Abstract
Ceftizoxime, a beta-lactam antibiotic with high selective affinity for penicillin-binding protein 2 (PBP2) of Staphylococcus aureus, was used to select a spontaneous resistant mutant of S. aureus strain 27s. The stable resistant mutant ZOX3 had an increased ceftizoxime MIC and a decreased affinity of its PBP2 for ceftizoxime and produced peptidoglycan in which the proportion of highly cross-linked muropeptides was reduced. The pbpB gene of ZOX3 carried a single C-to-T nucleotide substitution at nucleotide 1373, causing replacement of a proline with a leucine at amino acid residue 458 of the transpeptidase domain of the protein, close to the SFN conserved motif. Experimental proof that this point mutation was responsible for the drug-resistant phenotype, and also for the decreased PBP2 affinity and reduced cell wall cross-linking, was provided by allelic replacement experiments and site-directed mutagenesis. Disruption of pbpD, the structural gene of PBP4, in either the parental strain or the mutant caused a large decrease in the highly cross-linked muropeptide components of the cell wall and in the mutant caused a massive accumulation of muropeptide monomers as well. Disruption of pbpD also caused increased sensitivity to ceftizoxime in both the parental cells and the ZOX3 mutant, while introduction of the plasmid-borne mecA gene, the genetic determinant of the beta-lactam resistance protein PBP2A, had the opposite effects. The findings provide evidence for the cooperative functioning of two native S. aureus transpeptidases (PBP2 and PBP4) and an acquired transpeptidase (PBP2A) in staphylococcal cell wall biosynthesis and susceptibility to antimicrobial agents.
Although several observations have documented the importance of penicillin-binding protein 2 (PBP2) in the physiology of Staphylococcus aureus, the precise nature of its role(s) in cell wall synthesis and drug resistance is not well understood. PBP2 is the only bifunctional penicillin-binding protein in S. aureus (3, 8), and the transpeptidase (TPase) domain of the protein was found to be essential for the growth and survival of the bacteria (14, 17). In methicillin-resistant S. aureus (MRSA) strains the essential function of PBP2 may be replaced by PBP2A, the protein product of the resistance gene mecA, which functions as a surrogate transpeptidase (17). Thus, MRSA strains can grow without PBP2. Nevertheless, this protein becomes essential again once MRSA strains are challenged to grow in the presence of beta-lactam antibiotics. In this case, it is the transglycosylase (TGase) activity of PBP2 that appears to become critical for growth and cell wall biosynthesis (14). PBP2 also plays an important role in the vancomycin-intermediate S. aureus-type glycopeptide resistance (21), and PBP2 is essential for the expression of high-level vancomycin resistance from the vanA gene complex in vancomycin-resistant S. aureus (19). Direct biochemical evidence for the TGase activity of PBP2 is also available: S. aureus in which the active site of the transglycosylase domain of PBP2 was inactivated by a point mutation produced glycan strands in which the chain length was reduced (14). However, evidence for the TPase activity of PBP2 is based primarily on sequence homology with established transpeptidases.
In the study described here, we used a beta-lactam antibiotic, ceftizoxime, with highly selective inhibitory activity against the TPase domain of PBP2 and a spontaneous ceftizoxime-resistant mutant as experimental tools in combination with genetic, physiological, and biochemical approaches for the dissection of the role of PBP2 in staphylococcal cell wall synthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1. S. aureus was grown in tryptic soy broth (Difco, Detroit, Mich.) at 37°C with aeration. Growth was monitored by measuring optical density (at 620 nm) with an LKB spectrophotometer (Pharmacia LKB Biotechnology, Inc., Uppsala, Sweden). Antibiotic selection was applied when it was necessary for maintenance of plasmids or chromosomal inserts. The concentrations of antibiotics used as selective agents were as follows: erythromycin (Sigma, St. Louis, Mo.), 10 μg/ml; chloramphenicol (Sigma), 20 μg/ml. All of the S. aureus strains that contained derivatives of plasmid pSPT181 (pSTSW-2C, pMGP19, and pMC1) were cultured only at 30°C to prevent the loss of the plasmid, which relied on a temperature-sensitive origin of replication for its maintenance in the cell (24). Escherichia coli strains were propagated in Luria-Bertani medium (Difco) with addition of ampicillin (Sigma) at 100 μg/ml when appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Nitrosoguanidine-induced mutant of RN450 (NCTC 8325-4) that is able to accept E. coli DNA | 10 |
| 27s | Antibiotic-sensitive derivative of RN450 | R. Novick |
| 27s-ΔpbpD | 27s with pbpD insertionally inactivated; Eryr | This study |
| 27s-mecA | This study | |
| 27s-ΔpbpD-mecA | This study | |
| RN4220pPBP4-J | RN4220 with insertionally inactivated pbpD; Metr Eryr | M. G. Pinho |
| ZOX3 | 27s derivative with ceftizoxime MIC of 6 μg/ml | This study |
| ZOX3-ΔpbpD | ZOX3 with pbpD insertionally inactivated; Eryr | This study |
| ZOX3-mecA | This study | |
| ZOX3-ΔpbpD-mecA | This study | |
| ZOX12 | 27s derivative with ceftizoxime MIC of 25 μg/ml | This study |
| RN4220-pIB01-3 | RN4220 with plasmid pIB01 integrated into chromosome | I. Boncca |
| ZOX3-pbpB27s | ZOX3 with plasmid pIB01 integrated into chromosome | This study |
| RN-ZOX3-13.9 | RN4220 with plasmid pZOX3-13 integrated into chromosome | This study |
| 27s-pbpBzox3 | 27s with plasmid pZOX3-13 integrated into chromosome | This study |
| RN-pMC1-1 | RN4220 containing pMC1 | This study |
| COLpPBP2iC-2 | COL with spac controlled pbpB in the chromosome | 17 |
| 27s- pbpBspac | 27s derivative into which spac-controlled pbpB was transduced from COLpPBP2iC-2 | This study |
| 27s-pbpBspac(pMC1) | 27s-pbpBspac containing plasmid pMC1 | This study |
| 27s-pbpBspac(pMGP19) | 27s-pbpBspac containing plasmid pMGP19 | This study |
| E. coli | ||
| DH 5αa | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ | Invitrogen |
| XL1 Bluea | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pSP64E | pSP64 with erm marker from Tn551 | 15 |
| pMGP19 | Plasmid pSPT181 with cat gene and 3.2-kb insert comprising prfA and pbpB ORFs | 16 |
| pSTSW-2C | Plasmid pSPT181C with 3,737-bp PCR product of S. aureus mecA region | 24 |
| pZOX3-13 | pSP64E with pbpB fragment cloned in the AvaI site | This study |
| pMC1 | pMGP19 with C→T point mutation at position 1337 of pbpB ORF | This study |
Genotypes as published by manufacturer.
Population analysis profiles (PAPs).
Antibiotic susceptibilities of cultures were determined by population analysis, as described before (22). Briefly, the bacterial cells were grown overnight at 37°C, and then four different dilutions of the bacterial culture (100, 10−2, 10−4, and 10−6) were plated on control plates without antibiotic and on plates which contained a series of twofold dilutions of the appropriate antibiotic. Occasionally, when it was necessary for maintenance of plasmids or chromosomal inserts, a selective agent (erythromycin or chloramphenicol) was incorporated into the medium at a constant concentration. The plates were incubated at 37°C for 48 h, and colonies were counted. Methicillin and ceftizoxime powders were purchased from U.S.P.C. Inc. (Rockville, Md.)
Peptidoglycan purification and analysis by HPLC.
Peptidoglycan was purified from S. aureus cells by methods described previously (2). One-liter cultures of bacteria were incubated at 37°C, collected by centrifugation, and extracted with hot sodium dodecyl sulfate. The crude cell walls were washed with distilled water, mechanically ruptured with glass beads, and purified by stepwise treatments with DNase, RNase, protease, 8 M LiCl, and acetone and, finally, washing with distilled water. The lyophilized pellet of purified cell walls was next extracted with 49% hydrofluoric acid at 4°C for 48 h to remove wall teichoic acid. The insoluble peptidoglycan was washed extensively with distilled water, followed by enzymatic hydrolysis to generate muropeptides which were separated by reversed-phase high-pressure liquid chromatography (HPLC) as described previously (2), except that the step involving treatment with phosphatase was omitted.
Membrane purification and PBP affinity assays.
Membranes were prepared from cells grown to the late exponential stage as described previously (20). Proteins (80 μg per sample) for affinity assays were incubated with increasing concentrations of ceftizoxime (ranging from 0.1 to 20 μg/ml) at 30°C for 10 min and then labeled with 10 μg of benzyl-[14C]penicillin potassium salt (155 μCi per mg) (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) per ml for 10 min at 30°C. The reaction was stopped by the addition of an excess of nonlabeled benzylpenicillin. Labeled PBPs were resolved by the technique of Laemmli (7) and visualized by fluorography, as described before.
DNA techniques.
The plasmid constructs used in this study are listed in Table 1. DNA manipulations were performed according to standard procedures (18). The restriction enzymes were purchased from New England Biolabs (Beverly, Mass.). Digestions were conducted as recommended by the manufacturer. DNA ligations were done with a Rapid DNA Ligation kit (Roche Diagnostic, Indianapolis, Ind.) according to the manufacturer's instructions. DNA sequencing was performed in The Rockefeller University DNA Sequencing Resource Center by the BigDye terminator cycle sequencing method. Reaction mixtures were thermocycled in an ABI GeneAmp PCR system 9600/9700. Purified extension products were electrophoresed on either an ABI Prism 3700 DNA analyzer or a SpectruMedix 9610 Aurora DNA sequencer. PCRs were done with a DNA thermal cycler 480 (Perkin-Elmer) by standard methodology with either AmpliTaq polymerase (Applied Biosystems, Foster City, Calif.) for routine amplifications or PfuTurbo DNA polymerase (Stratagene, La Jolla, Calif.) for cloning when high-fidelity amplifications were necessary. Electroporation of S. aureus recipient strain RN4220 was done as described previously (6). Transduction with phage 80α was described previously (12).
Allelic replacement of pbpB.
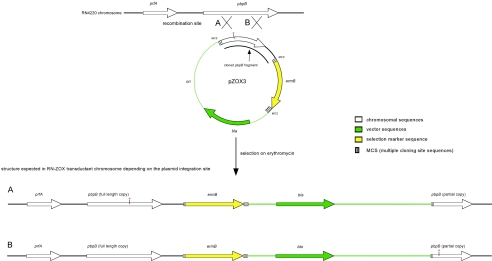
The allelic replacement of the wild-type pbpB gene with the ZOX3 mutant version of the gene was achieved by use of an insertion-duplication strategy. A fragment of the pbpB gene not including the promoter and membrane anchor was PCR amplified by using the primers F9-AvaI (5′-GGTTCCCGGGGTGCCTCAACATTAACAC-3′) and R9-AvaI (5-CTATCCCGGGTCGTAGGTGGATTTCTT-3′). The PCR product was cloned into AvaI site of plasmid pSP64E in such a way that the orientations of the pbpB and plasmid ermB open reading frames (ORFs) were the same. The integration plasmid pZOX3-13 was used to transform RN4220 by electroporation. Transformants were selected on erythromycin. Integration of pZOX3-13 into the chromosome created a structure in the chromosome in which the vector sequences were flanked by two copies of pbpB sequence: a full-length, functional 5′ copy which included all of the transcription signals and a partial, nonfunctional 3′ copy (Fig. 1). The existence of this structure in the transformants was confirmed by PCR. Depending on the site of the integration, one or the other copy of pbpB contained the characteristic point mutation. The sequences of pbpB of transformants were analyzed by sequencing, and one of the transformants, named RN-ZOX3-13.9, which contained the ZOX3 mutation in the functional pbpB copy, was selected. The structure obtained was then transduced into strain 27s. Transductants were selected on erythromycin. The pbpB copies of the transductants were analyzed by sequencing. One of the transductants, named 27s-pbpBZOX3, which contained the ZOX3 mutation in the functional pbpB copy, was selected for further characterization.
FIG. 1.
Strategy for the allelic replacement of pbpB. A fragment of pbpB from the ZOX3 mutant, not including the promoter and membrane anchor, was cloned into plasmid pSP64E to generate plasmid pZOX3, which was electroporated into the standard recipient strain RN4220, and transformants were selected on erythromycin (10 μg/ml). The plasmid integrated into the chromosome of RN4220, creating a structure in which vector sequences were flanked by a 5′ full-length functional copy of pbpB and a 3′ partial nonfunctional copy of this gene. There were two types of transformants expected, depending on the integration sites: strains with the mutation in the functional copy of pbpB (A) and strains with the mutation in the nonfunctional copy of the gene (B). One of the type A strains was used for transduction into the 27s background.
Allelic replacement of the mutated pbpB copy in strain ZOX3 with the wild-type sequence of strain 27s was performed in similar manner. In these experiments the source of the wild-type pbpB gene was strain RN4220pIB01-3. This strain was constructed by the integration of plasmid pIB01 into the RN4220 chromosome. Plasmid pIB01 is a derivative of pJF751; it carries a fragment of the pbpB gene without the promoter and membrane anchor from strain 27s. Sequences of both pbpB copies in this strain are wild type (unpublished data). Lysates of RN4220pIB01-3 were used to transduce the ZOX3 mutant. Transductants were selected on chloramphenicol. One of them, named ZOX3-pbpB27s, in which the functional copy of pbpB was confirmed to have the wild-type sequence by sequencing, was selected for further characterization.
Site directed mutagenesis of plasmid-borne pbpB and introduction of the mutated gene in the background of spac-controlled chromosomal pbpB.
In order to obtain plasmid pMC1 carrying a mutated version of the pbpB gene whose sequence is identical to the one present on the chromosome of the ZOX3 strain, we mutagenized plasmid pMGP19 (Table 1). pMGP19 contains the wild-type pbpB ORF together with sequences controlling its expression. The single nucleotide substitution of C to T at position 1373 of the pbpB open reading frame was introduced into pMGP19 by use of the QuikChange site-directed mutagenesis kit (Stratagene) according to manufacturer's instructions, using the primers ZOX3-MutF (5′-CGACAAAGTTTCAATATCCTAGCTTTAAAAGCTTGGCAATCAG-3′) and ZOX3-MutR (having the complementary sequence). The resulting plasmid, pMC1, was subsequently introduced into RN4220 by electroporation. Transformants were selected on chloramphenicol, and one of them was named RN-pMC1-1. The chromosomally located spac-controlled pbpB gene from strain COLpPBP2iC-2 was transferred into the 27s background by transduction followed by selection on erythromycin. The resulting strain was named 27s- pbpBspac. Next, plasmid pMC1 was introduced into 27s-pbpBspac by transduction with strain RN-pMC1-1 as a donor and selection of tranductants on plates containing both chloramphenicol and erythromycin. The resulting strain was named 27s-pbpBspac(pMC1).
In order to obtain a control strain carrying a plasmid with the wild-type pbpB sequence, the pMGP19 plasmid was introduced into strain 27s-pbpBspac by electroporation and transformants were selected on plates containing both chloramphenicol and erythromycin. The resulting strain was named 27s-pbpBspac(pMGP19). Strains were grown in medium containing 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (17).
Inactivation of pbpD.
An internal 0.67-kb DNA fragment of pbpD was amplified from the chromosome of strain COL by high-fidelity PCR with primers PBP4P22A (5′-TATCCCGGGTATGACACCATATGCACAAG-3′) and PBP4P23B (5′-GCAGGATCCGTGTAATACGTAACTGCTAG-3′). After digestion with AvaI and BamHI, the fragment was cloned into plasmid pSP64E and was electroporated into strain RN4220 (13). The inactivated pbpD was transduced into the 27s and ZOX3 backgrounds to generate strains 27s-ΔpbpD and ZOX3-ΔpbpD, respectively. Erythromycin was used as a selective agent in these transductions.
Introduction of plasmid-borne mecA into strains 27s and ZOX3.
Plasmid pSTSW-2C (Table 1), carrying a copy of mecA gene including its transcription signals, was transduced into strains 27s, 27s-ΔpbpD, ZOX3, and ZOX3-ΔpbpD. Transductants were selected on plates containing chloramphenicol or chloramphenicol and erythromycin (in the case of strains carrying the pbpD deletion).
RESULTS
Selection of mutant ZOX3.
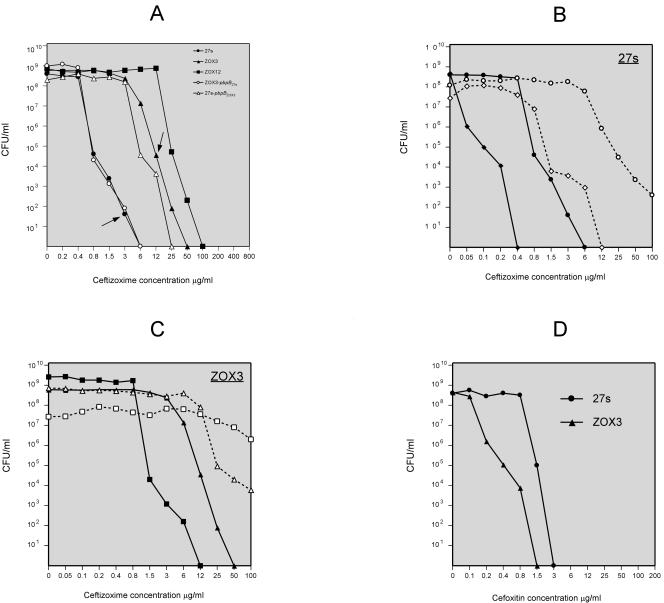
The relevant properties of strains and plasmids used in this study are shown in Table 1. Mutant ZOX3 was selected by plating an overnight culture of strain 27s on a series of agar plates containing increasing concentrations of the antibiotic ceftizoxime. A spontaneous ceftizoxime-resistant mutant was picked from the plate containing 3 μg of ceftizoxime per ml. The approximate frequency of the mutant was 10−7. The PAP of the ZOX3 mutant cultures showed elevated resistance to ceftizoxime for the majority of the bacteria, with an MIC of 6 μg/ml, an increase from the MIC of 0.8 μg/ml for the parental strain 27s (Fig. 2A). A second-stage mutant, ZOX12, with an additional increase in the ceftizoxime MIC to 12 μg/ml was also isolated from the subpopulation of ZOX3 colonies that grew on agar containing 6 μg of the antibiotic per ml. The mutant ZOX12 was not further characterized.
FIG. 2.
Antibiotic susceptibility profiles of S. aureus strain 27s and ZOX3 (carrying a mutated form of pbpB) and mutant derivatives of these strains. Antibiotic susceptibilities of strains were determined in overnight cultures plated on agar containing different concentrations of ceftizoxime and/or cefoxitin and were analyzed for PAPs as described in Materials and Methods. (A) Ceftizoxime susceptibility profiles of parental strain 27s and ceftizoxime-resistant mutants ZOX3 and ZOX12 (solid squares). Susceptibility profiles of transformant derivatives of strains 27s and ZOX3 carrying mutant and/or wild-type forms of pbpB (strain 27s-pbpBZOX3 [27s with the normal pbpB replaced by the mutant gene] and strain ZOX3-pbpB27s [ZOX3 in which the mutant pbpB was replaced by the wild-type form of the gene] are also shown. The arrow pointing to the line with solid circles indicates the source of ZOX3. The arrow pointing to the line with solid triangles indicates the source of the ZOX12. (B) Ceftizoxime susceptibility profiles of strain 27s (solid circles), strain 27s with inactivated pbpD (solid diamonds), strain 27s carrying the plasmid-borne mecA (empty circles), and 27s with inactivated pbpD carrying the plasmid-borne mecA (empty diamonds). (C) Ceftizoxime susceptibility profiles of strain ZOX3 (solid triangles), ZOX3 with inactivated pbpD (solid squares), ZOX3 carrying the plasmid-borne mecA (empty triangles), and ZOX3 with inactivated pbpD carrying the plasmid-borne mecA (empty squares). (D) Cefoxitin susceptibility profiles of strains 27s and ZOX3.
Mutant ZOX3 was stable: daily serial passage by picking and restreaking colonies on antibiotic-free tryptic soy agar for over 2 weeks did not cause any reduction in antibiotic resistance as determined by the PAP (not shown). The antibiotic resistance of ZOX3 was selective for ceftizoxime: there was no increase in the resistance of ZOX3 to methicillin, oxacillin, and a number of other beta-lactam antibiotics. The MICs of methicillin and oxacillin were actually slightly reduced, from 0.8 μg/ml (the MIC for the parental strain 27s) to 0.4 μg/ml and from 0.1 to 0.05 μg/ml (in ZOX3).
Change in ceftizoxime affinity of PBP2 in mutant ZOX3.
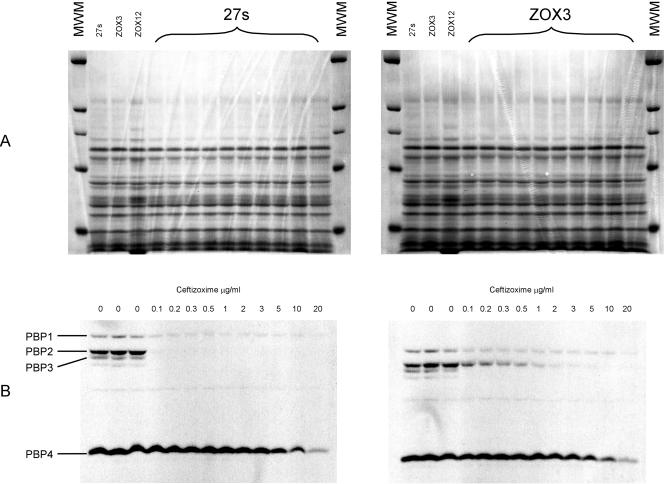
Figure 3 shows the results of a fluorographic competition assay in which the binding capacities (affinities) of PBPs for ceftizoxime in the parental strain 27s and mutant ZOX3 were compared. Membrane preparations containing the same amounts of protein (80 μg) from the two bacterial strains were preincubated with increasing amounts of ceftizoxime (0.1 to 20 μg), after which a single identical amount of radioactive penicillin was added to each test preparation and the concentration of ceftizoxime preventing binding of the radioactive reagent to various PBPs was determined. A concentration of ceftizoxime as low as 0.1 μg/ml inhibited the binding of radioactive penicillin to PBP2 in the parental strain 27s. A similar degree of inhibition required 1 to 2 μg of the drug per ml in the ZOX3 mutant. No differences in the affinity profiles of the other PBPs were apparent.
FIG. 3.
Ceftizoxime binding capacity of PBPs of susceptible and ceftizoxime-resistant S. aureus strains. Flurographic competition assays were used, with membrane preparations from the susceptible parental strain 27s and its ceftizoxime-resistant mutant ZOX3, to determine the ceftizoxime binding capacities of PBPs as described in Materials and Methods. Panel A shows the protein patterns of sodium dodecyl sulfate-polyacrylamide gels stained with Coomassie blue. Panel B shows the titration of the ceftizoxime binding capacity of PBPs by using the flurographic assay.
Analysis of the pbpB gene in mutant ZOX3.
The pbpB gene was PCR amplified from the parental and resistant strains and sequenced. The two sequences were identical except for a single point mutation detected in the transpeptidase domain of the mutant strain at position 1373 of the pbpB ORF, in which a cytosine residue was replaced by a thymine, causing a proline-to-leucine substitution at amino acid residue 458 in the protein. The point mutation was located in the immediate vicinity of the conserved motif SFN (8), which forms a part of the active site of the PBP2 TPase domain (Fig. 4).
FIG. 4.
Localization of the mutation in the pbpB gene of ceftizoxime-resistant mutant ZOX3. Deduced amino acid sequences of the PBP2 protein are shown for strains 27s and ZOX3. The TGase domain of the protein is shown in blue, the TPase domain is in green, conserved motifs are in yellow, and the proline (P)-to-leucine (L) mutation site is in red.
Decreased level of peptidoglycan cross-linking in the ZOX3 mutant.
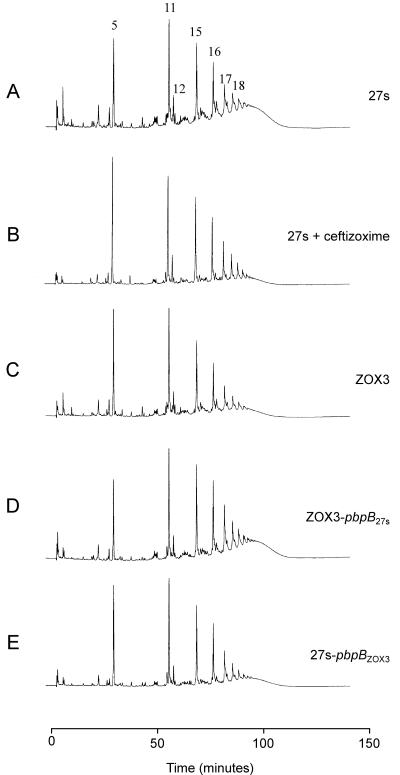
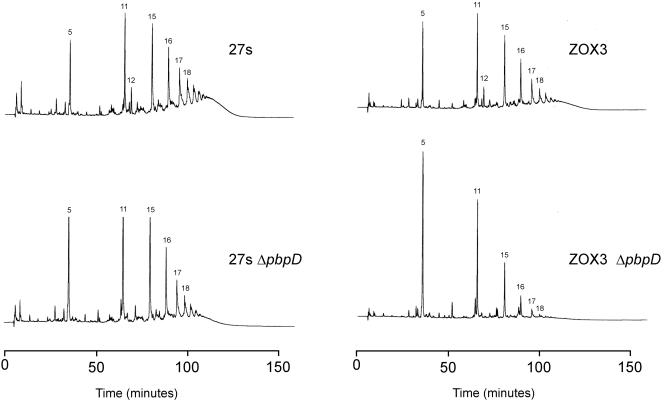
Cell wall peptidoglycan was isolated from the parental strain 27s and mutant ZOX3, and the muropeptide compositions were determined after enzymatic hydrolysis and HPLC separation. Comparison of the HPLC elution profiles of the two peptidoglycan preparations (Fig. 5A and C; see Fig. 7A) indicates a significant reduction in the highly cross-linked oligomeric muropeptides in the peptidoglycan of the mutant strain. A similar change in the elution profile of muropeptides was observed in the parental strain 27s grown in the presence of 1/10 times the MIC of ceftizoxime (Fig. 5B).
FIG. 5.
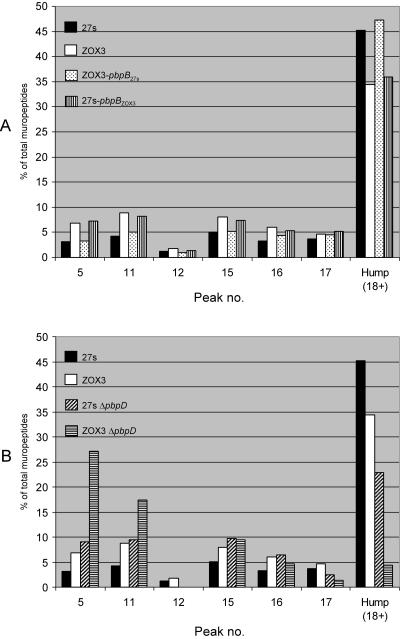
Differences in the peptidoglycan compositions of the ceftizoxime-resistant S. aureus mutant ZOX3 and its parental strain 27s. Peptidoglycans were prepared, and muropeptide patterns were analyzed by HPLC, as described in Materials and Methods. Panel A: parental strain 27s. Panel B: 27s grown in the presence of 1/10 times the MIC of ceftizoxime. Panel C: mutant ZOX3. Panel D: strain ZOX3-pbpB27s (mutant ZOX3 in which the mutated pbpB was replaced with the wild-type form by allelic replacement). Panel E: 27s pbpBZOX3 (parental strain in which the normal pbpB was replaced with the mutated form). Numbers above the HPLC peaks identify a structure of the respective muropeptides, as described earlier (2). Peaks 5, 11, 15, 16, and 17 represent the monomeric muropeptide (5) and its di-, tri-, tetra-, and pentameric derivatives. Peak 12 is the unique doubly cross-linked muropeptide dimer (1). The poorly resolved part (“hump”) of the HPLC profile eluting with retention times of greater than 100 min (i.e., after peak 18) contain highly cross-linked oligomeric components.
FIG. 7.
Quantitative differences in the peptidoglycan compositions of strains 27s and ZOX3 and their derivatives. The bars show the relative amounts of muropeptide species calculated from UV absorbances of the different muropeptide components.
Allelic replacement experiments.
In order to test whether the point mutation detected in the TPase domain of the resistant mutant was responsible for the antibiotic resistance and for the observed alterations in PBP2 affinity and cell wall composition, allelic replacement experiments were performed. The mutant form of pbpB from ZOX3 was replaced by the wild-type pbpB sequence to generate transformant ZOX3-pbpB27s. In the reciprocal cross, the wild-type pbpB gene of the parental strain 27s was replaced by the mutant pbpB sequence to generate transformant 27s-pbpBZOX3. Details of the methods used in the allelic replacement experiments are outlined in Fig. 1 (see Materials and Methods).
The PAP data show that ceftizoxime resistance was linked to the type of pbpB gene carried by the particular isolate (Fig. 2A): the ceftizoxime MIC for transformants carrying the wild-type pbpB was 0.8 μg/ml, and transformants carrying the mutated version of pbpB showed a ceftizoxime MIC of 6 μg/ml. The reduced affinity of PBP2 for ceftizoxime (not shown) and the reduced degree of peptidoglycan cross-linking (Fig. 5D and E; see Fig. 7A) was also reproduced in the reciprocal transformants.
Site-directed mutagenesis of pbpB and its expression from a plasmid in the absence of expression of its chromosomal copy.
To obtain additional confirmation that the point mutation in pbpB is indeed responsible for ceftizoxime resistance, the mutated pbpB copy (obtained by site-directed mutagenesis of the wild-type sequence) was expressed from a plasmid in strain 27s-pbpBspac, in which chromosomal copy of pbpB was put under the control of an IPTG-inducible promoter (see Materials and Methods).
Strain 27s-pbpBspac(pMC1), containing mutated pbpB, and control strain 27s-pbpBspac(pMGP19), containing wild-type pbpB, could grow in the absence of IPTG, indicating the successful transcription of the pbpB genes from the plasmids. The ceftizoxime MIC for the strain carrying the mutated pbpB was 4 to 6 μg/ml, while the strain carrying the wild-type pbpB on the plasmid had a ceftizoxime MIC of 0.38 μg/ml.
Effect of inactivation of pbpD and introduction of mecA on ceftizoxime resistance and cell wall composition.
The significant change in the degree of cross-linking of the peptidoglycan observed in the PBP2 transpeptidase mutant ZOX3 prompted us to test the effect of inactivation of PBP4 on the antibiotic resistance and cell wall structure of the bacteria, since PBP4 was shown earlier to participate in the cross-linking of the peptidoglycan of S. aureus (25). It was also of interest to test the effect of introducing the mecA determinant into these genetic backgrounds, since PBP2A, the protein product of mecA, is able to substitute for the TPase function of PBP2 (17).
Insertional inactivation of pbpD caused a drop in the ceftizoxime MIC for ZOX3 from 6 to 1.5 μg/ml, and inactivation of pbpD in the parental strain 27s decreased the ceftizoxime MIC from 0.4 to 0.05 μg/ml. Introduction of the plasmid-borne mecA gene caused the opposite effect: the ceftizoxime MIC for strain 27s increased from 0.8 to 12 μg/ml, and that for ZOX3 increased from 6 to about 25 μg/ml (Fig. 2B and C). Introduction of mecA into strain 27s caused no detectable change in the oxacillin MIC for the majority of cells (data not shown).
Inactivation of pbpD in 27s also caused a large reduction in the representation of highly cross-linked muropeptides, i.e., in the material eluting from the HPLC column with retention times of higher than 100 min (Fig. 6). The proportion of this muropeptide fraction dropped from 45 to 22% in strain 27s with the inactivated PBP4 (Fig. 7B). Inactivation of PBP4 in the resistant mutant ZOX3 had an even more pronounced effect: the highly cross-linked muropeptide fraction decreased from 35 to less than 5%, and there was a striking accumulation of monomeric muropeptides (peak 5 in Fig. 6) (from about 7 to 27%) and in the dimeric muropeptides (peak 11 in Fig. 6) (from about 8 to 17%) (Fig. 7B). In contrast to the highly significant increase in ceftizoxime resistance caused by the introduction of mecA into either strain 27s or ZOX3, the presence of the plasmid-borne mecA gene caused no detectable change in the muropeptide HPLC profiles of either strain 27S or mutant ZOX3 (data not shown).
FIG. 6.
Effect of inactivation of pbpD on the muropeptide compositions of S. aureus strains 27s and ZOX3. Strains 27s and ZOX3 and their derivatives with a disrupted pbpD gene (strains 27s-ΔpbpD and ZOX3-ΔpbpD) were grown, and HPLC analysis of peptidoglycan was performed as described in Materials and Methods.
DISCUSSION
The S. aureus pbpB gene encodes a two-domain protein carrying the characteristic motifs of a TPase as well as a TGase (3, 8). Direct biochemical evidence for the TGase activity of PBP2 has been obtained (14), and it was also shown that the TGase domain of PBP2 becomes essential for the growth of MRSA strains in the presence of methicillin in the medium (14).
Much less is known about the nature of the TPase activity, except that in methicillin-susceptible S. aureus strains it is essential for growth, and this essential function may be replaced by the activity of PBP2A in MRSA strains which carry the drug resistance determinant mecA (17). MRSA conditional mutants in which the transcription of the PBP2 gene was blocked grew and produced a cell wall peptidoglycan with close to a normal composition. However, this peptidoglycan must have been the product of the resistance protein PBP2A that has replaced the functions of the native PBP2 (17). Thus, direct biochemical evidence for the nature of the TPase activity of PBP2 in staphylococcal cell wall synthesis is not available.
In order to bypass this problem, we chose a strategy which involved the use of the beta-lactam antibiotic ceftizoxime, which, at low concentrations, has extremely selective inhibitory activity against PBP2 of S. aureus (11). According to Okonogi et al., the 50% inhibitory concentrations of ceftizoxime for the S. aureus PBPs were 2.74 μg/ml for PBP1, 0.0626 μg/ml for PBP2, and 3.61 and 50.8 μg/ml for PBP3 and PBP4, respectively (11). We used this antibiotic as an experimental tool to isolate a stable spontaneous ceftizoxime-resistant mutant, ZOX3, that showed an increased MIC only of ceftizoxime and not of other beta-lactams. The PBP2 of the mutant showed a striking and selective decrease in its binding capacity for ceftizoxime, and the mutant produced a peptidoglycan with an altered composition, the most prominent features of which were (i) a reduction in the percentage of the highly cross-linked muropeptides (from about 45% in the susceptible strain to about 35% in the mutant) and (ii) an increase in the percentage of monomeric (peak 5) and dimeric (peak 11) muropeptides. Reciprocal allelic replacement experiments and site-directed mutagenesis clearly showed that the three phenotypes, i.e., the increased ceftizoxime MIC, reduced affinity for PBP2, and altered cell wall composition, were the consequences of the proline-to-leucine point mutation in the structural gene of PBP2. The cell wall alterations appeared in the mutant bacteria growing in antibiotic-free medium, indicating that the replacement of a proline by a leucine residue in the mutant protein affected the normal functioning of PBP2 in the cross-linking of cell wall muropeptides. This interpretation is supported by the finding that parental cells grown in the presence of from 1/10 up to 1/3 the MIC of ceftizoxime also produced peptidoglycan with an altered muropeptide composition similar to that of ZOX3 grown in antibiotic-free medium (Fig. 5).
The reduced amounts of highly cross-linked oligomers in the cell walls of the ZOX3 mutant were surprising, since several previous studies showed that these oligomers are primarily the products of the activity of PBP4, a protein with known “secondary” transpeptidase activity (25). In order to clarify this issue, we examined the muropeptide composition of cell wall prepared from a mutant of strain 27s in which PBP2 was intact but PBP4 was interrupted. Inactivation of pbpD, the structural gene of PBP4, had relatively little effect on the relative proportions of monomeric (peak 5) and moderately cross-linked (peaks 11, 15, 16, and 17) muropeptides, but the percentage of highly cross-linked components was reduced from 45 to about 22%.
Next we analyzed the muropeptide composition of a double mutant carrying both the ZOX3 point mutation and the disrupted PBP4. The peptidoglycan of the double mutant showed a greatly increased proportion of monomeric muropeptides (an increase from 7 to 27%) and an increase in muropeptide dimers (from 9 to 17%). In addition, there was a drastic reduction in the highly cross-linked oligomers (a decrease from 35 to less than 5%) (Fig. 7B).
Peak 12 disappeared completely from the cell walls of bacteria with inactivated pbpD, suggesting that PBP4 participated in its biosynthesis. This unusually structured dimeric muropeptide has doubly cross-linked components by two pentaglycine bridges (1). Most interestingly, disruption of pbpD also affected ceftizoxime resistance: the ceftizoxime MIC for strain 27s decreased from 0.8 to 0.05 μg/ml, and that for mutant ZOX3 decreased from 6 to 0.8 μg/ml, when the pbpD gene was inactivated (Fig. 2B C). This finding was surprising, since the affinity titration experiments of Fig. 2 clearly demonstrated the selectivity of inhibition of PBP2 by ceftizoxime. This antibiotic showed no detectable inhibitory activity against PBP4 in the concentration range close to the MIC. Inactivation of pbpD had no effect on the methicillin MIC (data not shown).
Our findings provide direct biochemical evidence for the functioning of the TPase domain of PBP2 and also suggest that the two transpeptidases, PBP2 and PBP4, act in concert in the cross-linking of cell wall building blocks during the biosynthesis of the staphylococcal cell wall. While PBP4 cannot replace the essential TPase activity of PBP2, the hypersensitivity of PBP4 deletion mutants to ceftizoxime and the compositional changes in the cell walls of double mutants suggest that these two proteins function cooperatively to produce a staphylococcal peptidoglycan that carries the minimal numbers and types of cross-links essential for the survival and growth of the bacteria. Such cooperative functioning is also consistent with the observation that mutant ZOX3, in which PBP2 is suboptimal in function, becomes hypersensitive to cefoxitin, a specific inhibitor of PBP4 (9) (Fig. 2D).
Examination of the HPLC profiles in Fig. 6 and 7 suggests that the primary TPase function of PBP2 may be in producing muropeptide dimers (peptide 11) and trimers, tetramers, and pentamers (peptides 15 through 17) (with progressively decreasing efficiencies) and that the primary function of PBP4 appears to be in the production of muropeptides with a high degree of cross-linking.
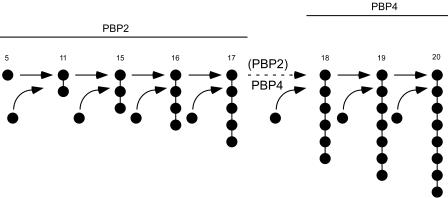
The modest but significant reduction in the highly cross-linked muropeptide fraction (from 45 to 35%) in the ZOX3 mutant may indicate the direct participation of PBP2 in the production of these oligomers. Alternatively, it may be the consequence of an indirect effect in which a suboptimal functioning of the mutant PBP2 affects the functioning of PBP4. One specific mechanism for this could be if the transpeptidase activity of PBP2 were to produce the substrates for PBP4, i.e., if the primary TPase activity of PBP2 were to be the production of dimers, trimers, and tetrameric muropeptides which then would be used by PBP4 to produce the more highly cross-linked oligomers by the addition of monomeric donor peptides to the more highly cross-linked acceptors. Basic features of this model are shown in Fig. 8.
FIG. 8.
Model for the cooperative functioning of PBPs in cross-linking the muropeptides in the S. aureus cell wall. Muropeptide monomers and multimers are identified by their peak numbers in the HPLC profile and by the bead diagram symbolizing the number of monomeric units cross-linked. Monomeric muropeptides are indicated by solid circles. The primary cross-linking functions of PBP2 (and PBP2A) are suggested to involve cross-linking of up to pentameric components, and cross-linking of higher multimers is catalyzed primarily by PBP4.
The accumulation of large amounts of un-cross-linked monomeric muropeptides in the double mutant is consistent with this proposal: it would be the result of the suboptimal TPase activity of the mutant form of PBP2, combined with the lack of PBP4 activity, which would stop the “consumption” of muropeptide monomers for the production of highly cross-linked wall components. Thus, under these conditions, the major contribution of PBP2 to cell wall composition would be through its TGase activity, producing a peptidoglycan enriched for un-cross-linked muropeptide monomers.
We introduced the mecA gene on a plasmid into the parental strain 27s and also into the mutant ZOX3. In each case the ceftizoxime MIC for the bacteria increased significantly. There was no effect on the methicillin MIC for the majority of the cells. This observation indicates that the mecA gene product PBP2A can effectively compensate for the loss of antibiotic resistance and at least partially replace the transpeptidase activity of PBP2, when the latter is inhibited by ceftizoxime or has a decreased efficiency of functioning as in the ZOX3 mutant. However, attempts to directly document such an effect have failed so far: strain 27s or mutant ZOX3 carrying the plasmid-borne mecA gene showed little if any increase in the proportion of muropeptide dimers and trimers (data not shown). Thus, the mechanism of the proposed “assistance” by PBP2A to the TPase activity of PBP2 remains to be determined.
Our studies exploited a combination of a selectively acting beta-lactam antibiotic and a specific mutant for the dissection of the functioning of PBP2 in staphylococcal cell wall synthesis. The cooperative functioning of PBP2 and PBP4 documented in our study is consistent with the notion that proteins involved with the complex task of bacterial cell wall assembly act as a coordinated multienzyme complex (4). How the activities of the other PBPs (the “essential” PBP1 [23] and the “nonessential” PBP3 [15]) of S. aureus interact in this system remains to be explored.
The significant effect of mecA on the ceftizoxime-resistant phenotype, in contrast to the lack of effect on peptidoglycan composition (or resistance to other beta-lactams such as oxacillin), requires comment. Introduction of mecA on a plasmid into the background of either 27s, ZOX3, or their pbpD-inactivated derivatives caused highly significant increases in the ceftizoxime MIC for the bacteria. In the mecA derivative of 27s, the ceftizoxime MIC increased from 0.8 up to 25 μg/ml; in strain 27s carrying the inactivated pbpD gene, the introduction of mecA caused an increase of the ceftizoxime MIC from 0.05 up to 0.8 to 1.5 μg/ml (Fig. 2B). Introduction of mecA into mutant ZOX3 or its pbpD-inactivated derivative also caused major increases in the ceftizoxime resistance level (Fig. 2C). Yet these major “corrections” of resistance level were specific for ceftizoxime; there was no change in the oxacillin MIC for the majority of bacteria. Also, the introduction of mecA did not cause any significant alteration in muropeptide composition as detectable by our HPLC system. These observations suggest that mecA (or PBP2A) interacts directly with the PBP2-PBP4 transpeptidation system which is targeted by the selective action of ceftizoxime. The biochemical mechanism by which mecA and/or its protein product PBP2A provides such a highly selective protection of staphylococci remains to be explained.
Acknowledgments
Partial support for this study was provided by a grant from the National Institutes of Health (RO1 AI37275) and by the American Austrian Foundation.
REFERENCES
- 1.Boneca, I. G., N. Xu, D. A. Gage, B. L. de Jonge, and A. Tomasz. 1997. Structural characterization of an abnormally cross-linked muropeptide dimer that is accumulated in the peptidoglycan of methicillin- and cefotaxime-resistant mutants of Staphylococcus aureus. J. Biol. Chem. 272:29053-29059. [DOI] [PubMed] [Google Scholar]
- 2.de Jonge, B. L., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248-11254. [PubMed] [Google Scholar]
- 3.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtje, J. V. 1996. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology 142:1911-1918. [DOI] [PubMed] [Google Scholar]
- 5.Katayama, Y., H. Z. Zhang, and H. F. Chambers. 2003. Effect of disruption of Staphylococcus aureus PBP4 gene on resistance to beta-lactam antibiotics. Microb. Drug Resist. 9:329-336. [DOI] [PubMed] [Google Scholar]
- 6.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:373-376. [Google Scholar]
- 7.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 8.Murakami, K., T. Fujimura, and M. Doi. 1994. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol. Lett. 117:131-136. [DOI] [PubMed] [Google Scholar]
- 9.Murakami, K., K. Nomura, M. Doi, and T. Yoshida. 1987. Increased susceptibility to cephamycin-type antibiotics of methicillin-resistant Staphylococcus aureus defective in penicillin-binding protein 2. Antimicrob. Agents Chemother. 31:1423-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novick, R., J. Kornblum, B. Kreiswirth, S. Projan, and H. Ross. 1990. Regulation of post-exponential-phase exoprotein synthesis in Staphylococcus aureus, p. 3-18. In T. J. Henry (ed.), Microbial determinants of virulence and host response. American Society for Microbiology, Washington, D.C.
- 11.Okonogi, K., Y. Noji, M. Nakao, and A. Imada. 1995. The possible physiological roles of penicillin-binding proteins of methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J. Infect. Chemother. 1:50-58. [Google Scholar]
- 12.Oshida, T., and A. Tomasz. 1992. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J. Bacteriol. 174:4952-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinho, M. G. 2001. Methicillin resistance in Staphylococcus aureus: the role of the native penicillin-binding proteins. Doctoral thesis. Universidade Nova de Lisboa, Lisbon, Portugal.
- 14.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 98:10886-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2000. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J. Bacteriol. 182:1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinho, M. G., H. de Lencastre, and A. Tomasz. 1998. Transcriptional analysis of the Staphylococcus aureus penicillin binding protein 2 gene. J. Bacteriol. 180:6077-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinho, M. G., S. R. Filipe, H. de Lencastre, and A. Tomasz. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 183:6525-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Severin, A., K. Tabei, F. Tenover, M. Chung, N. Clarke, and A. Tomasz. 2004. High level oxacillin and vancomycin resistance and altered cell wall composition in Staphylococcus aureus carrying the staphylococcal mecA and the enterococcal vanA gene complex. J. Biol. Chem. 279:3398-3407. [DOI] [PubMed] [Google Scholar]
- 20.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 21.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomasz, A., S. Nachman, and H. Leaf. 1991. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 35:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wada, A., and H. Watanabe. 1998. Penicillin-binding protein 1 of Staphylococcus aureus is essential for growth. J. Bacteriol. 180:2759-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, S. W., H. de Lencastre, and A. Tomasz. 2001. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J. Bacteriol. 183:2417-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyke, A. W., J. B. Ward, M. V. Hayes, and N. A. C. Curtis. 1981. A role in vivo for penicillin-binding protein 4 of Staphylococcus aureus. Eur. J. Biochem. 119:389-393. [DOI] [PubMed] [Google Scholar]