Abstract
A method for the detection of infectious human rotaviruses based on infection of CaCo-2 cells and detection of infected cells by indirect immunofluorescence and flow cytometry (IIF-FC) has been developed. The technique was validated by performing a seminested reverse transcription-PCR assay with sorted cell populations. The efficiency of the procedure has been compared with that of the standard method of infection of MA104 cells and ulterior detection by IIF and optical microscopy (IIF-OM) and with that of infection of MA104 cells and detection by IIF-FC. The limit of sensitivity for the detection of the cell-adapted strain Itor P13, expressed as the most probable number of cytopathogenic units, was established as 200 and 2 for MA104 and CaCo-2 cells, respectively, by the IIF-FC method. The ratio of infectious virus particles to total virus particles for a wild-type rotavirus was determined to be 1/2 × 106 and 1/2 × 104 for IIF-OM with MA104 cells and IIF-FC with CaCo-2 cells, respectively. The use of IIF-FC with CaCo-2 cells was tested with fecal and water samples and proved to be more effective than the standard procedure for rotavirus detection.
Human rotaviruses, the main cause of viral gastroenteritis in children, are transmitted by the fecal-oral route and may be acquired through ingestion of contaminated water and food (4). The most widely used techniques for the diagnosis of rotaviral diarrhea include electron microscopy (3), immunological methods such as latex agglutination or enzyme immunosorbent assays (5), and molecular techniques (6). However, these methods do not differentiate between infectious and noninfectious particles, and this difference is important from the point of view of public health, vaccine production, and antiviral research. The inclusion of an infectivity test prior to the specific detection solves not only this problem but also that of the lack of the sensitivity that is required for some kind of samples, such as environmental samples. Wild-type rotaviruses present difficulties in regard to their in vitro replication, although some of them may be adapted to grow in several cell lines, such as the human intestinal cell line CaCo-2 or the monkey kidney cell line MA104 (9). The standard methods for the diagnosis of specific infectious rotaviruses involve immunofluorescence tests and optical microscopy (OM) counting of infected foci in the culture (9). In the present work an alternative method for the quantification of infectious rotaviruses, based on the use of flow cytometry (FC), is described. The technique was applied to detect wild-type rotaviruses from fecal or water samples.
MATERIALS AND METHODS
Cell cultures and viral strains.
MA104 cells, derived from fetal monkey kidney, and CaCo-2 cells, derived from a human colon carcinoma, were propagated in Eagle’s minimal essential medium supplemented with fetal calf serum (FCS) at concentrations of 5 and 10%, respectively. The cytopathic strain Itor P13 (13) of group A human rotavirus, type 3, was used throughout these studies for the standardization of the technique.
Fecal samples.
Wild-type rotaviruses were obtained from stool samples from 2- to 12-month-old children with acute diarrhea from the Hospital de la Santa Creu i Sant Pau, Barcelona, Spain, and identified by latex agglutination (Rotalex; Orion Diagnostica, Espoo, Finland). All but one of these samples contained rotaviruses that were noncultivable in MA104 cells. Viruses were chloroform extracted from 0.1 g of stool sample and suspended in 1 ml of phosphate-buffered saline. The number of extracted viral particles was determined for some samples by the colloidal gold and negative-staining transmission electron microscopy method (8).
Water samples.
Wild-type rotaviruses were also isolated from water samples supplied by drinking water companies from different cities throughout Spain. Viruses were concentrated on location from 500 liters of river water by filtration through positively charged Zeta Plus MK II filters (AMF Cunó, Sefiltra, Alcobendas, Spain), elution with 500 ml of 3% beef extract–0.05 M glycine buffer, and organic flocculation to a final volume of 50 ml by the procedures described elsewhere (2). Rotavirus diagnosis was performed by infecting MA104 cells with water concentrates and detecting the infected cells by an indirect immunofluorescence (IIF) test.
Viral infections.
Wild-type rotavirus infections were performed with MA104 and CaCo-2 cells (11). Virus inocula were pretreated with trypsin (Difco, Detroit, Mich.; tissue culture grade 1:250) at a concentration of 10 μg/ml for 30 min at 37°C, and after adsorption a serum-free overlay medium containing 5 μg of trypsin per ml was added. Infections proceeded for 2 to 5 days, and cells were processed for IIF as described below. Viral enumerations of the cell-adapted strain Itor P13 were performed as previously described (11) and expressed as the most probable number of cytopathogenic units (MPNCU) per milliliter. All experiments involving detection and titration were performed at least in triplicate.
IIF-OM detection in MA104 cells.
The IIF-OM has been previously described (2) and has been applied to detect infectious rotavirus in water. Briefly, it consisted of infections of monolayers of cells grown in microtiter plates with 20 μl of inoculum per well, fixation of infected cells with cold (4°C) 80% acetone, incubation with human rotavirus antiserum (Institute Virion Ltd., Ruschlikon, Switzerland) diluted 1/100, and then staining with goat anti-human immunoglobulins conjugated to fluorescein isothiocyanate (Sigma-Aldrich, Alcobendas, Spain) diluted 1/32. As a blocking reagent, 2% FCS was used. The plates were examined inverted with a Nikon microscope fitted for epifluorescence. A total volume of 1.6 ml of each sample was assayed.
IIF-FC detection in MA104 and CaCo-2 cells.
The Itor P13 strain of human rotavirus was used to develop the IIF-FC detection procedure. In this procedure, cells grown in 60-mm-diameter dishes were inoculated with 200 μl of the sample. After the infection period, cells were mechanically detached, washed, and treated with different fixatives, such as cold (4°C) 80% acetone for 30 min, cold (−20°C) 70% ethanol for 30 min, cold (4°C) acetone-ethanol (4:6) for 30 min, and cold (4°C) acetone-methanol-formalin (1:1:1) for 1 min. An IIF assay was performed as described above but with the secondary antibody diluted 1/400. As blocking reagents, both 5% nonfat milk and 2% FCS were tested. The cellular suspensions were analyzed in a Coulter Epics Elite flow cytometer equipped with a 488-nm argon-ion laser at 15-mW power and with a combination of 550 DL and 525 BP filters in order to recover the green fluorescence of fluorescein. The forward-angle light scatter was used to select cell size, and the side-angle light scatter was used to select shape and structure, so as to restrict the readings to the population of intact eukaryotic cells and not the cell debris. Data obtained were represented in diagrams that displayed the distribution of the population of cells among 1,024 channels of fluorescence intensity. The height (y axis) indicated the number of cell counts with a particular level of fluorescence. Fluorescence intensity (x axis) was expressed on a log scale, which means that small differences in channel number represent large differences in the amount of dye per cell. The total number of cells counted was around 500,000. For quality assurance, 40 mock-infected samples were processed for IIF-FC, and an arbitrary cursor (A) was drawn at the right end of their fluorescence curves (channels 10 to 1024). This cursor included 2% of the negative cell population counts. The mean fluorescence of each of the A cursors from the 40 negative controls was calculated. These mean values followed a normal distribution. The mean and standard deviation of this curve were calculated, and a second cursor (B) was then defined, starting at the point obtained by adding two standard deviations to the mean fluorescence (channel 60) and ending at channel 1024. The ratio of cells present in cursor B to total counted cells was calculated for each negative sample, and the mean plus two standard deviations of these ratios in the 40 negative samples was established as the threshold of positivity (0.00112 and 0.00061 for MA104 and CaCo-2 cells, respectively). The procedure was validated by using 10-fold dilutions of the Itor P13 strain to infect monolayers of MA104 and CaCo-2 cells. Different infectious doses were assayed, and each of them was determined at least in triplicate for each cell type. When wild-type viruses were assayed, negative and positive control samples were included in each assay.
Nucleic acid detection in sorted cell populations by a seminested RT-PCR technique.
Nucleic acid detection was performed with sorted cell populations by a seminested reverse transcription-PCR (RT-PCR) technique. Infected CaCo-2 cells were processed by FC as described above and sorted by combining their scatter and fluorescence characteristics. The fluid stream was broken into droplets that were electrically charged and deflected into a collection vessel by passage through an electric field. Three different populations were sorted: 8,000 cells from the population included in cursor A, 800 cells from the population included in cursor B, and 80,000 cells from the rest of the population. A two-step RT-PCR was then performed by standard procedures as described elsewhere (7). Briefly, primers Beg9 and End9 were used for an initial RT-PCR to obtain the full-length gene 9 segment (1,062 bp). Primer RVG9 and primer aET3 were used to obtain, by a subsequent seminested PCR amplification, the characteristic 374-bp fragment of human rotavirus type 3. RT of RNA was carried out in 20-μl reaction mixtures. Ten microliters of eluates containing RNA extracted from the samples was heated at 99°C for 5 min and immediately placed on ice, and 10 μl of 2× RT reaction mix was added and incubated for 1 h at 43°C. The RT reaction mix consisted of 50 mM Tris hydrochloride (pH 8.3), 40 mM KCl, 7 mM MgCl2, 1 mM each primer (Beg9 and End9), 200 mM deoxynucleoside triphosphates (dNTPs), and 2 U of avian myeloblastosis virus reverse transcriptase. Forty microliters of PCR mix was added to 10 μl of the RT product, to yield a 50-μl mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 1 mM each primer (Beg9 and End9), 200 mM dNTPs, and 2 U of Taq polymerase. The mixture was initially denatured at 94°C for 4 min and then subjected to 40 cycles of amplification, each consisting of 1 min at 94°C, 2 min at 47°C, and 5 min at 72°C. A final extension was carried out at 72°C for 10 min. Ten microliters of the amplified product was used as the template in the seminested PCR. Primer and dNTP concentrations were 0.5 and 100 mM, respectively. Thirty cycles of amplification were performed under the same conditions as before but with an increase in the annealing temperature, to 50°C. Twenty microliters of each sample was analyzed in an agarose gel in order to observe the expected 374-bp band of the PCR product.
RESULTS
IIF-FC detection of the Itor P13 strain.
For the development of the IIF-FC detection method for rotavirus-infected cells, several fixatives were tested in order to permeabilize the cells to the different antibodies. Among all the fixatives, acetone-methanol-formalin (1:1:1) provided the best specific signal/background ratio. Two kinds of blocking reagents were assayed, and 5% nonfat milk was found to be the one of choice, since a better blocking activity in terms of background levels and more consistent results from experiment to experiment were achieved with nonfat milk than with FCS.
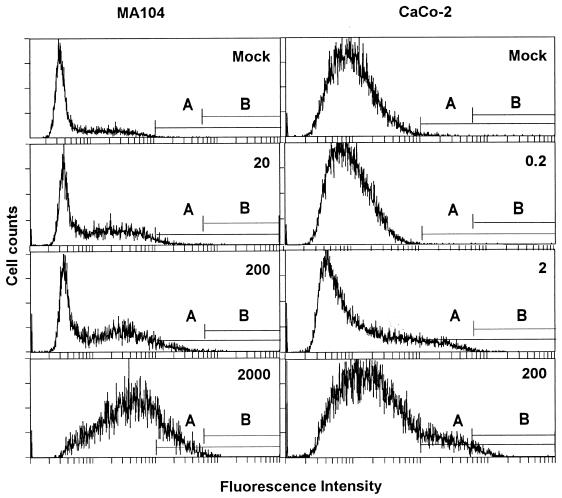
Tenfold dilutions of viral stocks of the Itor P13 strain containing 4 × 107 MPNCU/ml were assayed by IIF-FC after infection of monolayers of MA104 and CaCo-2 cells. CaCo-2 cells offered enhanced sensitivity in comparison with MA104 cells, as the minimum amounts of infectious rotavirus detected were 2 and 200 MPNCU, respectively (Fig. 1). The mean ratios, from three separate experiments, of the number of cells in cursor B (Fig. 1) to the total number of counted cells after infection with 200 MPNCU of virus were 0.003750 ± 0.004770 and 0.023123 ± 0.015138 for MA104 and CaCo-2 cells, respectively. This means that after infection at a multiplicity of infection of 0.0001, 0.375 and 2.3% of the MA104 and CaCo-2 cell populations, respectively, became positive.
FIG. 1.
Rotavirus detection by IIF-FC. Detection of Itor P13 virus in MA104 cells (left column) and CaCo-2 cells (right column) is shown. The inoculum used (MPNCU) is indicated in the upper right corner of each plot. Cursors A and B are defined in the text. Plots represent a typical data set.
Validation of the IIF-FC rotavirus detection method with CaCo-2 cells.
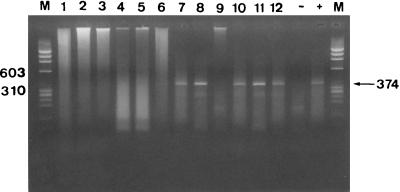
To confirm the validity of the IIF-FC method for the detection of rotavirus-infected CaCo-2 cells, viral RNA was detected by means of an RT-PCR method in sorted cell populations. After infection with 200 MPNCU of the Itor P13 strain, all three sorted populations (cells from cursor A, cells from cursor B, and cells from the rest of the population) contained rotavirus RNA (Fig. 2), while after infection with 2 MPNCU, only the cells included in cursors A and B were positive (Fig. 2). None of the three populations was positive for the presence of rotavirus RNA after infection with 0.2 MPNCU (Fig. 2).
FIG. 2.
Rotavirus RNA detection by a seminested RT-PCR after infection of CaCo-2 cells with different inocula of the Itor P13 strain and cell sorting. Lanes: 1, mock-infected cells from cursor A; 2, mock-infected cells from cursor B; 3, mock-infected cells from the rest of the population; 4, cells from cursor A after infection with 0.2 MPNCU; 5, cells from cursor B after infection with 0.2 MPNCU; 6, cells from the rest of the population after infection with 0.2 MPNCU; 7, cells from cursor A after infection with 2 MPNCU; 8, cells from cursor B after infection with 2 MPNCU; 9, cells from the rest of the population after infection with 2 MPNCU; 10, cells from cursor A after infection with 200 MPNCU; 11, cells from cursor B after infection with 200 MPNCU; 12, cells from the rest of the population after infection with 200 MPNCU; M, molecular size marker; −, negative control RT-PCR; +, positive control. Cursors A and B are defined in the text. Numbers on the left and right indicate base pairs.
IIF-FC detection of rotaviruses in feces.
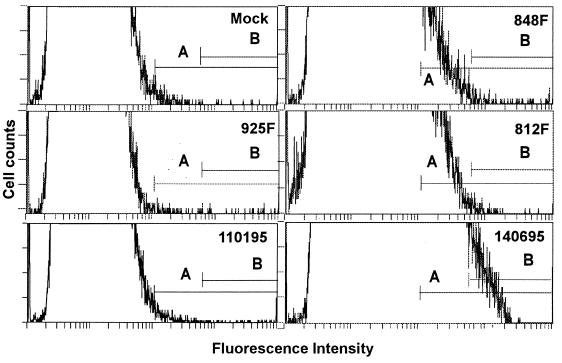
Eight fecal samples containing noncultivable rotaviruses and one fecal sample with rotaviruses growing in MA104 cells were assayed by IIF-FC. Two of the eight noncultivable samples became positive by IIF-FC after infection of CaCo-2 cells (Table 1). The sample with cultivable rotaviruses was also positive by this procedure. The number of physical particles present in this sample (812F) was evaluated and found to be 108 particles per g of extracted feces. The limit of sensitivity of IIF-FC for this sample was at a stool suspension dilution of 10−2, which means that the ratio of infectious particles to physical particles is 1/2 × 104. The same sample (812F) evaluated by IIF-OM detection with MA104 cells showed 50 fluorescent foci per g of extracted feces, which means a ratio of infectious particles to physical particles of 1/2 × 106. The characteristic profiles for positive and negative stool samples are shown in Fig. 3.
TABLE 1.
Detection of wild-type rotavirus in fecal and water samples
| Sample | Latex agglutination | IIF-OM, MA104 cellsa (FF/g or FF/mlb) | IIF-FC, CaCo-2 cellsa,c |
|---|---|---|---|
| Fecal | |||
| 180F | +++ | 0 | 0.00026 |
| 198F | +++ | 0 | 0.00037 |
| 812F | +++ | 50 | 0.00067 |
| 846F | +++ | 0 | 0.00068 |
| 848F | +++ | 0 | 0.00080 |
| 900F | +++ | 0 | 0.00026 |
| 925F | +++ | 0 | 0.00027 |
| 939F | +++ | 0 | 0.00026 |
| 964F | +++ | 0 | 0.00013 |
| Mock-infected cells | NTd | 0 | 0.00061 |
| Water | |||
| 131094 | NT | 1.25 | 0.001497 |
| 110195 | NT | 0 | 0.000168 |
| 180195 | NT | 0 | 0.000198 |
| 150295 | NT | 0.63 | 0.000335e |
| 080295 | NT | 0 | 0.000128 |
| 080395 | NT | 0 | 0.000578 |
| 100395 | NT | 0 | 0.000155 |
| 150395 | NT | 0 | 0.000310 |
| 181095 | NT | 0 | 0.000234 |
| 081195 | NT | 0 | 0.000137 |
| TER02 | NT | 0 | 0.003728 |
| OT07 | NT | 0.63 | 0.001463 |
| GP04 | NT | 0.63 | 0.007076 |
| GP05 | NT | 1.25 | 0.003735 |
| 130396 | NT | 0 | 0.000222 |
| 170496 | NT | 0 | 0.000198 |
| 030595 | NT | 0 | 0.002360 |
| 140695 | NT | 0 | 0.003996 |
| 210695 | NT | 0 | 0.000243 |
| 120795 | NT | 0 | 0.001138 |
| 240895 | NT | 0 | 0.000120 |
| 300895 | NT | 0 | 0.000120 |
| ABR04 | NT | 0 | 0.001635 |
| PF0506 | NT | 0.63 | 0.001230 |
| Mock-infected cells | NT | 0 | 0.000610 |
Boldface indicates a positive result.
FF/g and FF/ml, fluorescent foci per gram of feces and fluorescent foci per milliliter of water concentrate, respectively.
See text for explanation of units.
NT, not tested.
This sample became positive (cytometric value of 0.0035982) after the sample volume inoculated on CaCo-2 cells was increased (see text).
FIG. 3.
Detection of wild-type rotavirus in CaCo-2 cells. The sample (Table 1) is indicated in the upper right corner of each plot. Cursors A and B are defined in the text.
IIF-FC detection of waterborne rotaviruses.
Twenty-four water sample concentrates were analyzed by IIF-FC after infection of CaCo-2 cells, and the results were compared with the data provided by IIF-OM detection after infection of MA104 cells (Table 1 and Fig. 3). Ten and six of these samples were positive by IIF-FC and IIF-OM, respectively. Five samples were positive by both methods. The one discordant sample became positive by the IIF-FC method with CaCo-2 cells after ultracentrifugation of a 1.6-ml sample (the volume used in the MA104 system), resuspension in 200 μl, and cell inoculation of the latter amount. The characteristic profiles for positive and negative water samples are shown in Fig. 3.
DISCUSSION
The clinical diagnosis of rotaviral gastroenteritis can be performed on the basis of detection of physical particles. However, the determination of infectious viruses is of critical importance in some situations, such as the monitoring of water for the prevention of waterborne outbreaks of rotaviral gastroenteritis (1, 2). Other situations where control of the infectivity of samples is also required include vaccine production and the development of antiviral and disinfectant agents. The poor replicating capacity of rotaviruses in cell culture has prompted the use of cell-adapted strains in most studies involving antiviral (10, 15) and disinfection (14) research. The method described here could be of relevant application in these areas and enables the use of cell culture-adapted strains and some wild-type strains as well. The development of this method relied on the use of the CaCo-2 cell line and ulterior FC for the detection of infected cells. CaCo-2 cells showed higher sensitivity to rotavirus infection than MA104 cells, either with the cell-adapted strain Itor P13 or with wild-type viruses. The sensitivity of CaCo-2 cells was 2 log units higher than that of MA104 cells. With the wild-type strains the higher sensitivity was determined from the higher number of positive stool and water samples detected and from the higher ratio of infectious particles to total virus particles observed in the 812 stool samples. The higher sensitivity of CaCo-2 cells to rotavirus infections has also been previously described (9, 11). The sensitivity of IIF-FC with CaCo-2 cells could be increased by concentrating the sample prior to cell inoculation. The use of FC for the detection of virus-infected cells has several advantages over OM detection and has been used for different viruses (12, 16). The IIF-OM detection method, although efficient, is cumbersome and requires well-trained personnel, while IIF-FC is an automatable procedure. Most major hospitals and public health institutions in developed countries own a flow cytometer. The standardization of the IIF-FC methodology requires the study of several critical points, such as fixation and minimization of the background noise. As a fixative, the best choice in our assay was the mixture of acetone, methanol, and formalin, and as a blocking reagent, the solution of 5% nonfat milk was the most effective in reducing background fluorescence. The antibody concentration also played an important role in lowering the background, and the working dilutions were adjusted in the IIF-FC procedure with respect to those in the IIF-OM detection method in order to achieve a good ratio of specific labeling to background. The method described here has been found to be highly useful and reproducible for the specific detection of infectious wild-type rotaviruses and enables the processing of a large number of samples. Highly sensitive detection procedures are also required for the detection of very low numbers of rotaviruses in some environmental situations in order to accurately assess the level of risk posed by their presence.
ACKNOWLEDGMENTS
We acknowledge the skillful assistance of J. Comas of the Flow Cytometry Unit of the Scientific and Technical Services of the University of Barcelona. We are grateful to N. Margall, Servei de Microbiologia, Hospital de la Santa Creu i Sant Pau, for providing stool samples.
This work was supported in part by grant 1995SGR00197 from the CIRIT, Generalitat de Catalunya. F. X. Abad has a PQS contract from the Generalitat de Catalunya.
REFERENCES
- 1.Ansari S A, Springthorpe V S, Sattar S A. Survival and vehicular spread of human rotaviruses: possible relation to seasonality of outbreaks. Rev Infect Dis. 1991;13:448–461. doi: 10.1093/clinids/13.3.448. [DOI] [PubMed] [Google Scholar]
- 2.Bosch A, Lucena F, Diez J M, Gajardo R, Blasi M, Jofre J. Human enteric viruses and indicator microorganisms in a water supply associated with an outbreak of infectious hepatitis. J Am Water Works Assoc. 1991;83:80–83. [Google Scholar]
- 3.Brandt C D, Kim H W, Rodriguez W J, Arrobio J O, Jeffries B C, Stallings E P, Lewis C, Miles A J, Chanock R M, Kapikian A Z, Parrot R H. Pediatric viral gastroenteritis during eight years of study. J Clin Microbiol. 1983;18:71–78. doi: 10.1128/jcm.18.1.71-78.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craun G F. Causes of waterborne outbreaks in the United States. Water Sci Tech. 1991;24:17–20. [Google Scholar]
- 5.Doern G V, Herrman J E, Henderson P, Stobbs-Walro D, Perron D M, Blacklow N R. Detection of rotavirus with a new polyclonal antibody enzyme immunoassay (Rotazyme II) and a commercial latex agglutination test (Rotalex): comparison with a monoclonal antibody enzyme immunoassay. J Clin Microbiol. 1986;23:226–229. doi: 10.1128/jcm.23.2.226-229.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores J, Sears J, Perez Schael I, White L, Garcia D, Lanata C, Kapikian A Z. Identification of human rotavirus serotype by hybridization to polymerase chain reaction-generated probes derived from a hyperdivergent region of the gene encoding outer capsid protein VP7. J Virol. 1990;64:4021–4024. doi: 10.1128/jvi.64.8.4021-4024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gajardo R, Bouchriti N, Pintó R M, Bosch A. Genotyping of rotaviruses isolated from sewage. Appl Environ Microbiol. 1995;61:3460–3462. doi: 10.1128/aem.61.9.3460-3462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grabow W O K, Puttergill D L, Bosch A. Propagation of adenovirus types 40 and 41 in the PLC/PRF/5 primary liver carcinoma cell line. J Virol Methods. 1992;37:307–313. doi: 10.1016/0166-0934(92)90047-h. [DOI] [PubMed] [Google Scholar]
- 9.Kitamoto N K, Ramig R F, Matson D O, Estes M K. Comparative growth of different rotavirus strains in differentiated cells (MA104, HepG2 and CaCo-2) Virology. 1991;184:729–737. doi: 10.1016/0042-6822(91)90443-f. [DOI] [PubMed] [Google Scholar]
- 10.Kitaoka S, Konno T, De Clerc E. Comparative efficacy of broad-spectrum antiviral agents as inhibitors of rotavirus replication in vitro. Antiviral Res. 1986;6:57–65. doi: 10.1016/0166-3542(86)90039-2. [DOI] [PubMed] [Google Scholar]
- 11.Pintó R M, Diez J M, Bosch A. Use of the colonic carcinoma cell line CaCo-2 for in vivo amplification and detection of enteric viruses. J Med Virol. 1994;44:310–315. doi: 10.1002/jmv.1890440317. [DOI] [PubMed] [Google Scholar]
- 12.Qvist P, Aasted B, Bloch B, Mayling A, Rosholt L, Houe H. Comparison of flow cytometry and virus isolation in cell culture for identification of cattle persistently infected with bovine viral diarrhea virus. J Clin Microbiol. 1991;29:660–661. doi: 10.1128/jcm.29.3.660-661.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato K, Inaba Y, Shinizaki T, Fujii R, Matumoto M. Isolation of human rotavirus in cell culture. Arch Virol. 1981;69:155–160. doi: 10.1007/BF01315159. [DOI] [PubMed] [Google Scholar]
- 14.Sattar S A, Jacobsen H, Rahman H, Cusack T M, Rubino J R. Interruption of rotavirus spread through chemical disinfection. Infect Control Hosp Epidemiol. 1994;15:751–756. doi: 10.1086/646852. [DOI] [PubMed] [Google Scholar]
- 15.Smee D F, Sidwell R W, Clark S M, Barnett B B, Spendlove R S. Inhibition of rotaviruses by selected antiviral substances: mechanisms of viral inhibition and in vivo activity. Antimicrob Agents Chemother. 1982;21:66–73. doi: 10.1128/aac.21.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snoeck R, Schols D, Sadzot-Delvaux C, Cloes J M, Andrei G, De Clercq E, Piette J, Rentier B. Flow cytometric method for the detection of gpI antigens of varicella zoster virus and evaluation of anti-VZV agents. J Virol Methods. 1992;38:243–254. doi: 10.1016/0166-0934(92)90114-s. [DOI] [PubMed] [Google Scholar]