Abstract
The 14-kDa BlaI protein represses the transcription of blaZ, the gene encoding β-lactamase. It is homologous to MecI, which regulates the expression of mecA, the gene encoding the penicillin binding protein PBP2a. These genes mediate resistance to β-lactam antibiotics in staphylococci. Both repressors can bind either bla or mec DNA promoter-operator sequences. Regulated resistance genes are activated via receptor-mediated cleavage of the repressors. Cleavage is induced when β-lactam antibiotics bind the extramembrane sensor of the sensor-transducer signaling molecules, BlaR1 or MecR1. The crystal structures of BlaI from Staphylococcus aureus, both in free form and in complex with 32 bp of DNA of the mec operator, have been determined to 2.0- and 2.7-Å resolutions, respectively. The structure of MecI, also in free form and in complex with the bla operator, has been previously reported. Both repressors form homodimers, with each monomer composed of an N-terminal DNA binding domain of winged helix-turn-helix topology and a C-terminal dimerization domain. The structure of BlaI in complex with the mec operator shows a protein-DNA interface that is conserved between both mec and bla targets. The recognition helix α3 interacts specifically with the conserved TACA/TGTA DNA binding motif. BlaI and, probably, MecI dimers bind to opposite faces of the mec DNA double helix in an up-and-down arrangement, whereas MecI and, probably, BlaI dimers bind to the same DNA face of bla promoter-operator DNA. This is due to the different spacing of mec and bla DNA binding sites. Furthermore, the flexibility of the dimeric proteins may make the C-terminal proteolytic cleavage site more accessible when the repressors are bound to DNA than when they are in solution, suggesting that the induction cascade involves bound rather than free repressor.
β-Lactam antibiotics are becoming less effective therapy for treating staphylococcal infections as resistance to them increases. Resistance is mediated by a β-lactamase (encoded by blaZ) that hydrolyzes penicillins (4) and an alternate penicillin binding protein target (PBP2a, encoded by mecA) to which β-lactam antibiotics bind poorly (17). The transcription of blaZ and mecA is corepressed by related regulators (encoded by blaI and mecI). Both BlaI and MecI bind to palindromic promoter-operator sequences, presumably as dimers, and can interchangeably repress the transcription of either target gene (8, 26, 36). Signal transduction through either of two transmembrane inducers, BlaR1 or MecR1, leads to proteolytic autocleavage of the cytoplasmic domains of these molecules. Autocleavage of the signal transducer is followed by cleavage of the cognate repressor (BlaI or MecI, respectively) and induction of the transcription of blaZ or mecA (42). The exact mechanism of cleavage is unclear at present. The current hypothesis (1, 42) is that β-lactam binding to the extramembrane sensor of BlaR1 or MecR1 triggers autocleavage of its cytoplasmic domain at a single site, leaving the putative cytoplasmic protease tethered to the membrane. Sequence-specific repressor cleavage then ensues, either through direct interaction with the activated BlaR1 or MecR1 cytoplasmic protease domain or, more likely, through interaction with an intermediary molecule (BlaR2 or MecR2). Candidate R2 molecules have not yet been identified.
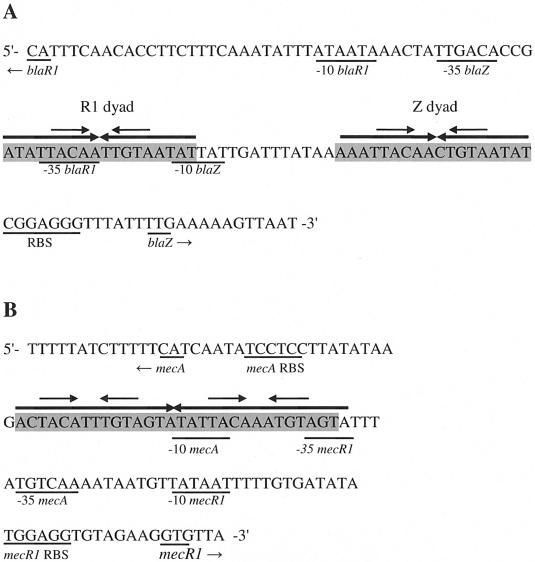
Genes for the signal transducers (blaR1 and mecR1) are part of two-gene operons that also contain genes for each repressor (blaI or mecI). Each regulon is divergently transcribed from its regulated gene, and promoter-operators for each resistance gene and its regulator overlap on opposite DNA strands. Thus, not only does binding of BlaI or MecI repress transcription of blaZ or mecA, but the regulatory operon is autorepressed as well. BlaI or MecI dimers are believed to bind specifically to two regions of dyad symmetry in the bla operator, the Z and R1 dyads located in the blaZ-blaR1 intergenic region (8). Each dyad contains an 18-bp palindrome, and they are separated from each other by a 13-bp linker (Fig. 1A). These two repressors also bind the mec operator, which consists of a single 30-bp palindrome with two 15-bp half-sites (36). Within each 15-bp half-site of the mec operator is 12 bp of dyad symmetry (Fig. 1B). Fingerprint studies show that each repressor binds to both the bla and mec operators and that each repressor protects the same sequences (8, 26, 36). The molecular masses of the monomers are about 14 kDa for both BlaI and MecI, with 61% of the residues identical, as shown in Fig. 2A. The high degree of sequence homology suggests that the two structures should have the same fold. Extensive characterization of the activities of the repressors also suggests that the structures have two domains, one for dimerization and another for DNA binding. However, it is not clear how the structures of BlaI and MecI change to allow binding to two different targets that vary markedly in the number and spacing of contacted nucleotides.
FIG. 1.
bla and mec DNA promoter-operator sequences (7, 36). (A) The bla operator consists of the R1 dyad and Z dyad (shaded). Each dyad contains an 18-bp palindrome, and they are separated from each other by a 13-bp linker. (B) The mec operator (shaded) consists of a single 30-bp palindrome with two 15-bp half-sites. Both the bla and mec operators contain identical 4-bp inverted repeats (TACA/TGTA DNA binding motif) separated by two nucleotides (identified with smaller arrows) within the larger palindromes.
FIG. 2.
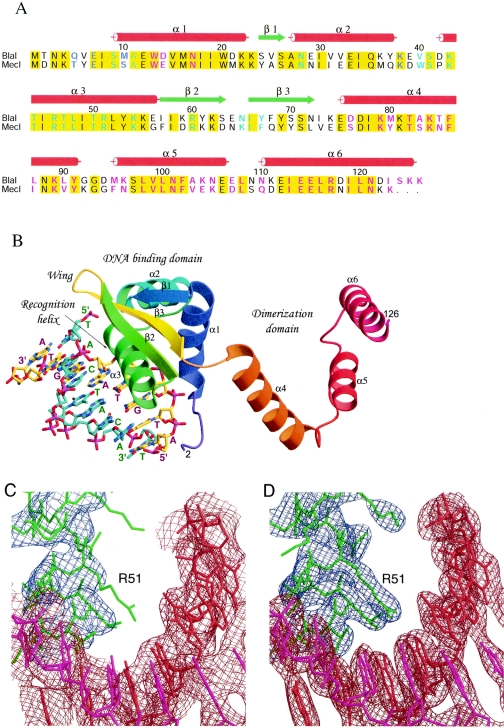
Sequence alignment, structure, and electron density maps of the BlaI-DNA complex. (A) Sequence alignment of BlaI and MecI repressor proteins. The proteins used in the alignment are as follows: BlaI, S. aureus N395 (Q9AC78), and MecI, S. aureus N395 (P26598). The red cylinders and green arrows indicate the secondary structural elements observed in the BlaI and BlaI-DNA crystals. Identical residues in the two sequences are shaded in yellow. Residues involved in DNA binding and dimerization are highlighted in cyan and magenta, respectively; those involved in both are in blue. (B) Overall structure of the BlaI monomer bound to DNA in the asymmetric unit. The protein molecule is shown as a ribbon representation, and the DNA is shown as sticks. The N-terminal domain has a winged helix topology that interacts with the TACA/TGTA motif of DNA via the recognition helix α3. Upon dimer formation, the three helices of the C-terminal domain intertwine with those from another monomer. (C) The refined model using the in-house data at 3.0-Å resolution is superimposed on the initial composite omit map based on the molecular replacement solution. The contour level is 1.0 σ centered about the residue Arg51. The model is shown as sticks in red and magenta for the double-stranded DNA and in green for the protein molecule. The DNA densities are shown in brown, and those for the protein are in blue. (D) The final model is superimposed on the 2Fo-Fc map calculated at 2.7-Å resolution from the synchrotron data set, contoured at a level of 1.5 σ.
The three-dimensional structure of the N-terminal DNA binding domain of BlaI from Bacillus licheniformis was determined from heteronuclear nuclear magnetic resonance (NMR) spectroscopy (27). It is composed of three α-helices and three β-strands typical of the winged helix protein family, and helix α3 makes specific contacts with the TACA motif of the target DNA. The crystal structure of MecI (Protein Data Bank [PDB] code 1OKR) determined recently by Garcia-Castellanos et al. (14) shows a dimer of two independent N-terminal winged helix domains and two intertwined C-terminal dimerization domains of spiral helical architecture. Further elucidation of interactions between the MecI dimer and the blaZ operator DNA (PDB code 1SAX) confirmed the involvement of helix α3 of the protein and the GTAXT motif of the DNA for specific recognition (13) (Fig. 2B). However, Garcia-Castellanos et al. (14) did not investigate the binding of MecI to its homologous target, the mec promoter-operator, and did not address the question of how related repressors could bind to two different targets. In this paper we report the crystal structures of BlaI from Staphylococcus aureus, both in free form and in complex with 32 bp of DNA of the mec operator, determined to 2.0- and 2.7-Å resolutions, respectively. Comparison of the BlaI and MecI structures suggests that the proteins may assume different quaternary conformations, a closed form and an open form. Based on the crystal structures, as well as data from DNase protection experiments, we propose a model for binding of the repressors to both bla and mec operators.
MATERIALS AND METHODS
Protein expression and purification.
The blaI gene was amplified by PCR from plasmid pCN with primers blaI-NcoI (5′-ACTGCCATGGGCCAATAAGCAAGTTGAAATATCTATGG-3′) and blaI-BamHI (5′-CTAGGGATCCCTAATTTAATAAGAGTCAAGC-3′) (restriction sites are underlined). The ca. 500-bp PCR products were digested with NcoI and BamHI, and the DNA fragments were cloned into pET3 (Novagen). This construct was transferred into Escherichia coli strain B834(DE3) (methionine auxotroph; Novagen) competent cells, and DNA sequencing was performed to confirm the appropriate orientation. BlaI was then overexpressed in the E. coli B834(DE3) in 6 liters of M9 defined medium (2 mM MgSO4, 0.4% glucose, 0.1 mM CaCl2, 1 g of thiamine hydrochloride per liter, 40 mg of all amino acids except methionine per liter, 100 μg of ampicillin per ml, and 40 mg of seleno-methionione [SeMet] per liter) at 37°C with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) as an inducer. The harvested cells were solubilized in 20 mM phosphate buffer (pH 6.8) by sonification. The crude lysate was precipitated with 60 to 90% ammonium sulfate. Following dialysis against 20 mM phosphate buffer (pH 6.8) overnight, the protein was purified with a Mono-S affinity column (Amersham-Pharmacia). About 15 mg of BlaI was obtained, with a purity of greater than 99% as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Crystallization of unbound BlaI and data collection.
Initial crystallization trials with the SeMet-incorporated BlaI (in 20 mM potassium phosphate buffer [pH 7.6] and 200 mM NaCl) were set up by using the hanging-drop vapor diffusion technique with Crystal Screens I and II from Hampton Research (Laguna Niguel, Calif.). After optimization, high-quality single crystals were obtained within a week, using 7 mg of protein per ml and precipitant solution containing 1.8 to 2.0 M ammonium sulfate and 10% glycerol. The crystals were flash cooled in a cryoprotectant solution containing 20 mM potassium phosphate [pH 7.4], 200 mM NaCl, 2.5 M ammonium sulfate, and 23% glycerol prior to X-ray data collection. Three-wavelength multiple anomalous dispersion data were collected on beamline x12c at the National Synchrotron Light Source at 100 K. The data were processed and scaled by using the HKL2000 package (32). Statistics are shown in Table 1.
TABLE 1.
Data collection and phasing parameters of SeMet BlaI and BlaI-DNA crystals
| Crystal parameter | Valuea for:
|
|
|---|---|---|
| SeMet BlaI | BlaI-DNA | |
| Space Group | P21212 | I4122 |
| Unit cell (a, b, c) | 64.41, 116.36, 40.17 | 72.1, 72.1, 243.5 |
| Data collection statisticsb | ||
| Wavelength (Å) | 0.9792, 0.9795, 0.9611 | 1.1000 |
| Resolution (Å) | 30.0-2.0 (2.07-2.00), 30-2.0 (2.07-2.00), 30.0-2.0 (2.07-2.00) | 30.0-2.7 (2.8-2.7) |
| No. of measurements | 288,556 289,048 285,421 | 127,462 |
| No. of unique reflections | 20,927 20,908 20,786 | 9,316 |
| Redundancy | 13.8 (10.8), 13.8 (10.6), 13.7 (10.3) | 13.7 (11.8) |
| Average I/σ(I) | 54.5 (4.2), 55.8 (4.3), 51.9 (3.6) | 44.0 (4.5) |
| Completeness(%) | 98.5 (95.5), 98.5 (95.6), 98.4 (94.5) | 100.0 (99.8) |
| Rmerge (%)c | 7.7 (48), 6.3 (47), 6.1 (51) | 6.5 (55) |
| MAD phasing stats | ||
| Resolution (Å) | 30-2.0 | |
| FOM before/after DMd | 0.65/0.69 | |
Numbers in parentheses refer to the outermost resolution bins.
For SeMet BlaI, the three values are peak, inflection, and remote, respectively.
Rmerge = Σ|〈I〉 − I|/ΣI.
DM, density modification.
Crystallization of the BlaI-DNA complex and data collection.
Several complementary oligonucleotides from the mec palindromic operator, including 24-, 26-, 28-, and 32-bp sequences, were used in the initial crystallization trials. X-ray-quality crystals were obtained only from the 32-bp sequence (5′-GACTACATTTGTAGTATATTACAAATGTAGTA-3′ and its complement 5′-TACTACATTTGTAATATACTACAAATGTAGTC-3′). To crystallize the complex, the double-stranded DNA at a concentration of 0.57 mM was mixed with BlaI solution (1.08 mM in 20 mM potassium phosphate [pH 7.6] and 200 mM NaCl) at a molar ratio of 1.5 to 1 and then incubated for 2 days at 4°C. After optimization, the best-diffracting BlaI-DNA cocrystals were obtained from drops containing 1.2 μl of protein-DNA solution and 1.2 μl of a reservoir that contained 12% polyethylene glycol 8000, 6% ethylene glycol, 10 mM MgCl2, and 100 mM Na HEPES [pH 7.5]. Before flash cooling in liquid nitrogen, crystals were soaked in solutions containing 100 mM Na HEPES [pH 7.5], 10 mM MgCl2, 20% polyethylene glycol 8000, and 10 to 30% ethylene glycol, reaching the final concentration of 30% ethylene glycol in three steps of 1 min each. An in-house data set was collected at 100 K for the BlaI 32-bp DNA cocrystals (3.0 Å), using a Molecular Structure Corp. (The Woodlands, Tex.) X-Stream cryogenic cooler system, an R-axis II image plate detector equipped with OSMIC confocal mirrors, and a Rigaku RU-200 generator operating at 50 kV and 100 mA. The data were processed with the Molecular Structure Corp. BIOTEX software program. Later, another data set for the same BlaI-DNA cocrystal (2.7 Å) was collected at the National Synchrotron Light Source at 100K. The intensity data were processed and scaled by using the HKL2000 package (32). Statistics are also shown in Table 1.
Determination and refinement of the structure of unbound BlaI.
The unbound BlaI crystallized in the orthorhombic space group of P21212. The Matthews coefficient of 2.73 Å3/Da (25) and water content of 54.5% were consistent with one dimer per asymmetric unit. Eight of the 10 expected selenium sites were found by using Solve Version 2.02 (38), and the phases were improved by use of maximum-likelihood density modification in Resolve (29, 37). An initial model consisting of 227 residues in four chains was built by using ARP/wARP version 6.0 (33, 39). After one cycle of manual fitting, two continuous polypeptide chains of 119 and 117 residues were obtained. Side chains were docked onto these polypeptides by using ARP/wARP, and the model that contained two monomers A and B was subsequently refined against the 2.0-Å data set (remote wavelength) by using the program CNS (5). Subsequent conjugate gradient minimization and simulated annealing resulted in an R value of 0.312 and an Rfree value of 0.345 (for 5% randomly selected data). Repeated refinement with CNS and model building yielded a model that included residues 5 to 126 for each monomer, 4 sulfate molecules, and 225 water molecules, and further refinement with REFMAC (29) led to final R and Rfree of values 0.203 and 0.237, respectively, for all reflection in the resolution range of 30 to 2.0 Å.
Determination and refinement of the structure of the BlaI-DNA complex.
The BlaI-DNA complex crystal belongs to the tetragonal space group of I4122. Molecular replacement with AMoRe (30) using the unbound BlaI dimer and the in-house data failed. Instead, using the MecI model of 1OKR, a solution was obtained with a final correlation coefficient of 0.28 and an R factor of 0.52 after rigid-body refinement. The next peak has a corresponding correlation coefficient of 0.25 and an R factor of 0.55. The asymmetric unit contains one monomer, constituting about 23% of the unit cell volume. The molecular dyad axis coincides with the crystallographic twofold axis. The initial Fourier map showed good densities for the protein and some extra densities near the N-terminal domain, presumably for the DNA. By using CNS and the in-house data, the model was refined to 3.0 Å, with R and Rfree values of 0.43 and 0.49, respectively.
A composite map was then calculated by density modification, with a figure of merit (FOM) of 0.36. This map showed stronger densities for the DNA, and an 8-bp model of arbitrary sequence was constructed. The model was also modified by substitution of all side chains for BlaI and inclusion of 80 water molecules. This model was refined to R and Rfree values of 0.28 and 0.35, respectively, and the FOM was 0.69. When the synchrotron data set was used, it gave R and Rfree values of 0.27 and 0.33, respectively, at 2.7 Å for all reflections. The new map showed clear densities for the DNA, and the DNA sequence was modified accordingly. The refinement proceeded with minor adjustments of the loop regions and addition of more water molecules. It concluded with a BlaI model of residues 2 to 126, a single-stranded DNA of 16 nucleotides, and 181 water molecules, with final R and Rfree values of 0.230 and 0.282, respectively. Figure 2C and D show the initial composite omit and final 2Fo-Fc electron density maps, respectively, at the protein-DNA interface.
Manual adjustments of the models were made with the programs TOM (6) and O (18). O was also used in model comparison and crystal packing analyses. Surface areas and cavities were examined by using CCP4 (10) and GRASP (31). All color figures were produced by using MOLSCRIPT (20) and Raster3D (28), except Fig. 2A, which was made by using ALSCRIPT (3), and Fig. 2C and D, which were made by using BOBSCRIPT (11).
Protein structure accession numbers.
The structure factors and atomic coordinates of the unbound BlaI and its DNA complex have been deposited in the Protein Data Bank with accession codes 1SD4 and 1XSD, respectively.
RESULTS
Overall structures of BlaI and the BlaI-DNA complex.
The crystal structure of a dimeric BlaI in the asymmetric unit was determined to 2.0-Å resolution by the MAD-phasing technique with an orthorhombic SeMet derivative crystal. Refinement statistics are shown in Table 2. The high-temperature factors of the sulfate molecules in the BlaI structure are due to partial occupancies. The BlaI and MecI proteins share 76% sequence similarity, with 61% strictly conserved (Fig. 2A), and as expected, the two proteins share a very similar fold, except there is an additional β-strand between helices α1 and α2 in BlaI. Each monomer has α1-β1-α2-α3-β2-wing-β3-α4-α5-α6 fold topology and consists of N-terminal and C-terminal domains. The N-terminal residues before Gln5 are disordered and not visible in either subunit, whereas the C terminus continues to Lys126 in both subunits. The dimeric structures of the BlaI and MecI proteins are triangular, as shown in Fig. 3. The C-terminal domains (helices α4-α5-α6) from both monomers are spirally intertwined at one of the vertices of the triangle to hold the dimer together. The N-terminal domains (α1-β1-α2-α3-β2-wing-β3), which are disposed at the other two vertices, are very compact, with extensive hydrophobic interactions in the core regions. The N-terminal domain has been classified as a winged helix DNA binding motif based on structure prediction, with helix α3 as the DNA recognition helix (9). Such winged helix architecture has also been identified in a number of DNA binding proteins (see reference 12 for a comprehensive review).
TABLE 2.
Refinement statistics for the SeMet BlaI and BlaI-DNA crystals
| Crystal parameter | Valuea for:
|
|
|---|---|---|
| SeMet BlaI | BlaI-DNA | |
| Resolution limits (Å) | 30-2.0 (2.05-2.00) | 30-2.7 (2.8-2.7) |
| No. of reflectionsb | 19,686 (1,414) | 9,023 (822) |
| R for 95% working data set | 0.203 (0.277) | 0.230 (0.405) |
| Rfree for 5% test data set | 0.237 (0.380) | 0.282 (0.528) |
| RMSD from ideal geometry | ||
| Bond lengths (Å) | 0.017 | 0.016 |
| Bond angles (°) | 1.4 | 1.7 |
| Average B values (Å2)/no. of atoms | ||
| Protein atoms | 33.6/2,036 | 52.1/1,042 |
| Sulfate/DNA atoms | 70.5/20 (sulfate) | 52.9/328 (DNA) |
| Water atoms | 49.1/225 | 61.6/181 |
| Ramachandran plot, % of residues: | ||
| In most favored regions | 91.4 | 81.8 |
| In additional allowed | 8.1 | 17.4 |
| Estimated coordinate errors (Å) | 0.19 | 0.36 |
Numbers in parentheses refer to the outermost resolution bins.
All reflections were used in the refinements.
FIG. 3.
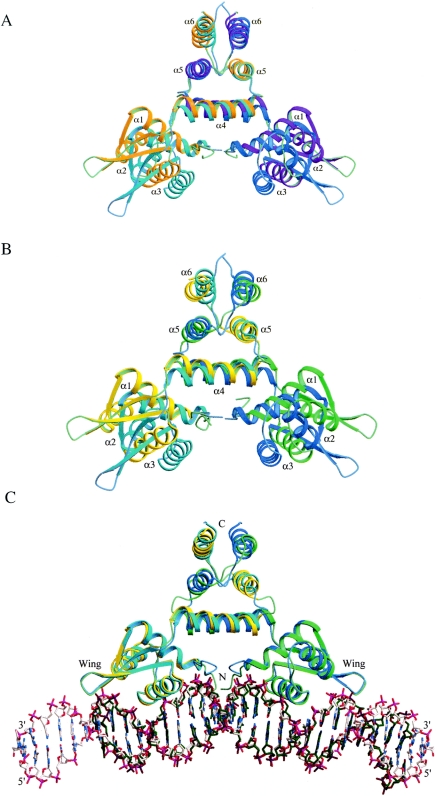
Structural comparison. (A) Superposition of unbound BlaI and DNA-bound BlaI dimers. (B) Superposition of unbound BlaI and unbound MecI dimers (1OKR). (C) Superposition of DNA-bound BlaI and DNA-bound MecI (1SAX) complexes. The structures were superimposed as described in the text. Protein molecules are shown as ribbons, and DNA molecules are shown as stick models in which the carbon atoms are in white and green for the DNA bound to BlaI and MecI, respectively. In (A), the unbound BlaI monomers are in cyan and blue, whereas the DNA-bound BlaI monomers are in orange and magenta. In (B) and (C), the two BlaI monomers are in cyan and blue, whereas MecI is in yellow and green.
The protein model of the BlaI-DNA cocrystal contains residues Thr2 to Lys126. The two BlaI monomers in a dimer are related by a crystallographic dyad axis parallel to the c axis. Each BlaI monomer is presumed, based on symmetry constraints, to bind to a crystallographically identical site on the DNA. The refined DNA model comprises a single-stranded 16-base palindrome, TACTACATATGTAGTA, of residues Thy201 to Ade216 that contains a phosphate group at the 5′ end, equivalent to a double-stranded TACTACAT/ATGTAGTA DNA with 8 bp, shown in Fig. 2B. However, the DNA used in crystallization contains 32 bp, and each one-fourth of the DNA should be equivalent due to the symmetry of the tetragonal crystal. Therefore, the single-stranded DNA model of 16 nucleotides represents an average of the four different but similar half-length DNA strands of GACTACATTTGTAGTA, TATTACAAATGTAGTA, TACTACATTTGTAATA, and TACTACAAATGTAGTC. In the BlaI-DNA cocrystal, the tetragonal unit cell contains eight BlaI dimers in which all of the monomers have identical conformation. The DNA forms a virtual double helix throughout the unit cell (see below). The corresponding electron densities for all base pairs are clear, consistent with most of the sequences. Nevertheless, this model is not perfect, especially in the central TA base pairs, which could also be the reverse. Disorders in DNA bound to proteins have been observed before in a number of other crystal structures, including in the recent work on KorB, a member of the ParB family of bacterial partitioning proteins (19).
Comparison of BlaI structures.
When the dimer structures of the unbound and DNA-bound BlaI are superimposed, the root mean square deviation (RMSD) calculated for all 2,036 equivalent atoms is 4.5 Å. This significant structural difference is mostly due to a large variation in the orientations of the N- and C-terminal domains in the two structures. Superposition of 71 pairs of Cα atoms in the N-terminal domains (residues 5 to 75) of individual unbound and DNA-bound BlaI monomers are 0.75 and 0.70 Å, respectively, resulting in a rotation of 15.6 and 24.8°, respectively, of the C-terminal domains. Superposition of the 98 pairs of Cα atoms in the C-terminal domains (RMSD of ∼0.85 Å) also results in similar N-terminal domain rotations. The large conformational changes occur as a result of a hinge-bending movement, which is pivoted at the domain linker connecting strand β3 to helix α4 (residues Ile73, Asn74, and Glu75). Figure 3A shows the conformational differences of the N-terminal domains when the C-terminal domains of the unbound BlaI and DNA-bound BlaI dimers are superimposed, which clearly indicate that the N-terminal domains in the unbound BlaI are closer to each other than those of the bound protein. The rotations are about two axes perpendicular to the molecular dyad axis. The two BlaI structures in the unbound and DNA-bound forms thus assume different quaternary conformations: a closed form and an open form, respectively. Unlike the case for BlaI, it has been reported that the conformation of MecI bound to DNA is identical to that of the unbound protein (13). Such conformational flexibilities and hinge-bending movements are also observed in the bacterial transcriptional regulators MexR (22) and MtaN (15), which share structural similarity with BlaI and MecI.
The average B values are about 34 and 52 Å2 for the BlaI molecules in the orthorhombic and tetragonal crystals, respectively (Table 2). In the orthorhombic crystal the average B values for both the N-terminal domains (residues 5 to 73) and C-terminal domains (residues 74 to 126) show no significant variation from each other. However, in the tetragonal crystal, the average B values for the N- and C-terminal domains are about 47 and 56 Å2, respectively. In general, the C-terminal domain seems to become more flexible than the N-terminal domain when the repressor is bound to DNA. Similar results can also be obtained by analyzing the structures of MecI (PDB codes 1OKR and 1SAX).
Comparison of BlaI and MecI structures.
Interestingly, we observed similar conformational differences (as described above for the unbound and DNA-bound BlaI) when the unbound BlaI was compared with both the unbound MecI (PDB code 1OKR) and the MecI complexed to DNA (PDB code 1SAX). The RMSDs in coordinates calculated for all 237 pairs of equivalent Cα atoms in the two respective dimers are 4.76 and 4.65 Å. Superposition of the C-terminal domains of individual monomers of unbound BlaI with unbound and bound MecI also results in extensive rotation of the N-terminal domains, as previously described for the two BlaI structures. Figure 3B shows the conformational differences of the N-terminal domains when the C-terminal domains of the unbound BlaI and unbound MecI dimers are superimposed, which clearly indicate that the unbound MecI is in an open conformation.
Figure 3C shows the dimeric structures of BlaI and MecI (PDB code 1SAX) in complexes with DNA when superimposed, which indicates that both repressors bind DNA with similar open conformations. It should be noted that the targets to which the repressors are bound are different. BlaI is bound to the mec promoter-operator, while the published data on MecI have it bound to the bla promoter-operator (13). In Fig. 3C the DNA molecule bound to BlaI has been extended by crystallographic symmetry to encompass that bound to MecI. The RMSD for all 240 pairs of equivalent Cα atoms in the dimeric proteins is 1.51 Å. The 48 C1′ atoms in the 24 bp of 1SAX have an RMSD of 2.02 Å from the equivalents in the BlaI-DNA complex. The more bent conformation of the unbound DNA segment in the MecI-DNA complex may be caused by lattice contact of the DNA molecule with neighboring symmetry-related protein molecules or by the different combinations of the repressor protein and target DNA sequence. Analysis of the DNA bound to BlaI dimer by using 3DNA (23) shows that most base pairs have the B-DNA conformation, with only limited deviations in those covered by the wings.
The different structures of unbound and DNA-bound BlaI may explain why the molecular replacement search failed to yield a correct solution for the BlaI-DNA complex crystal but had to use the MecI model of 1OKR. We have also determined two different crystal structures of unbound MecI at 2.65-Å resolution (PDB codes 1SD6 and 1SD7). They belong to the same orthorhombic space group of P212121 but have slightly different cell dimensions than 1OKR. Some variations of the rotation angle between the N- and C-terminal domains are seen in all of the MecI structures; however, they are all in an open conformation.
The closed conformation observed in the unbound BlaI structure is stabilized by strong interactions between helices α6, as well as by a unique salt bridge between Asp14 and Lys89. On the other hand, the open conformation observed in the MecI structures and the DNA-bound BlaI structure is stabilized by interactions between Lys79 and Val90, Asp77 and Lys105, Ser76 and Lys105, Ser76 and Asn101, Tyr80 and Glu106, Ser83 and Tyr91, and Lys84 and Glu106 across the helix α4 and α5 dimer interface. It is likely that the free forms of BlaI and MecI can exist in closed and open conformations, due to the fact that the residues involved in stabilizing either conformation are highly conserved in both repressors. Consistent with this conclusion is the fact that the conformational differences between the unbound BlaI and DNA-bound BlaI, as well as the MecI structures, are also observed even between the two monomers in the unbound BlaI, as well as those of the MecI structures, although on a much smaller scale. In addition, the two MecI structures of 1SD6 and 1SD7 show a gradation of open conformation to closed conformation, which could be attributed to differences in the overall strength of the interactions of the helices α4 and α5. The existence of both open and closed conformations reflects the intrinsic flexibility of the repressors, which has been addressed in the NMR studies of BlaI (27). A crystal packing effect may have resulted in the crystallization of the closed and open forms of unbound BlaI and MecI, respectively.
Protein-protein and protein-DNA interactions.
The dimer interface of the unbound BlaI in the orthorhombic crystal buries 2,150 and 2,130 Å2 of the 9,540- and 9,260-Å2 surface areas of the two monomers, A and B, respectively. In the bound tetragonal crystal it buries 2,000 Å2 of the 9,640-Å2 surface, which is typical of homodimeric proteins (2). A similar repertoire for dimerization of BlaI is employed in both crystal structures, either alone or in complex with DNA. As described for MecI (14), most of the interactions are between the three C-terminal α-helices, and part of helix α1 is also involved. Besides, in the orthorhombic crystal, the free carboxyl group of the C-terminal Lys126 forms a salt bridge with the side chain of Arg117 in the counter monomer. The side chain of Lys38 in monomer A is hydrogen bonded to that of Gln5 in monomer B. These additional interactions are not seen in the tetragonal crystal structure, because the two N-terminal domains are farther apart.
Cavities in the associated dimerization domains have been observed in MecI (14), which may account for certain flexibility. In the orthorhombic BlaI crystal structure, there are two dyad-related cavities of 23 and 24 Å3 between helices α4 and α5. They are surrounded by the hydrophobic side chains of Ala83, Phe86, Leu87, Met95, Leu98, and Phe102 from both monomers. In the tetragonal crystal, these two cavities merge into one, with a volume of 45 Å3, but are contributed by the same residues. There are three additional cavities in the tetragonal crystal between helices α5 and α6. Two of them, each having a volume of 42 Å3, are related by the molecular dyad and are formed by Val99, Ala103, Leu108, Ile113, Leu116, Leu120*, and Ile123*, where * indicates residues from the counter subunit. The third cavity of 35 Å3 is located on the dyad axis, formed by Met95, Val99, Leu116, and Leu120 from both monomers. Unlike the unbound BlaI molecule, the helices α4, α5, and α6 of the DNA-bound BlaI molecule show significant differences in their average B values, which are 37, 52, and 77 Å2, respectively. These are consistent with the additional cavities in the C-terminal domain observed in the DNA-bound form. As shown in Fig. 3, the helices have slightly different dispositions in the bound and unbound forms of BlaI, which is also true for MecI. As discussed below, such flexibility may be related to the cleavage site of Asn101-Phe102 located in helix α5.
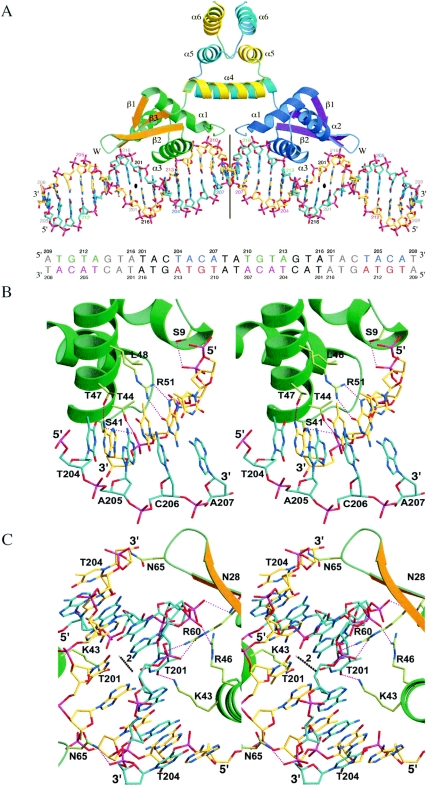
By crystallographic symmetry operators, the DNA model forms a virtual double helix in the BlaI-DNA tetragonal crystal, but it is actually made of the 32-bp DNA molecules joined end to end by base-stacking interactions. When the repressor binds to the DNA, the buried areas are 830 Å2 on both the BlaI monomer and on the DNA, comprising only about 11% of their surfaces. However, this small interface contains a number of specific interactions, which are listed in Table 3. Figure 4A shows the overall contact between BlaI and the DNA; Fig. 4B and C show detailed interactions between two regions of the protein-DNA interface. In the first region, helix α3 of the BlaI protein interacts with the conserved motif TACA/TGTA of the DNA. In the second region it interacts with the distal ATA/TAT sequence as well as the DNA dyad. In addition to the hydrogen bond between the side chain OG atom of Ser9 and the phosphate of Gua211 shown in Fig. 4B, the backbone N atoms of Met10 and Ala11 also form two hydrogen bonds with the same phosphate group. These two residues are located at the N-terminal end of helix α1, which contains a positive dipole directed toward the phosphate group of DNA. The observed DNA contact by helix α1 is consistent with studies that show the importance of the first eight N-terminal residues of B. licheniformis BlaI in binding to DNA (40) but does not explain why Lys4 is also found to be important in S. aureus BlaI (16). From the helix α3, the side chain OG and OG1 atoms of Ser41 and Thr44 are hydrogen bonded to the phosphate group of Ade213. The side chain of Arg51 (NH1 and NH2) makes sequence-specific hydrogen bonds with O6 and N7 of Gua211. In addition, OG1 of Thr47 forms a purine-specific hydrogen bond with Ade213. There is a hydrophobic interaction between the methyl group in the base of Thy212 and a pocket formed by the side chains of Thr44, Thr47, and Leu48.
TABLE 3.
Specific bonds between BlaI and DNA
| Protein | Atom 1 | DNAa | Atom 2 | Distance | Commentsb |
|---|---|---|---|---|---|
| Ser9 | OG | Gua211 | O2P | 3.18 | H.B., helix α1-TACA/TGTA |
| Met10 | N | Gua211 | O1P | 3.29 | H.B., helix α1-TACA/TGTA |
| Ala11 | N | Gua211 | O1P | 2.91 | H.B., helix α1-TACA/TGTA |
| Asn28 | ND2 | Ade216+ | O1P | 3.18 | H.B., helix α2-dyad strand |
| Ser41 | OG | Ade213 | O2P | 3.08 | H.B., helix α3-TACA/TGTA |
| Lys43 | NZ | Thy201* | O4 | 2.61 | H.B., helix α3-opposite distal strand |
| Thr44 | OG1 | Ade213 | O2P | 2.97 | H.B., helix α3-TACA/TGTA |
| Arg46 | NH1 | Thy201* | O1P | 2.74 | S.B., helix α3-opposite distal strand |
| Thr47 | OG1 | Ade213 | N6 | 2.85 | H.B., helix α3-TACA/TGTA |
| Arg51 | NH1 | Gua211 | N7 | 2.83 | H.B., helix α3-TACA/TGTA |
| Arg51 | NH2 | Gua211 | O6 | 2.87 | H.B., helix α3-TACA/TGTA |
| Arg60 | NH1 | Ade202* | O2P | 3.14 | S.B., strand β2-opposite distal strand |
| Arg60 | NH2 | Ade202* | O1P | 2.93 | S.B., strand β2-opposite distal strand |
| Asn65 | ND2 | Thy204+* | O1P | 2.73 | H.B., wing-opposite dyad strand |
| Tyr67 | N | Thy201* | O2P | 3.06 | H.B., strand β3-opposite distal strand |
The interactions are centered at the DNA strand that contains the TGTA motif, whereas * indicates the opposite strand that contains the TACA motif. In the distal region, the protein also interacts with the dyad-related DNA strands, indicated by + and +*.
S.B. and H.B., salt bridges and hydrogen bonds, respectively.
FIG. 4.
Interaction between BlaI and DNA. (A) Overall view of BlaI bound to DNA. (B) Specific interactions between BlaI and DNA in the region of the TACA/TGTA motif. (C) Interactions in the distal dyad region of DNA. The proteins are shown as ribbons, in green, orange, and yellow, respectively, for the N-terminal helices, the β-strands, and the C-terminal helices of one monomer and in blue, magenta, and cyan, respectively, for those of another monomer. The DNA molecules and the specific amino acid side chains are shown as sticks, with carbon bonds in yellow and cyan for different DNA strands and in pale green for the protein. Hydrogen bonds are shown as strings of red beads. In (A) the DNA sequence is shown in colors and the model is labeled with the same colors. The dyad axes of DNA are indicated by a line (proximal dyad) and a bullet (distal dyad). In (C) the distal DNA dyad axis is represented as a chain of alternating black and white beads.
In Fig. 4C, the ND2 atom of Asn28 from the helix α2 makes hydrogen bond interaction with O1P of Ade216 in the opposite strand of DNA. The NZ atom of Lys43 from the helix α3 is hydrogen bonded to the O4 atom of the base of Thy201 in the opposite strand of DNA, and so is the NH1 atom of Arg46 salt bridged to the O1P of the phosphate of the same nucleotide, Thy201. The side chain of Arg60 forms a bidentate salt bridge with the phosphate of Ade202 in the opposite strand. The backbone N atom of Tyr67 forms a hydrogen bond with the O2P of Thy201 in the opposite strand (not shown). These two residues are in strands α2 and α3 of the wing of BlaI. Across the crystallographic dyad axis, the side chain ND2 atoms of Asn28 and Asn65 in the α2 helix and the α2-α3 loop also form hydrogen bonds to the phosphate groups of Ade216 and Thy204 of the opposite DNA strand, respectively. Most of the above-mentioned residues were identified as being important in binding DNA in the NMR study of BlaI (27). There are at least six water molecules located at the interface between the BlaI monomer and DNA. However, none of them is involved in specific hydrogen bonding with both BlaI and DNA. Presumably, for high selectivity and affinity, BlaI must bind specifically to the operator, and the interactions between the protein and DNA molecules should be direct, i.e., with no mediating solvents.
The crystallographic analysis also explains why the missense mutations A2T, G21D, Y37N, S63Y, E64N, and N72I, identified in BlaI among wild-type isolates, have no effect on repressor function (4). Even though all of the mutant residues are located in the N-terminal domain, none is capable of making any contact with DNA. Corresponding residues in MecI also make no contact with DNA and are unlikely to have any effect on repressor function if mutated.
DISCUSSION
Models for repressor binding to DNA.
Any model proposed to explain the mechanism of repression by BlaI and MecI must take into account the ability of each repressor to bind to both mec and bla operator sequences and of each repressor to protect the same DNA sequences in each target. DNase I footprint studies show that BlaI and MecI bind to the bla operator, equally protecting 24- and 25-bp Z and R1 dyad sequences, respectively, with 12 bp of unprotected DNA between the two dyads (8). The repressors also bind to the mec operator, protecting a single 43- to 46-bp sequence that contains two 15-bp half-sites, each of which has 12 bp of dyad symmetry (36). The model should also account for the fact that binding to the mec operator results in oligomerization of molecules, as well as cooperative DNA binding (36).
Garcia-Castellanos et al. (13) solved the crystal structure of MecI bound to the bla promoter-operator, while we have solved the structure of BlaI bound to mec DNA sequences. Since the two repressors bind and protect the same DNA sequence in each of the two operons, they are likely to assume the same quaternary structure at each binding site. In addition, the DNA binding mode of BlaI should be very similar to that of MecI. All of the residues that interact directly with DNA are conserved in the two repressors, except Asn65 in BlaI, which is replaced by Lys65 in MecI. Finally, we have solved the structure of MecI bound to mec DNA at a low resolution (3.6 Å), showing that this repressor assumes a conformation and DNA binding mode similar to those of BlaI bound to these sequences (M. K. Safo et al., unpublished observations). Thus, we will assume the same binding model for both proteins at each site.
Although both proteins are flexible in solution, capable of conformational changes between open and close forms, conformational adaptation is not a likely explanation for the ability of the two repressors to bind to different DNA targets. There are 24 bp between two phosphate groups of Thy204 bound to the side chains of Asn65 (Lys65 in MecI) from the two wings of either BlaI or MecI dimer bound to either the mec or bla operator, as shown in Fig. 3C. The close conformation would contact only 18 bp of DNA, and therefore both repressors must bind to both DNA targets in the open conformation, consistent with the crystal structures of both BlaI and MecI complexed to DNA. Moreover, a DNA-docking experiment using the closed BlaI conformation does not show optimal repressor-DNA binding because of steric contacts between the recognition helices and the DNA dyad axis. It is interesting that the crystal structure of MexR has four dimers in the asymmetric unit and one has significantly shorter spacings between the DNA binding domains than the others (22). Docking of this dimer onto DNA also resulted in steric contacts with the DNA, although more severe than those observed with the closed BlaI structure.
What is not obvious is whether a closed or open form of BlaI initiates binding to DNA or what triggers the transition from a closed to an open conformation. It has been suggested that an effector modulates the closed and open conformations in MexR and that only the open form is capable of binding DNA (22). A different mechanism is proposed for MtaN, where the closed form is believed to first bind to DNA, followed by structural adjustment that induces formation of the open form to optimally bind the DNA (15). No effector has been reported for either BlaI or MecI. Perhaps, as proposed for MtaN, the closed form of BlaI first binds to a B-like DNA, followed by breaking of the salt bridge between Asp14 and Lys89, which would allow additional structural and conformational changes of the repressor to assume an open form to maximize its DNA contacts. This assumes that free BlaI occurs entirely in a closed form. On the other hand, if free BlaI and MecI can occur in both a closed and an open form, as suggested earlier, then both conformations must coexist in solution. In this case, the open form may preferentially bind DNA as it is in a more favorable DNA binding conformation.
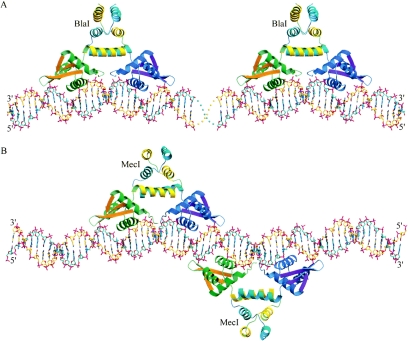
Since the separation between the 18-bp DNA dyads of the adjacent R1 and Z palindromes in bla operators is 13 bp (Fig. 1A), there would be no steric conflict if two dimers of the repressors bind to the same side of the DNA double helix (Fig. 5A). This model is consistent with the 24 to 25 bp of DNA of each of the two bla operators protected by the repressors (8). However, at the mec operator the spacing between the centers of each of the two local dyads that contain the TACA/TGTA binding motif is only 16 bp (Fig. 1B), equivalent to about 1.5 turns of B-DNA helix. Thus, unlike the bla operator, two repressors should bind to the opposite sides of the DNA to avoid steric contacts. This model, in which BlaI dimers bind to DNA in an up-and-down arrangement, was confirmed by our tetragonal crystal structure, in which the DNA employed in cocrystallization with BlaI corresponded to the exact sequence of the mec operator (Fig. 5B). This is corroborated by the low-resolution structure of MecI bound to mec DNA, which also shows two repressor dimers bound to the mec operator in an up-and-down arrangement (Safo et al., unpublished observations). This model can also explain the single 43- to 46-bp DNA sequence protected from DNase I digestion, because the separation between the left wing of the upper dimer and the right wing of the lower dimer is 40 bp. The cooperative DNA binding observed in an earlier study (36) could be due to perturbation of the DNA double helix by binding of the first dimer, making the opposite side of the DNA more accessible to a second dimer. Specifically, interactions across the dyad axis may produce slight deviations from the canonical B-form DNA in this region. Such cooperative binding has also been observed in the diphtheria toxin repressor, DtxR (34), and multidrug binding protein, QacR (35). In contrast, the cooperative binding exhibited by lambda repressors is due to direct protein-protein interactions. Lambda DNA dyads are spaced about 20 bp apart, so the repressors can bind to the same side of the double helix (21).
FIG. 5.
Models of repressor bound to operator DNA. (A) Proposed model of two repressor dimers bound to the bla Z and R1 dyads. (B) Proposed model of two repressor dimers bound to the mec operator. The protein models are shown as ribbons, in green, orange, and yellow, respectively, for the N-terminal helices, the β-strands, and the C-terminal helices of one monomer and in blue, magenta, and cyan, respectively, for those of another monomer. In (A), the two BlaI repressors bind to the same side of DNA and are well separated, consistent with the lack of cooperativity when bound to the bla operator. In (B), the two MecI repressor dimers bind to opposite sides of the mec operator as observed in the tetragonal crystal. This avoids steric contact between the repressors and is consistent with the protection of a single 43- to 46-bp sequence within the mecA regulon, as well as cooperativity. Cooperative binding of MecI to opposite sides of DNA may occur via the protein-DNA-protein interactions through the dyad axis in the center.
Implications for repressor cleavage.
The proteolytic cleavage site of Asn101-Phe102 is highly conserved among the members of the BlaI-MecI family. Interestingly, the cleavage site, which is located in the middle of helix α5, is not easily accessible to the exterior. It has been suggested that another, yet-unidentified protein, BlaR2, may be an intermediary in the cleavage process (1). It could act by unwinding the helix into a less-ordered conformation to allow access to a BlaR1- or MecR1-associated protease. There are many precedents for protease directly cleaving buried α-helix. The membrane-embedded sequence of site 2 protease (a zinc metalloprotease) is believed to unwind an α-helix of the membrane-bound sterol regulatory element binding protein into a less-ordered structure to facilitate proteolytic cleavage (41). Calcium binding in calmodulin is also known to induce flexibility in the central helix of calmodine for proteolytic attack (24).
The observed flexibility in the C-terminal domain and especially in helix α5 of BlaI as well as MecI could therefore be essential for proteolysis and may suggest molecular models that explain the C-terminal proteolytic cleavage, mediated by the BlaR1 or MecR1 receptors, that leads to derepression. Because the TACA/TGTA motifs are separated by 2 bp at the dyad, when the repressors are bound to the DNA operators, the N-terminal domains should adopt the same interdomain spacing. However, the C-terminal domains still retain mobility, and a dynamic interchange between several different conformations should make the interior of helix α5 exposed and accessible for binding to other proteins. The four cavities observed between helices α4 and α5 (one cavity) and helices α5 and α6 (three cavities) in the DNA-bound BlaI dimer, compared to two between helices α4 and α5 in the unbound BlaI, indicate that the dimer interface may contain more faults when bound to the DNA, making the repressor more prone to protease attack at the specific site of Asn101-Phe102 in helix α5. This would suggest that derepression takes place while BlaI (or MecI) is bound to the operator rather than free in solution. The precise mechanism is currently under investigation.
Conclusions.
On the basis of the BlaI-mec DNA structure presented here, the reported MecI-bla DNA structure, and the virtual identity of the binding domains of the two repressors, we have extrapolated from the repressor-heterologous binding site data to establish a model of binding of both repressors to their cognate DNA operator sites. The orthorhombic and tetragonal crystal structures of BlaI show closed and open conformations of the unbound and DNA-bound repressor dimers, respectively, displaying the flexibility of the molecule due to hinge bending at Ile73-Glu75, the junction between the N-terminal binding and the C-terminal dimerization domains. This flexibility, as well as cavities within the C-terminal dimerization domain, may allow the repressors to interact with other regulator proteins, including BlaR1 and MecR1.
The protected numbers of base pairs in the operator DNA are explained by the wingspan and the up-and-down binding mode of the repressors. The cooperativity and noncooperativity of MecI and BlaI binding mechanisms, respectively, to their cognate operators are also proposed based on the crystal structures (Fig. 5). The MecI-mec model shows cooperative binding of regulatory protein to DNA mediated by protein-DNA interactions rather than direct protein-protein contacts.
Acknowledgments
This work was supported by NIH grant AI035705-11. Data for this study were measured at beamline x12c of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and from the National Center for Research Resources of the National Institutes of Health.
REFERENCES
- 1.Archer, G. L., and J. M. Bosilevac. 2001. Signaling antibiotic resistance in staphylococci. Science 9:1915-1916. [DOI] [PubMed] [Google Scholar]
- 2.Bahadur, R. P., P. Chakrabarti, F. Rodier, and J. Janin. 2003. Dissecting subunit interfaces in homodimeric proteins. Proteins 53:708-719. [DOI] [PubMed] [Google Scholar]
- 3.Barton, G. J. 1993. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 6:37-40. [DOI] [PubMed] [Google Scholar]
- 4.Brown, D. F., and P. E. Reynolds. 1980. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 122:275-278. [DOI] [PubMed] [Google Scholar]
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Cambillau, C., and E. Horjales. 1987. TOM: a Frodo subpackage for protein-ligand fitting with interactive energy minimization. J. Mol. Graph. 5:174-177. [Google Scholar]
- 7.Clarke, S. R., and K. G. Dyke. 2001. The signal transducer (BlaRI) and the repressor (BlaI) of the Staphylococcus aureus beta-lactamase operon are inducible. Microbiology 147:803-810. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, S. R., and K. G. H. Dyke. 2001. Studies of the operator region of the Staphylococcus aureus beta-lactamase operon. J. Antimicrob. Chemother. 47:377-389. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412-420. [DOI] [PubMed] [Google Scholar]
- 10.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 11.Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. 15:132-134. [DOI] [PubMed] [Google Scholar]
- 12.Gajiwala, K. S., and S. K. Burley. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10:110-116. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Castellanos, R., G. Mallorqui-Fernandez, A. Marrero, J. Potempa, M. Coll, and F. X. Gomis-Ruth. 2004. On the transcriptional regulation of methicillin resistance: MecI repressor in complex with its operator. J. Biol. Chem. 279:17888-17896. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Castellanos, R., A. Marrero, G. Mallorqui-Fernandez, J. Potempa, M. Coll, and F. X. Gomis-Ruth. 2003. Three-dimensional structure of MecI. Molecular basis for transcriptional regulation of staphylococcal methicillin resistance. J. Biol. Chem. 278:39897-39905. [DOI] [PubMed] [Google Scholar]
- 15.Godsey, M. H., N. N. Baranova, A. A. Neyfakh, and R. G. Brennan. 2001. Crystal structure of MtaN, a global multidrug transporter gene activator. J. Biol. Chem. 276:47178-47184. [DOI] [PubMed] [Google Scholar]
- 16.Gregory, P. D., R. A. Lewis, S. P. Curnock, and K. G. Dyke. 1997. Studies of the repressor (BlaI) of beta-lactamase synthesis in Staphylococcus aureus. Mol. Microbiol. 24:1025-1037. [DOI] [PubMed] [Google Scholar]
- 17.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with beta lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 19.Khare, D., G. Ziegelin, E. Lanka, and U. Heinemann. 2004. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 11:656-663. [DOI] [PubMed] [Google Scholar]
- 20.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 21.Lewin, B. 2000. Genes VII, p. 333-339. Oxford University Press, Oxford, United Kingdom.
- 22.Lim, D., K. Poole, and N. C. Strynadka. 2002. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Biol. Chem. 277:29253-29259. [DOI] [PubMed] [Google Scholar]
- 23.Lu, X. J., and W. K. Olson. 2003. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31:5108-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackall, J., and C. B. Klee. 1991. Calcium-induced sensitization of the central helix of calmodulin to proteolysis. Biochemistry 30:7242-7247. [DOI] [PubMed] [Google Scholar]
- 25.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491-497. [DOI] [PubMed] [Google Scholar]
- 26.McKinney, T. K., V. K. Sharma, W. A. Craig, and G. L. Archer. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate MecA and beta-lactamase regulators. J. Bacteriol. 183:6862-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melckebeke, H. V., C. Vreuls, P. Gans, P. Filee, G. Llabres, B. Joris, and J. P. Simorre. 2003. Solution structural study of BlaI: implications for the repression of genes involved in beta-lactam antibiotic resistance. J. Mol. Biol. 333:711-720. [DOI] [PubMed] [Google Scholar]
- 28.Merrit, E. A., and M. E. P. Murphy. 1994. Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. D 50:869-873. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 30.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. D 50:157-163. [Google Scholar]
- 31.Nicholls, A., K. A. Shart, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307-326. In C. W. Carter and R. M. Sweet (ed.), Methods in enzymology: macromolecular crystallography. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 33.Perrakis, A., T. K. Sixma, K. S. Wilson, and V. S. Lamzin. 1997. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. D 53:448-455. [DOI] [PubMed] [Google Scholar]
- 34.Pohl, E., R. K. Holmes, and W. G. Hol. 1998. Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR. J. Biol. Chem. 273:22420-22427. [DOI] [PubMed] [Google Scholar]
- 35.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, V. K., C. J. Hackbarth, T. M. Dickinson, and G. L. Archer. 1998. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J. Bacteriol. 180:2160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger, T. C. 2001. Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr. D 57:1755-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Asselt, E. J., A. Perrakis, K. H. Kalk, V. S. Lamzin, and B. W. Dijkstra. 1998. Accelerated X-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP. Acta Crystallogr. D 54:58-73. [DOI] [PubMed] [Google Scholar]
- 40.Wittman, V., H. C. Lin, and H. C. Wong. 1993. Functional domains of the penicillinase repressor of Bacillus licheniformis. J. Bacteriol. 175:7383-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye, J., U. P. Dave, N. V. Grishin, J. L. Goldstein, and M. S. Brown. 2000. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl. Acad. Sci. USA 97:5123-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science 291:1962-1965. [DOI] [PubMed] [Google Scholar]