Abstract
The global anaerobic regulator FNR is a DNA binding protein that activates transcription of genes required for anaerobic metabolism in Escherichia coli through interactions with RNA polymerase (RNAP). Alanine-scanning mutagenesis of FNR amino acid residues 181 to 193 of FNR was utilized to determine which amino acid side chains are required for transcription of both class II and class I promoters. In vivo assays of FNR function demonstrated that a core of residues (F181, R184, S187, and R189) was required for efficient activation of class II promoters, while at a class I promoter, FF(−61.5), only S187 and R189 were critical for FNR activation. Site-directed mutagenesis of positions 184, 187, and 189 revealed that the positive charge contributes to the function of the side chain at positions 184 and 189 while the serine hydroxyl is critical for the function of position 187. Subsequent analysis of the carboxy-terminal domain of the α subunit (αCTD) of RNAP, using an alanine library in single copy, revealed that in addition to previously characterized side chains (D305, R317, and L318), E286 and E288 contributed to FNR activation of both class II and class I promoters, suggesting that αCTD region 285 to 288 also participates in activation by FNR. In conclusion, this study demonstrates that multiple side chains within region 181 to 192 are required for FNR activation and the surface of αCTD required for FNR activation is more extensive than previously observed.
A common mechanism of activation by transcription factors employs a series of macromolecular interactions, from binding DNA to multiple contacts with RNA polymerase (RNAP), which allows target promoters to overcome intrinsic defects in transcription initiation (21). This study focused on the Escherichia coli global anaerobic regulator FNR and the protein requirements essential for transcription activation. In particular, this study further investigated the residues of FNR that have been proposed to interact with the α subunit of RNAP.
FNR, which regulates transcription in response to O2 deprivation, belongs to a family of transcriptional regulators related to the cyclic AMP receptor protein, CRP (13). In its active form, FNR is a homodimer (16) which protects an ∼22-bp sequence in DNase I footprinting experiments (9, 14). The vast majority of FNR-activated promoters contain an FNR binding site centered approximately 41.5 bp upstream of the transcriptional start site and are termed class II promoters (Fig. 1A) (6). At this position, FNR is able to make multiple contacts with RNAP through three domains: activating region 1 (AR1), AR2, and AR3 (3, 5, 14, 15, 31, 33, 34). FNR-AR3 is active in the downstream subunit (3) and is likely to contact the σ70 subunit of RNAP (23). FNR-AR2, which plays a minor role in activation, is also active in the downstream subunit (5), but its interaction partner is proposed to be the amino-terminal domain of the α subunit of RNAP (5). FNR-AR1 is active in the upstream subunit (3, 33) and is proposed to interact with the carboxy-terminal domain of the α subunit (αCTD) of RNAP (32). In contrast, at a class I promoter, the FNR binding site is located further upstream, centered at 61.5 or 71.5 bp upstream of the transcription start site (Fig. 1A) (34), although few naturally occurring class I promoters exist. At the class I position, FNR appears to make only one contact with RNAP through interaction of FNR-AR1 with the αCTD of RNAP (19, 32). Thus, FNR-AR1 is critical to activation of both class I and class II FNR-dependent promoters.
FIG. 1.
(A) Model of FNR-activated promoters. At class I FNR-dependent promoters (FNR binding site at −61.5 in this example), FNR is proposed to contact αCTD through AR1 in the downstream subunit (filled circle) (3, 32). At class II FNR-dependent promoters (FNR binding site at −41.5), FNR is in position to make multiple contacts with RNAP through three activating regions (AR1, AR2, and AR3; filled circles) (3, 5, 15). (B) Model of FNR-AR1. The ribbon diagram of the CRP monomer (30) was modified by extending the N terminus to include the 29-amino-acid extension (thin line) of FNR (17), which contains three of the four cysteines required for ligation of the O2-sensitive [4Fe-4S]2+ cluster. The locations of residues that constitute AR1 of FNR are marked with arrows. FNR residues 181 to 193, which are the focus of this study, are highlighted with an asterisk.
From characterization of FNR mutants defective in transcription of class I promoters, FNR-AR1 was localized to three predicted surface-exposed loops on one face of FNR (residues 71 to 75, 116 to 121, and 181 to 191) (Fig. 1B) (3, 20, 32). This FNR-AR1 surface is predicted to be larger than the AR1 of CRP (analogous to FNR residues 181 to 191) observed in the cocrystal structure of CRP with αCTD (4), indicating some unique aspects of the interaction of FNR-AR1 with αCTD. To identify the site of interaction of FNR-AR1 within the α subunit, Lee et al. (19) screened an alanine library of αCTD and found that five residues (D305, G315, R317, L318, and E319) were important for transcription activation of FF(−71.5). While two of the residues (G315 and R317) are part of the same region shown to contact CRP (4, 27, 28), D305 is outside of this region. In addition, suppression genetic studies to determine the interacting partners between FNR-AR1 and αCTD (19) suggest that AR1 residue FNR-T118, which is predicted to be outside the region equivalent to CRP-AR1, targets D305 of αCTD, consistent with the proposal that a larger surface of both FNR and αCTD interact. In contrast, residue FNR-S187, which is predicted to be in a region analogous to CRP-AR1, appears to interact with αCTD-R317 and αCTD-L318. In the CRP-αCTD model (4), αCTD residue R317 appears to be in close proximity to the C terminus of CRP (R209), which is equivalent to D236 in FNR. Furthermore, while αCTD residues 285 to 289 make direct side chain interactions with the CRP region analogous to FNR residues 184 to 192, these αCTD residues were not found to be required for FNR activation of a class I promoter (19). These differences suggest that not only is the surface of AR1 larger in FNR but the interactions required for activation by FNR-AR1 are different than in CRP. Thus, to enhance our understanding of the interaction between the FNR-AR1 determinant and αCTD, we chose to further study FNR-AR1 region 181-193, which is well conserved among other members of the FNR family and encompasses the region in CRP that directly contacts αCTD (4, 13). While random mutagenesis studies previously indicated that various residues in FNR region 181-191 are important for transcription activation (32), only the side chain of Ser187 had been shown to be specifically required by replacing it with an alanine residue (32, 33).
The goal of our study was to further characterize FNR-AR1 residues 181-193 and the αCTD subunit of RNAP by utilizing alanine scanning and site-directed mutagenesis. In vivo assays of FNR function at three different class II promoters, FF(−41.5) (33), PdmsA (14), and PnarG (14), and a class I promoter, FF(−61.5) (32), were used to investigate which amino acid side chains of FNR-AR1 region 181-193 and αCTD are specifically required for FNR-dependent activation. The results of these studies increased our understanding of the flexibility in the requirement of side chains within FNR-AR1, provided additional perspective on the determinants within αCTD required for FNR activation, and allowed for further comparison between FNR and CRP.
MATERIALS AND METHODS
Strain construction.
Site-directed mutagenesis of pRZ7411 (Cmr, a pACYC184derivative containing sequence from −521 to + 1115 of fnr) (15) and pPK823 (Apr, +1 to + 1115 of fnr in pET11a) (15) were carried out by using the QuikChange Site-Directed Mutagenesis kit (Stratagene) to create fnr alanine mutations from residues 181 to 193. Mutations were confirmed by DNA sequencing performed at the University of Wisconsin Biotechnology Center. Plasmids were transformed into strains containing null alleles of fnr (Δfnr::Spr) and chromosomal lacZ fusions to various FNR-regulated genes (PK3293 [dmsA′-lacZ] [14], PK7024 [FF(−41.5)-lacZ], RZ8480 [narG′-lacZ] [14], PK3289 [ndh′-lacZ] [15], and PK7028 [FF(−61.5)-lacZ]) for measurement of in vivo β-galactosidase activity. PK7024 and PK7028 were created by replacing the chromosomal lac promoter with either FF(−41.5) (−80 to + 40 relative to the transcriptional start site) or FF(−61.5) (−103 to + 40 relative to the transcriptional start site) (32, 33) and an adjacent upstream Knr cassette by recombination as previously described (12).
Chromosomally encoded rpoA mutations were transferred to RZ4500 by P1 transduction, selecting for tetracycline resistance from the nearby zhc::Tn10 of WAM123 (1). To confirm the presence of the appropriate alanine codon in the chromosomal rpoA gene, the rpoA alleles from the transductants were sequenced from PCR-amplified chromosomal DNA using primers 1413 (5′-TTCTGATCTGTCTGCGGACATTAAC-3′) and 1414 (5′-ATATTGCGGAACATAGCCTGGC-3′) (1). Strains containing the correct alanine substitution encoded in rpoA were then P1 transduced with lysates carrying FF(−41.5) or FF(−61.5), with selection for kanamycin resistance.
β-Galactosidase assays.
Assays of β-galactosidase from whole cells were carried out as described previously (14, 15). Activity was measured by the method of Miller (24) from strains grown anaerobically to an A600 of 0.3 to 0.5 in M9 minimal medium containing 0.2% (wt/vol) glucose, 20 μg of chloramphenicol/ml, 10 μM ferric ammonium citrate, and 2 μM ammonium molybdate.
Western blots.
Cells containing either wild-type (WT) FNR or mutant FNR encoded on pACYC184 (7) were grown anaerobically to an A600 of ∼0.3. Aliquots (125 μl) of cells were collected and resuspended in a total volume of 10 μl of 75 mM Tris (pH 6.8), 10% glycerol, 2% sodium dodecyl sulfate, 0.01% bromophenol blue, and 2.2 M β-mercaptoethanol and heated at 95°C for 10 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto nitrocellulose membranes (Fisher Scientific). FNR protein was detected by using antisera raised against FNR as a primary antibody and a fluorescein-conjugated secondary antibody (BD-Pharmigen). Proteins were then detected with a fluoroimager (FMBIOII; Hitachi).
Protein purification.
WT FNR or FNR-RA184 proteins were purified anaerobically from PK22 containing WT fnr or fnr-RA184 under control of T7 RNAP on the pET11a-derived plasmid pPK824 (WT FNR) or pPK7234 (FNR-RA184) and stored as described previously (29). FNR protein concentration was determined with the Coomassie Plus Protein Reagent (Pierce) with bovine serum albumin as the protein standard. The occupancy of the protein (defined as the percentage of the FNR molecules containing a [4Fe-4S]2+ cluster) was determined by Fe2+ and S2− analysis (29). RNAP holoenzyme was purchased from Epicentre Technologies.
In vitro transcription.
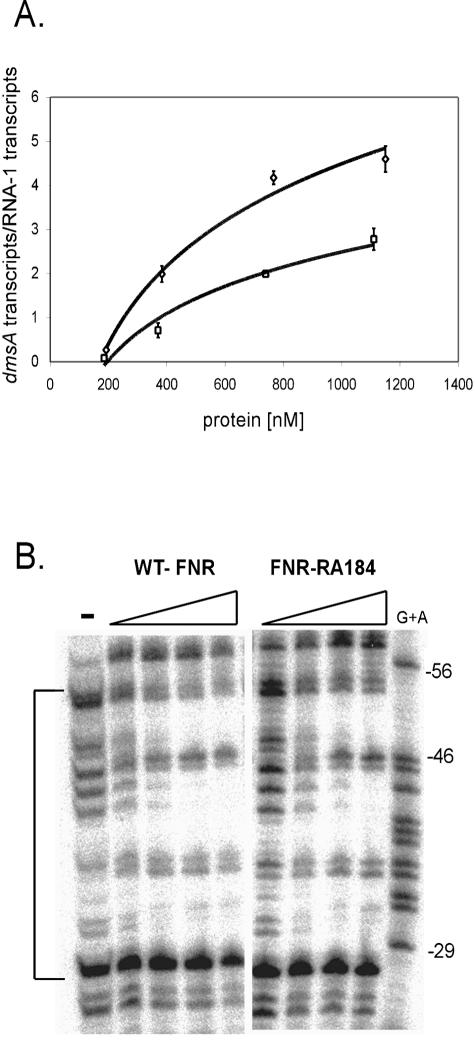
Single-round in vitro transcription assays were performed anaerobically (29) using a supercoiled DNA template carrying the dmsA promoter (pPK1034) and the FNR-independent control promoter RNA-1 (8). The assays were performed in a final volume of 16 μl using 2 nM DNA, 50 nM holo-RNAP, and increasing amounts of FNR (WT or RA184 as required) in 40 mM Tris-HCl (pH 7.9)-30 mM KCl-10 mM MgCl2-1 mM dithiothreitol-1% (vol/vol) glycerol-50 μg of bovine serum albumin/ml. Template DNA was preincubated for 10 min with FNR at 37°C. RNAP was then added, and the mixture was incubated for an additional 5 min. Transcription was initiated by addition of 2.5 μCi of [α-32P]UTP (3,000 Ci/mmol), unlabeled UTP (6.25 μM), ATP, CTP, and GTP to a final concentration of 250 μM, and heparin to a final concentration of 100 μg/ml. After 5 min, reactions were terminated by adding 8 μl of a 95% formamide-10 mM NaOH-0.05% bromophenol blue-0.05% xylene cyanol FF solution (U.S. Biochemicals). Samples were heated at 90°C for 2 min and analyzed by denaturing gel electrophoresis. The RNA transcripts were quantified by using a Molecular Dynamics PhosphorImager and ImageQuant software and plotted as a ratio of dmsA transcripts to RNA-1 transcripts.
Footprinting assays.
DNA fragments carrying the dmsA promoter region (from + 40 to −419 relative to the transcriptional start site) were isolated as previously described (14). Klenow fragment (New England Biolabs) was used to radiolabel the BamHI end of the fragment with [α-32P]dGTP (∼4,000 Ci/mmol) (14). Footprinting assays were performed anaerobically in a Coy chamber (Coy Laboratory Products Inc., Ann Arbor, Mich.) in a total volume of 20 μl by incubating 1 nM DNA and 120 to 720 nM FNR for 15 min at 37°C in 40 mM Tris-HCl (pH 7.9)-30 mM KCl-1 mM dithiothreitol-1% glycerol (vol/vol)-50 μg of bovine serum albumin/ml, followed by the addition of DNase I (1.6 μg/ml) and 10 mM MgCl2 for 30 s. DNase I reactions were terminated by the addition of 80 μl of 3 mM sodium acetate-25 mM EDTA. Reaction mixtures were then ethanol precipitated at room temperature and resuspended in 6 μl of 8 M urea-0.5× Tris-borate-EDTA-0.05% bromophenol blue-0.05% xylene cyanol. Samples were electrophoresed for 3 h at 1,400 V in 0.5× Tris-borate-EDTA buffer and visualized with a Molecular Dynamics PhosphorImager.
RESULTS
Several amino acid side chains in addition to S187 are required for FNR-AR1 function at class II promoters in vivo.
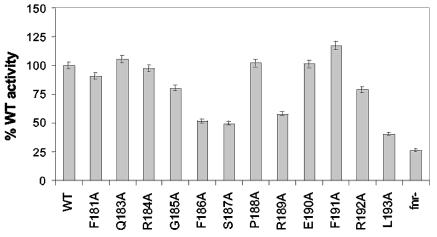
To further assess the role of each amino acid side chain within the FNR-AR1 loop region 181-193 in transcription activation, we determined the effect of alanine substitutions on expression of a class II FNR-dependent promoter, FF(−41.5) (Fig. 2A). This semisynthetic promoter was used in previous studies of FNR-AR1 (33) and thus allowed a direct comparison to previously published data. FNR function was assayed by measuring β-galactosidase expression from cells grown under anaerobic conditions containing an FF(−41.5)-lacZ fusion. In agreement with previous observations (33), FNR-SA187 showed a decrease in transcription of the class II FF(−41.5) promoter (55% of the β-galactosidase activity in strains containing WT FNR). However, in addition to position 187, alanine substitutions at positions F181, R184, G185, F186, R189, R192, and L193 also led to a greater than 25% loss in activity compared to WT FNR (Fig. 2A). Of all the residues tested, FNR-RA184, and not the previously characterized FNR-SA187, caused the largest reduction in activity (38% of WT FNR activity).
FIG. 2.
Activity of FNR-AR1 alanine mutants. β-Galactosidase activities from three independent isolates of strains carrying WT FNR or mutant FNR encoded on pACYC184 were measured. All strains were grown under anaerobic conditions in glucose-minimal medium. β-Galactosidase activities are normalized to WT FNR activity (100%). (A) Expression from FF(−41.5)′-lacZ reporter strain (average activity of WT FNR, ∼3,300 Miller units; activity from FNR− strain, ∼25 Miller units). (B) Expression from dmsA′-lacZ reporter strain (average activity of WT FNR, ∼2,400 Miller units; activity from FNR− strain, ∼21 Miller units). (C) Expression from narG′-lacZ reporter strain (average activity of WT FNR, ∼70 Miller units; activity from FNR− strain, ∼2 Miller units). (D) Expression from ndh′-lacZ reporter strain (average activity of WT FNR, ∼8.5 Miller units; activity from FNR− strain, ∼55 Miller units). The x axes are the same for panels A through D.
FIG. 3.
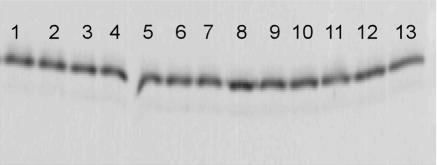
Western blot analysis of strains containing FNR-AR1 alanine mutant proteins. The amount of FNR present in strains containing the following mutant or WT protein encoded on pACYC184 was determined by analyzing equivalent cell numbers standardized by culture optical density at 600 nm: lane 1, WT FNR; lane 2, FNR-FA181; lane 3, FNR-QA183; lane 4, FNR-RA184; lane 5, FNR-GA185; lane 6, FNR-FA186; lane 7, FNR-SA187; lane 8, FNR-PA188; lane 9, FNR-RA189; lane 10, FNR-EA190; lane 11, FNR-FA191; lane 12, FNR-RA192; lane 13, FNR-LA193.
To test whether these mutants had similar effects on transcription of naturally occurring class II promoters, we assayed expression of the well-characterized narG and dmsA promoters (14). Expression of PdmsA-lacZ was less sensitive to changes in FNR-AR1 than FF(−41.5) (Fig. 2B) since only five (FNR-FA181, FNR-RA184, FNR-SA187, FNR-RA189, and FNR-LA193) of the seven mutants that decreased FF(−41.5) expression showed a greater than 25% loss in activity at PdmsA. However, the positions (R184 and L193) that showed the largest decrease in expression of FF(−41.5) also showed the largest defect in expression of PdmsA. Surprisingly, the Q183A mutant was twice as active as WT FNR at PdmsA, suggesting that removal of the Gln side chain relieves an inhibitory interaction.
On the other hand, the narG promoter was more sensitive to changes in FNR-AR1 than FF(−41.5) (Fig. 2C). We assayed activity of PnarG in the absence of nitrate to eliminate contributions from other protein factors, such as NarL (14). In this case, a total of 10 mutants had at least a 25% reduction in WT FNR transcription activation, including the core residues that were defective at both FF(−41.5) and PdmsA. The two mutants with the greatest defect at PnarG were FNR-RA184 and -FA186, as alanine substitutions at these positions resulted in 23 and 13% of WT FNR activity, respectively.
The ability of the FNR-AR1 alanine mutants to repress Pndh was assessed to determine if any of the defects seen in transcription activation result from decreased DNA binding (14, 15). All but two mutants, FNR-FA191 and FNR-LA193, were able to repress Pndh at WT FNR levels (Fig. 2D). Since FNR-LA193 was a mutant that showed a decrease in activation at all three promoters tested, we directly tested whether this was a result of a DNA binding defect, rather than a specific repression defect (10). For ease of analysis, in vitro DNA binding was assayed using a well-characterized O2-stable mutant of FNR, FNR-DA154 (15, 17, 36). Analysis of the O2-stable variant of FNR-LA193 (FNR-LA193-DA154) demonstrated that FNR-LA193 had a 10-fold defect in binding DNA compared to FNR-DA154, indicating that its transcription defect results from a loss of DNA binding (data not shown). Protein levels for each alanine mutant were also measured to determine if changes in the amount of protein account for the loss of activation by some mutants. The levels of all 13 FNR-AR1 alanine mutant proteins were within 10% of WT FNR levels, ruling out the possibility that defects in transcription were due to changes in protein expression (Fig. 3). Thus, taken together, these data indicate that in addition to the previously described S187 residue (32, 33), a core of residues (F181, R184, and R189) was required for efficient activation by FNR at all three class II promoters tested.
FNR-RA184 is defective in transcription activation in vitro.
We chose to study the properties of FNR-RA184 in vitro, since this mutant exhibited one of the largest defects at all three promoters tested. To assess whether FNR-RA184 displayed similar transcription defects in vitro, we purified both the WT FNR protein and FNR-RA184 anaerobically to ensure that the O2-sensitive [4Fe-4S]2+ cluster remained intact (29). Transcription activation was measured by single-round in vitro transcription reactions performed anaerobically at a variety of FNR concentrations, utilizing PdmsA template DNA. In comparison with the WT FNR protein, FNR-RA184 exhibited an approximately twofold defect in transcription (Fig. 4A). To exclude the possibility that decreases in transcription activation were caused by defects in DNA binding by FNR-RA184, DNase I footprinting (14, 15) was performed anaerobically. Using the same protein concentrations as in the in vitro transcription assays, we observed no differences in DNA binding between WT FNR and FNR-RA184 (Fig. 4B). Thus, these in vitro results agree with the in vivo data, demonstrating that R184 has a specific role in transcription activation by FNR-AR1.
FIG. 4.
In vitro analysis of FNR-RA184. (A) Transcription activation of PdmsA by WT FNR (diamonds) and FNR-RA184 (squares). The corrected dmsA transcript levels were normalized by the amount of RNA-1 transcript obtained from triplicate assays containing RNAP (50 nM) and WT FNR (192, 383, 766, and 1,150 nM) or FNR-RA184 (185, 370, 739, and 1,110 nM). No observable dmsA transcripts were observed in the absence of FNR. (B) Footprinting of WT FNR and FNR-RA184 at PdmsA. The area of protection from DNase I by FNR is indicated by the brackets and is analogous to the FNR binding site located at −41.5. FNR proteins were included at increasing concentrations identical to those in the in vitro transcription assays.
The positive charge of R184 makes a minor contribution to transcription activation by FNR.
To evaluate the type of interactions that R184 contributes to transcription activation, we replaced the Arg with other side chains by site-directed mutagenesis and measured FNR-dependent transcription of the class II FF(−41.5) promoter (Fig. 5A). The side chain substitution resulting in the largest defect was the negatively charged Glu (25% of WT activity). However, replacement of Arg184 with residues lacking any charged side chains, Ile and Gln, resulted in only a small decrease in FNR activation (∼75% of WT FNR activity), and replacement with another positively charged residue, Lys, resulted in a twofold defect. Thus, these data suggest that the positive charge of Arg184 is not the most critical feature of this side chain and that polar and/or hydrophobic interactions may also be important. In addition, the Gln and Lys substitutions at position 184 showed normal repression of Pndh, whereas RE184 showed a minor decrease (15%) in expression compared to WT FNR (data not shown). Western blots demonstrated that FNR-RE184 is produced at similar levels to the WT (data not shown), suggesting then that this substitution may cause a small DNA-binding defect.
FIG. 5.
Probing the side chain requirements of FNR-R184, FNR-S187, and FNR-R189 at FF(−41.5). β-Galactosidase activities from three independent isolates of strains containing WT FNR or mutant FNR encoded on pACYC184 were measured and plotted as described for Fig. 2. (A) Activities of R184 mutants. (B) Activities of S187 mutants. (C) Activities of R189 mutants.
Residue 187 specifically requires the serine side chain to support transcription activation.
Previous studies indicated that FNR-S187 was an important residue within FNR-AR1 (32, 33). To determine whether the hydroxyl was critical to S187 function, we substituted several other residues at this position. All polar and nonpolar residues tested (Thr, Val, Gln, Glu, and Lys) had the same activity as alanine (Fig. 5B), demonstrating the importance of the OH group on the β carbon. The addition of a methyl group (Thr) interfered with activation, suggesting that the Ser side chain is critical at position 187. All derivatives of FNR-S187 repressed Pndh at WT FNR levels and produced protein similarly to WT FNR (data not shown), suggesting that these mutants are specifically defective in activation by FNR.
Residue 189 requires a positively charged side chain for its transcription activation function.
We also studied the side chain requirements at R189, since alanine substitution at this position resulted in a decrease in FNR activity at all three promoters tested. Substitution of a negatively charged Glu for Arg resulted in a 60% decrease in FNR-dependent activity, while substitution of Lys resulted in WT FNR levels of expression (Fig. 5C), suggesting that the positive charge at position 189 is critical for efficient activation. Both mutants were able to repress Pndh at WT FNR levels, suggesting that the defect in activation by FNR-RE189 was not a result of a loss in DNA binding (data not shown). Also, both R189 derivatives were produced at similar levels compared to WT FNR as determined by Western blot analysis (data not shown).
Arg184 is not required for activation of a class I promoter.
We also determined the effect of the AR1 alanine library on expression of the class I FF(−61.5) promoter. This promoter was used in the original analysis (32) to identify AR1 mutants. Mutants that showed greater than a 25% loss in activity included FNR-FA186, FNR-SA187, and FNR-R189A. Surprisingly, alanine substitutions at positions F181 and R184, which were part of the core of residues required for activation of class II promoters, showed little to no decrease in FNR activation (Fig. 6). Thus, these data indicate that for the FF promoter, the requirements for activation differ based on the location of the FNR site.
FIG. 6.
Activities of FNR-AR1 alanine mutants at the class I FF(−61.5) promoter. β-Galactosidase activities from three independent isolates of strains containing WT FNR or mutant FNR encoded on pACYC184 were measured and plotted as described for Fig. 2. Expression the from FF(−61.5)-lacZ strain yielded ∼ 90 Miller units in cells containing WT FNR and ∼ 20 Miller units in FNR− cells.
Identification of additional residues of αCTD required for FNR-dependent activation.
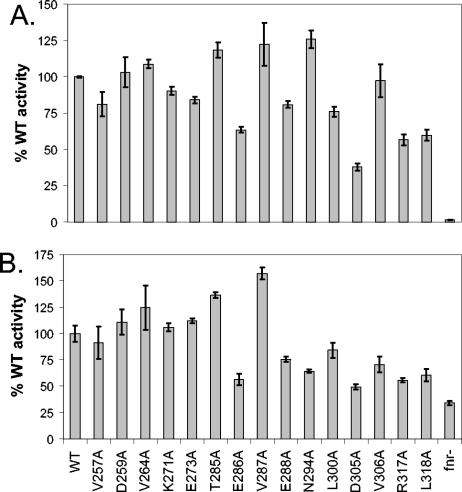
Previous studies of the requirements for side chains within αCTD necessary for FNR activity (19) were performed at the class I FF(−71.5) promoter, using αCTD mutants expressed from a plasmid in a strain containing WT αCTD on the chromosome since rpoA is an essential gene. In this study, we utilized a recently created library of 15 nonessential rpoA alanine mutations recombined into the chromosome (1) to investigate their effect on FNR transcription at the class II FF(−41.5) promoter or the class I FF(−61.5) promoter. The absence of a WT rpoA copy allows for greater sensitivity of this in vivo assay. For both the class II and class I promoters, we observed a reduction in FNR-dependent transcription with alanine substitutions of D305, R317, and L318, as was previously observed with FF(−71.5) (Fig. 7) (19). In addition, we found that αCTD-E286 and -E288 also appear to be important for FNR-dependent activation, since substitution of alanine results in a decrease in transcription activation of FF(−41.5) and FF(−61.5) (60 and 80% WT α activity, respectively) (Fig. 7). The defects in transcription observed with αCTD-E286 and -E288 appear to be specific for FNR activation, since in the absence of FNR, these αCTD mutants had no effect on transcription (data not shown). The other mutants demonstrating defects in our studies (αCTD-VA257, -NA294, -LA300, and -VA306) have previously been implicated in DNA binding by αCTD or interactions with the rest of RNAP (27, 28).
FIG. 7.
Effects of single alanine substitutions in αCTD on expression of different FNR-dependent promoters. (A) Expression of the FF(−41.5)-lacZ reporter strain. β-Galactosidase activities from three independent isolates of strains containing WT αCTD or mutant αCTD expressed in single copy from the chromosomal rpoA allele are shown. β-Galactosidase activities are expressed as percentages of WT αCTD activity (∼ 2,100 miller units). (B) Expression of the FF(−61.5)-lacZ reporter strain. β-Galactosidase activities from three independent isolates of strains containing WT αCTD or mutant αCTD expressed in single copy from the chromosomal rpoA allele are shown. β-Galactosidase activities are expressed as percentages of WT αCTD activity (∼ 78 miller units).
To investigate whether the two positively charged arginines within FNR-AR1 (R184 and R189) target the two negatively charged glutamates within αCTD (E286 and E288), we utilized a charge reversal approach. We substituted positively charged Lys and Arg at αCTD positions 286 and 288. While all four derivatives (αCTD-EK286, αCTD-ER286, αCTD-EK288, and αCTD-ER288) demonstrated a defect in FNR-dependent transcription, neither FNR-RE184 nor FNR-RE189 was able to restore activity to these αCTD mutants (data not shown). However, a negative result does not rule out these interactions since contacts between side chains may not be simple 1-to-1 interactions and substitution of opposite charges at these positions may cause steric hindrance by perturbing the local structure. Therefore, more work needs to be done to address the targets for interaction for both FNR-R184/R189 and αCTD-E286/E288 in mediating FNR-dependent transcription activation.
DISCUSSION
The results reported here extend previous studies (32, 33) by identifying additional residues in both FNR and the C-terminal domain of the α subunit of RNAP that participate in FNR-dependent transcription activation. These results also suggest that while a common core of residues participates in transcription activation, there is also a flexible array of determinants within FNR-AR1 whose importance varies among different FNR-dependent promoters.
A core of residues participates in transcription activation of class II promoters.
By examining the effects of FNR-AR1 alanine mutants on several class II promoters, we found that a core of side chains within AR1 region 181-192 (F181, R184, S187, and R189) is required for FNR activation of class II promoters. In the case of CRP, the core residues, identified by alanine scanning mutagenesis to participate in activation of class II promoters, are T158, P160, G162, and M163 (Fig. 8) (25, 35). Interestingly, there is no overlap in the residues between FNR-AR1 and CRP-AR1. In fact, the determinants required for FNR activation appear to be dispersed over a slightly larger region than those for CRP (Fig. 8).
FIG. 8.
Alignment of FNR-AR1 region 181-193 with the analogous region in CRP. The core of residues, determined by this work and others (25, 35), required for efficient activation of class II promoters by FNR and CRP are highlighted by asterisks
Requirements within FNR-AR1 are flexible.
In contrast to these core residues, the requirements for other side chains within FNR-AR1 at class II promoters appear to vary based on the promoter studied. For example, in the case of F186, replacement of Phe with Ala rendered FNR almost inactive (13% WT FNR activity) at the narG promoter; in contrast, at the dmsA promoter, FNR-F186A had no effect on activation. This suggests that some promoters require additional contacts to those mediated by the core residues.
The variability in the requirements for transcription activation within AR1 is also exhibited at the class I promoter FF(−61.5). Activation of this promoter by FNR is weak, as it is induced only fivefold in the presence of FNR in vivo and attempts to reconstitute transcription of this promoter in vitro have been unsuccessful (19). The positively charged R184 side chain, the most critical at a class II promoter, is not required for efficient activation of the class I FF(−61.5) promoter. Perhaps, R184 of FNR is unable to make contacts with RNAP from this upstream position, which could explain the low levels of activation by FNR at this promoter.
CRP-AR1 requirements do not appear to vary quantitatively between different class I promoters (35) since the patterns of transcriptional defects observed with alanine substitutions of T158, P160A, G162A, and M163 of CRP were similar for two different class I CRP-dependent promoters, CC (−61.5) and CC (−72.5). In contrast, analysis of the same set of CRP mutants at the class II CC(−41.5) promoter showed that while most of the same side chains are required for transcription activation, the pattern of the transcriptional defects differed in comparison to that of the class I promoters. In particular, CRP-GA162, rather than CRP-TA158, resulted in the largest defect in transcription and CRP-PA160 and CRP-MA163 showed larger defects than CRP-TA158. In addition, studies of two CRP-AR1 mutants (CRP-TA158 and CRP-HL159) showed that between class II promoters [CC(−41.5), pmelR, and pmelRcon] the pattern of defects varied unlike the effects of AR1 mutants at class I promoters (26). In summary, while the side chains of CRP-AR1 that contribute to activation of class II and class I promoters are similar, there are some quantitative differences in their effects.
Additional residues within αCTD contribute to activation by FNR.
By using rpoA mutations present in single copy in the chromosome, we discovered that two additional residues within the “287 determinant” of αCTD (27, 28), E286 and E288, in concert with previously implicated residues D305, R317, and L318 (19, 22), are required for FNR activation at both class I and class II promoters. These results indicate that mutant proteins expressed from the chromosomal rpoA locus provided a more sensitive measure of alanine substitution defects than alanine substitutions of rpoA expressed from a plasmid in an rpoA+ background; however, some positions that were previously shown to be defective in FNR activation (GA315 and DA319) could not be tested since our attempts to construct these derivatives in single copy were unsuccessful (data not shown). αCTD-K304A, shown previously to be more active than WT αCTD (19), was also not included in our analysis due to the presence of a second mutation in rpoA at position 265 (αCTD-RL265). These studies demonstrate that the requirements for αCTD at a class II FNR-dependent promoter are the same as the requirements at a class I FNR-dependent promoter. In addition to our finding that alanine substitutions at E286 and E288 appear to remove contacts necessary for activation, alanine substitutions at T285 and V287 appear to remove inhibitory contacts with FNR, suggesting that region 285-289 of αCTD is in close proximity to FNR. This result agrees with recent work (2) using an artificial chemical nuclease, which demonstrated that a single subunit of αCTD binds immediately upstream of FNR at a class II promoter in a position and orientation that would place αCTD region 285-288 in close proximity to FNR-AR1 region 184-192. Taken together, these data suggest that the region of αCTD required for activation by FNR and CRP overlap since CRP also requires region 285-288 of αCTD to activate transcription (27, 28) and that αCTD binds in a similar orientation relative to both FNR and CRP at a class II promoter (18).
Nevertheless, it is clear that some interactions between FNR-AR1 and αCTD show subtle differences from those of CRP-AR1 and αCTD. While in FNR, αCTD residues T285 and V287 play an inhibitory role, in the case of CRP these residues contribute to activation. Residues E286 and E288, which play a significant role in FNR activation, play a secondary role in activation by CRP. Minor differences in αCTD requirements for activation have also been observed with the CRP homolog CooA from Rhodospirillum rubrum (11). While CooA requires αCTD-V287 for efficient activation, other αCTD variants important for CRP-dependent transcription (T285A, E288A, and R317A) had little to no effect (11). Lastly, based on suppression genetic studies (19), it was postulated that S187 of FNR targets a region of αCTD in close proximity to αCTD-R317 and αCTD-L318. In the CRP-αCTD cocrystal structure (4), it appears that the C terminus of CRP is in close proximity to αCTD region 317-318. This suggests that FNR-AR1 region 181-192 may target multiple surfaces of αCTD (285-288 and 317-318).
Determining which residues within FNR-AR1 region 181-192 directly contact αCTD remains an open question, since no crystal structure is available for FNR in complex with αCTD. Nevertheless, it is possible that multiple residues contribute to transcription activation since substitution of no one residue eliminated most FNR activity. Based on genetic data, as well as the CRP-αCTD cocrystal structure (4, 25, 35), CRP-T158 appears to play a major role in activation, since it is the only side chain within AR1 to participate in direct contact with αCTD. Other residues of CRP-AR1 (M157, H159, P160, and Q164) participate in van der Waals interactions with αCTD. Therefore, more work needs to be done to further model not only the face of FNR-AR1 region 181-192 but also the rest of FNR-AR1 (regions 71-75 and 116-121) to determine how this extended AR1 interacts with αCTD to activate transcription.
In summary, this study enhances our understanding of the side chain determinants within FNR-AR1 and αCTD required for efficient activation. It demonstrates that there is flexibility within the side chain requirements in AR1, which may be important for allowing different promoters to overcome intrinsic limitations in transcription initiation. We also conclude that the FNR determinant within αCTD is more extensive than previously hypothesized and overlaps with the αCTD determinant for contact with CRP-AR1. This study also suggests that determinants within αCTD required for activation of a class II FNR-dependent promoter are the same as those required at class I promoters. This study will allow for further comparison between FNR and CRP and will aid in our understanding of the interface between FNR-AR1 and αCTD.
Acknowledgments
We thank Steve Busby for pRW50 plasmids containing either FF(−41.5) or FF(−61.5). We also thank Marc Thomas and Sarah Aiyar for strains containing alanine substitutions encoded in rpoA in single copy. We also thank Wilma Ross for technical advice and Victoria Sutton for providing purified WT FNR and FNR-RA184 proteins.
This work was supported by National Institutes of Health grant GM-45844.
REFERENCES
- 1.Aiyar, S. E., S. M. McLeod, W. Ross, C. A. Hirvonen, M. S. Thomas, R. C. Johnson, and R. L. Gourse. 2002. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol. 316:501-516. [DOI] [PubMed] [Google Scholar]
- 2.Barnard, A. M., G. S. Lloyd, J. Green, S. J. Busby, and D. J. Lee. 2004. Location of the Escherichia coli RNA polymerase alpha subunit C-terminal domain at an FNR-dependent promoter: analysis using an artificial nuclease. FEBS Lett. 558:13-18. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A., and S. Busby. 1994. Location and orientation of an activating region in the Escherichia coli transcription factor, FNR. Mol. Microbiol. 11:383-390. [DOI] [PubMed] [Google Scholar]
- 4.Benoff, B., H. Yang, C. L. Lawson, G. Parkinson, J. Liu, E. Blatter, Y. W. Ebright, H. M. Berman, and R. H. Ebright. 2002. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297:1562-1566. [DOI] [PubMed] [Google Scholar]
- 5.Blake, T., A. Barnard, S. J. Busby, and J. Green. 2002. Transcription activation by FNR: evidence for a functional activating region 2. J. Bacteriol. 184:5855-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browning, D., D. Lee, and S. Busby. 2002. Secrets of bacterial transcription initiation taught by the Escherichia coli FNR protein, p. 127-142. In D. Hodgson and C. Thomas (ed.), Signals, switches, regulons, and cascades. Cambridge University Press, Cambridge, United Kingdom.
- 7.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson, J. W., and C. A. Gross. 1989. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 9.Green, J., A. S. Irvine, W. Meng, and J. R. Guest. 1996. FNR-DNA interactions at natural and semi-synthetic promoters. Mol. Microbiol. 19:125-137. [DOI] [PubMed] [Google Scholar]
- 10.Green, J., and F. A. Marshall. 1999. Identification of a surface of FNR overlapping activating region 1 that is required for repression of gene expression. J. Biol. Chem. 274:10244-10248. [DOI] [PubMed] [Google Scholar]
- 11.He, Y., T. Gaal, R. Karls, T. J. Donohue, R. L. Gourse, and G. P. Roberts. 1999. Transcription activation by CooA, the CO-sensing factor from Rhodospirillum rubrum. The interaction between CooA and the C-terminal domain of the alpha subunit of RNA polymerase. J. Biol. Chem. 274:10840-10845. [DOI] [PubMed] [Google Scholar]
- 12.Kang, Y., K. D. Weber, Y. Qui, P. J. Kiley, and F. R. Blattner. 2005. Genomewide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 87:1135-1160. [DOI] [PMC free article] [PubMed]
- 13.Korner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559-592. [DOI] [PubMed] [Google Scholar]
- 14.Lamberg, K. E., and P. J. Kiley. 2000. FNR-dependent activation of the class II dmsA and narG promoters of Escherichia coli requires FNR-activating regions 1 and 3. Mol. Microbiol. 38:817-827. [DOI] [PubMed] [Google Scholar]
- 15.Lamberg, K. E., C. Luther, K. D. Weber, and P. J. Kiley. 2002. Characterization of activating region 3 from Escherichia coli FNR. J. Mol. Biol. 315:275-283. [DOI] [PubMed] [Google Scholar]
- 16.Lazazzera, B. A., D. M. Bates, and P. J. Kiley. 1993. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7:1993-2005. [DOI] [PubMed] [Google Scholar]
- 17.Lazazzera, B. A., H. Beinert, N. Khoroshilova, M. C. Kennedy, and P. J. Kiley. 1996. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271:2762-2768. [DOI] [PubMed] [Google Scholar]
- 18.Lee, D. J., S. J. Busby, and G. S. Lloyd. 2003. Exploitation of a chemical nuclease to investigate the location and orientation of the Escherichia coli RNA polymerase alpha subunit C-terminal domains at simple promoters that are activated by cyclic AMP receptor protein. J. Biol. Chem. 278:52944-52952. [DOI] [PubMed] [Google Scholar]
- 19.Lee, D. J., H. J. Wing, N. J. Savery, and S. J. Busby. 2000. Analysis of interactions between Activating Region 1 of Escherichia coli FNR protein and the C-terminal domain of the RNA polymerase alpha subunit: use of alanine scanning and suppression genetics. Mol. Microbiol. 37:1032-1040. [DOI] [PubMed] [Google Scholar]
- 20.Li, B., H. Wing, D. Lee, H. C. Wu, and S. Busby. 1998. Transcription activation by Escherichia coli FNR protein: similarities to, and differences from, the CRP paradigm. Nucleic Acids Res. 26:2075-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd, G., P. Landini, and S. Busby. 2001. Activation and repression of transcription initiation in bacteria. Essays Biochem. 37:17-31. [DOI] [PubMed] [Google Scholar]
- 22.Lombardo, M. J., D. Bagga, and C. G. Miller. 1991. Mutations in rpoA affect expression of anaerobically regulated genes in Salmonella typhimurium. J. Bacteriol. 173:7511-7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lonetto, M. A., V. Rhodius, K. Lamberg, P. Kiley, S. Busby, and C. Gross. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J. Mol. Biol. 284:1353-1365. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. 1972. Experiments in molecular genetics, p. 48. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Niu, W., Y. Zhou, Q. Dong, Y. W. Ebright, and R. H. Ebright. 1994. Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP). I. Saturation and alanine-scanning mutagenesis. J. Mol. Biol. 243:595-602. [DOI] [PubMed] [Google Scholar]
- 26.Rhodius, V. A., D. M. West, C. L. Webster, S. J. Busby, and N. J. Savery. 1997. Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res. 25:326-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savery, N. J., G. S. Lloyd, S. J. Busby, M. S. Thomas, R. H. Ebright, and R. L. Gourse. 2002. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase alpha subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J. Bacteriol. 184:2273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savery, N. J., G. S. Lloyd, M. Kainz, T. Gaal, W. Ross, R. H. Ebright, R. L. Gourse, and S. J. Busby. 1998. Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 17:3439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutton, V. R., and P. J. Kiley. 2003. Techniques for studying the oxygen-sensitive transcription factor FNR from Escherichia coli. Methods Enzymol. 370:300-312. [DOI] [PubMed] [Google Scholar]
- 30.Weber, I. T., and T. A. Steitz. 1987. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolutioin. J. Mol. Biol. 198:311-326. [DOI] [PubMed] [Google Scholar]
- 31.Williams, R., A. Bell, G. Sims, and S. Busby. 1991. The role of two surface exposed loops in transcription activation by the Escherichia coli CRP and FNR proteins. Nucleic Acids Res. 19:6705-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, S. M., N. J. Savery, S. J. Busby, and H. J. Wing. 1997. Transcription activation at class I FNR-dependent promoters: identification of the activating surface of FNR and the corresponding contact site in the C-terminal domain of the RNA polymerase alpha subunit. Nucleic Acids Res. 25:4028-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wing, H. J., J. Green, J. R. Guest, and S. J. Busby. 2000. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoters. J. Biol. Chem. 275:29061-29065. [DOI] [PubMed] [Google Scholar]
- 34.Wing, H. J., S. M. Williams, and S. J. Busby. 1995. Spacing requirements for transcription activation by Escherichia coli FNR protein. J. Bacteriol. 177:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, Y., T. J. Merkel, and R. H. Ebright. 1994. Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP). II. Role at class I and class II CAP-dependent promoters. J. Mol. Biol. 243:603-610. [DOI] [PubMed] [Google Scholar]
- 36.Ziegelhoffer, E. C., and P. J. Kiley. 1995. In vitro analysis of a constitutively active mutant form of the Escherichia coli global transcription factor FNR. J. Mol. Biol. 245:351-361. [DOI] [PubMed] [Google Scholar]