Ivanov, and N. P. A. Huner. 2002. Stoichiometry of the photosynthetic apparatus and phycobilisome structure of the cyanobacterium Plectonema boryanum UTEX 485 are regulated by both light and temperature. Plant Physiol. 130:1414-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomsawai, P., N. Tandeau de Marsac, J.-C. Thomas, M. Tanticharoen, and S. Cheevadhanarak. 1999. Light regulation of phycobilisome structure and gene expression in Spirulina platensis C1 (Arthrospira sp. PCC 9438). Plant Cell Physiol. 40:1194-1202. [Google Scholar]
- 28.Noubir, S., I. Luque, J. de Alda, I. Perewoska, N. Tandeau de Marsac, J. G. Cobley, and J. Houmard. 2002. Co-ordinated expression of phycobiliprotein operons in the chromatically adapting cyanobacterium Calothrix PCC 7601: a role for RcaD and RcaG. Mol. Microbiol. 43:749-762. [DOI] [PubMed] [Google Scholar]
- 29.Ong, L. J., and A. N. Glazer. 1991. Phycoerythrins of marine unicellular cyanobacteria. I. Bilin types and locations and energy transfer pathways in Synechococcus spp. phycoerythrins. J. Biol. Chem. 266:9515-9527. [PubMed] [Google Scholar]
- 30.Ong, L. J., and A. N. Glazer. 1987. R-phycocyanin II, a new phycocyanin occurring in marine Synechococcus species. J. Biol. Chem. 262:6323-6327. [PubMed] [Google Scholar]
- 31.Ong, L. J., A. N. Glazer, and J. B. Waterbury. 1984. An unusual phycoerythrin from a marine cyanobacterium. Science 224:80-83. [DOI] [PubMed] [Google Scholar]
- 32.Palenik, B. 2001. Chromatic adaptation in marine Synechococcus strains. Appl. Environ. Microbiol. 67:991-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 34.Rippka, R., T. Coursin, W. Hess, C. Lichtlé, D. J. Scanlan, K. A. Palinska, I. Iteman, F. Partensky, J. Houmard, and M. Herdman. 2000. Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll a2/b2-containing cyanobacterium (Oxyphotobacteria). Int. J. Syst. Evol. Microbiol. 50:1833-1847. [DOI] [PubMed] [Google Scholar]
- 35.Ritter, S., R. G. Hiller, P. M. Wrench, W. Welte, and K. Diederichs. 1999. Crystal structure of a phycourobilin-containing phycoerythrin at 1.90-angstrom resolution. J. Struct. Biol. 126:86-97. [DOI] [PubMed] [Google Scholar]
- 36.Ritz, M., J. C. Thomas, A. Spilar, and A. L. Etienne. 2000. Kinetics of photoacclimation in response to a shift to high light of the red alga Rhodella violacea adapted to low irradiance. Plant Physiol. 123:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sidler, W. A. 1994. Phycobilisome and phycobiliprotein structure, p. 139-216. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 38.Six, C., J. C. Thomas, B. Brahamsha, Y. Lemoine, and F. Partensky. 2004. Photophysiology of the marine cyanobacterium Synechococcus sp. WH8102, a new model organism. Aquat. Microb. Ecol. 35:17-29. [Google Scholar]
- 39.Sobczyk, A., G. Schyns, N. Tandeau de Marsac, and J. Houmard. 1993. Transduction of the light signal during complementary chromatic adaptation in the cyanobacterium Calothrix sp. PCC 7601: DNA-binding proteins and modulation by phosphorylation. EMBO J. 12:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storf, M., A. Parbel, M. Meyer, B. Strohmann, H. Scheer, M. G. Deng, M. Zheng, M. Zhou, and K. H. Zhao. 2001. Chromophore attachment to biliproteins: specificity of PecE/PecF, a lyase-isomerase for the photoactive 3(1)-Cys-alpha 84-phycoviolobilin chromophore of phycoerythrocyanin. Biochemistry 40:12444-12456. [DOI] [PubMed] [Google Scholar]
- 41.Tandeau de Marsac, N., and J. Houmard. 1988. Complementary chromatic adaptation: physiological conditions and action spectra. Methods Enzymol. 167:318-328. [Google Scholar]
- 42.Thomas, J. C., and C. Passaquet. 1999. Characterization of a phycoerythrin without alpha-subunits from a unicellular red alga. J. Biol. Chem. 274:2472-2482. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tooley, A. J., and A. N. Glazer. 2002. Biosynthesis of the cyanobacterial light-harvesting polypeptide phycoerythrocyanin holo-alpha subunit in a heterologous host. J. Bacteriol. 184:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trinquet, E., F. Maurin, M. Preaudat, and G. Mathis. 2001. Allophycocyanin 1 as a near-infrared fluorescent tracer: isolation, characterization, chemical modification, and use in a homogeneous fluorescence resonance energy transfer system. Anal. Biochem. 296:232-244. [DOI] [PubMed] [Google Scholar]
- 46.Waterbury, J. B., S. W. Watson, F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can. Bull. Fish. Aquat. Sci. 472:71-120. [Google Scholar]
- 47.Wilbanks, S. M., R. de Lorimier, and A. N. Glazer. 1991. Phycoerythrins of marine unicellular cyanobacteria. III. Sequence of a class II phycoerythrin. J. Biol. Chem. 266:9535-9539. [PubMed] [Google Scholar]
- 48.Wilbanks, S. M., and A. N. Glazer. 1993. Rod structure of a phycoerythrin II-containing phycobilisome. I. Organization and sequence of the gene cluster encoding the major phycobiliprotein rod components in the genome of marine Synechococcus sp. WH8020. J. Biol. Chem. 268:1226-1235. [PubMed] [Google Scholar]
- 49.Wilbanks, S. M., and A. N. Glazer. 1993. Rod structure of a phycoerythrin II-containing phycobilisome. II. Complete sequence and bilin attachment site of a phycoerythrin γ subunit. J. Biol. Chem. 268:1236-1241. [PubMed] [Google Scholar]