Abstract
The nucleoid-associated protein H-NS is thought to play an essential role in the organization of bacterial chromatin in Escherichia coli. Homologues, often with very low sequence identity, are found in most gram-negative bacteria. Microscopic analysis reveals that, despite limited sequence identity, their structural organization results in similar DNA binding properties.
The chromosomal DNA of bacteria is organized into a compact structure, the nucleoid. A number of factors contribute to maintaining this compact state: macromolecular crowding (or depletion), DNA supercoiling, and so-called nucleoid-associated proteins (28). It is unclear how much each of these factors contributes to nucleoid compactness, but nucleoid-associated proteins play an essential role. Important members of this group of proteins are H-NS, HU, Fis, and integration host factor (IHF) (12, 18). In addition to a role in compaction, these proteins act as specific regulators (for instance in transcription). Both HU (27) and IHF (1) reduce the effective volume occupied by DNA molecules by inducing (strong) local bends. Interestingly, the effect of HU is very concentration dependent, and higher concentrations of this protein (locally) induce the formation of rigid filaments of as-yet-undefined significance (27). H-NS employs a different mechanism: this protein has the ability to bridge adjacent tracts of DNA. Furthermore, H-NS can locally induce some type of higher-order compaction by an as-yet-unknown mechanism (7). The ability to bridge is exploited when H-NS—which does not exhibit sequence-specific binding—exerts its specific regulatory role in transcription. This ability is the molecular basis for the recognition of curved DNA (8), which is an essential feature of many genes that are repressed by H-NS. Also, bridging provides a direct explanation for the mechanism underlying trapping of RNA polymerase in the open initiation complex at the rrnB P1 (9).
In recent years, H-NS homologues have been identified in most gram-negative bacteria (3, 4). The first homologue identified, StpA, was found in Escherichia coli and bears about 60% identity with H-NS at the amino acid level (29). Many recently discovered homologues have low sequence identity with E. coli H-NS and cannot be identified on the basis of sequence homology. Nevertheless, these proteins can be identified by screening genomic libraries for reversal of the H-NS deficiency in E. coli mutant strains (17). Recently, a novel class of H-NS like proteins was found in Pseudomonas species (19, 25, 26). These so-called MvaT proteins do not exhibit any significant homology with any known H-NS-related protein (e.g., 18% identity with E. coli H-NS). Functionally, these H-NS homologues have not yet been extensively characterized, but a role as a repressor of transcription has recently been attributed to them (11, 19).
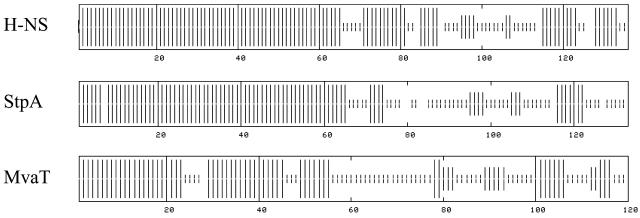
At the structural level all H-NS like proteins are similar: the proteins can be divided into a C-terminal DNA binding domain and an N-terminal oligomerization domain, which are separated by a flexible linker region (13, 20, 25). Nuclear magnetic resonance spectroscopy has shown that the N-terminal domain of E. coli H-NS is mainly α-helical and that, due to the formation of a coiled coil, H-NS exists as a dimer (5, 14). On top of this, the dimer has the ability to self-associate and to form large oligomers (6, 22). As a consequence of the strong homology with H-NS, StpA is predicted to be very similar in its overall structure. Although MvaT bears little homology to E.coli H-NS, the C-terminal domains of these proteins have been predicted to be structurally very similar (26). In addition, the N-terminal domain of MvaT, like that of H-NS, is predicted to be α-helical and capable of dimerization (26). The large structural similarity among H-NS like proteins (Fig. 1) suggests that they employ mechanisms similar to those of E. coli H-NS to modulate DNA architecture. In this paper we describe the scanning force microscopic (SFM) analysis of the DNA binding properties of StpA and MvaT.
FIG. 1.
Secondary structure of H-NS, StpA, and MvaT proteins. These structures were predicted using a consensus prediction method (10) available on the NPSA website (http://npsa-pbil.ibcp.fr). Vertical bars represent the probability of α-helix (full size), β-strand (intermediate size), and random coil (small size) in these H-NS-like proteins.
Preparation of E. coli StpA and Pseudomonas sp. MvaT.
StpA was overexpressed and purified from E. coli cells in an hns mutant background. To this aim the strain BL21(DE3)hns::kan was constructed by transduction of BL21(DE3) with a mutant hns allele from strain M182hns::kan (30). The strain was transformed with the multicopy plasmid pUC18-StpA, which was obtained by insertion of a 1,273-bp HindIII-NdeI fragment obtained from the vector pT7-StpA (30) containing the complete stpA gene under the control of the phage T7 promoter. Cells were grown in M9 medium-0.2% glucose-0.2% Casamino Acids supplemented with ampicillin (50 μg/ml) to an optical density at 600 nm of 0.5 and further incubated for 3 h after induction with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were disrupted by sonication in 50 mM Tris-HCl (pH 7.5)-200 mM NaCl-2 mM EDTA-0.2 mM phenylmethylsulfonyl fluoride-1 mM leupeptin-1 mM pepstatin (the presence of protease inhibitors was maintained throughout the purification), and the cleared lysate was separated from ribosomes by high-speed centrifugation. The supernatant was precipitated at 4°C with 0.3% polyethylenimine, and the pellet was extracted with a buffer containing 50 mM Tris-HCl (pH 7.5) and 500 mM NaCl. The protein suspension was cleared by centrifugation, and the supernatant was precipitated with 40% (NH4)2SO4. The pellet was extracted with 50 mM Tris-HCl, pH 7.5, and dialyzed against 50 mM Tris-HCl (pH 7.5)-1 mM EDTA-0.5 mM dithiothreitol (DTT)-10% glycerol. Precipitated StpA was concentrated by centrifugation and redissolved in 50 mM Tris-HCl (pH 7.5)-1 mM EDTA-0.5 mM DTT-0.3 M NaCl-10% glycerol. The protein fraction was further purified to homogeneity by DNA cellulose chromatography employing a linear gradient of 300 mM to 1 M NaCl. Pooled StpA fractions were dialyzed against 50 mM Tris-HCl (pH 7.5)-1 mM EDTA-0.5 mM DTT-10% glycerol, and the precipitated StpA was redissolved in the presence of 300 mM NaCl. The final concentration was determined by Bradford analysis. The purified protein was routinely assayed for its capacity to bind DNA in a standard binding assay (2 nM 32P-labeled DNA containing the rrnB P1 promoter upstream region, 5 μM protein, 20 ng of heparin/μl in 50 mM Tris-HCl [pH 7.4], 70 mM KCl, 15 mM NaCl, 1 mM EDTA, 10 mM 2-mercaptoethanol). Under those conditions generally half of the DNA fragment was found in a specific complex.
MvaT was overexpressed and purified from E. coli cells. The strain BL21(DE3) carrying the lacI-expressing plasmid pDIA17 (18a) was transformed with the multicopy plasmid pDIA591, which was obtained by insertion of the mvaT gene (26) under the control of the phage T7 promoter into the NdeI-XhoI sites of the pET22 vector (Novagen). Cells were grown in Hyperbroth (Athena) supplemented with 200 μg of ampicillin/ml and 20 μg of chloramphenicol/ml to an optical density at 600 nm of 2.5 and further incubated for 2 h after induction with 3 mM IPTG. Cells were disrupted using FastPrep FP120 (Bio 101) in buffer A (31.6 mM NaH2PO4, 68.4 mM Na2HPO4 [pH 7.2], 300 mM NaCl). Cell debris was removed by centrifugation, and the supernatant was loaded onto an 0.6-ml NiSO4 chelation column. After adsorption to the column and washing with buffer A containing 5 mM imidazole, the protein was eluted with a 200 to 500 mM imidazole gradient in buffer A. Buffer A was then changed to 500 mM NaCl and 40 mM NaH2PO4 (pH 8) by using a PD 10 desalting column (Pharmacia). The final concentration was determined by Bradford analysis.
Visualization of StpA- and MvaT-DNA complexes.
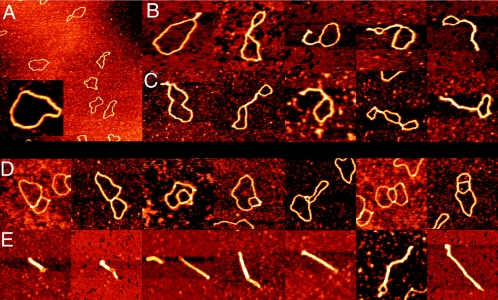
Complexes of pUC19 and StpA or MvaT were prepared at different protein/DNA ratios and visualized using SFM (following a method described previously [7]). Typical complexes as observed in these experiments are shown in Fig. 2. The images in Fig. 2B indicate that, like H-NS, StpA has the ability to bridge DNA. Interestingly, unlike H-NS (7), the structural effects of StpA seem to be limited to bridging. The formation of local higher-order complexes on molecules that are extensively bridged, as observed with H-NS, does not occur in the range of StpA concentrations tested (relative amount of one StpA dimer per 200 [Fig. 2B], 400, and 600 bp [data not shown]). At the highest protein concentrations tested (one StpA dimer per 100 bp and higher) globular aggregates are observed (data not shown). Control experiments with high StpA concentrations in the absence of DNA do not show such aggregates.
FIG. 2.
SFM images of circular nicked pUC19 DNA molecules with and without E. coli StpA or Pseudomonas strain Y1000 MvaT. (A) Bare relaxed pUC19 DNA molecules. Image shows a 3- by 3-μm surface area. (Inset) Close-up of a bare relaxed pUC19 DNA molecule at the same magnification as used in panels B to E. (B) DNA molecules after incubation with StpA (one dimer per 200 bp). Complexes are laterally compacted by antiparallel DNA bridging. (C) DNA molecules after incubation with MvaT (one dimer per 160 bp). Complexes are laterally compacted by antiparallel DNA bridging. (D) DNA molecules after incubation with MvaT (one dimer per 160 bp). Complexes are compacted by parallel DNA bridging. (E) DNA molecules after incubation with MvaT (one dimer per 80 bp). Two types of complexes (reflected in clearly different sizes) are formed. Large complexes are laterally fully compacted by regular antiparallel DNA bridging, and small complexes are more strongly compacted through extensive parallel loop-loop bridging. Images in panels B to E show an 0.5- by 0.5-μm surface area. Color represents height ranging from 0 to 2 nm.
The images in Fig. 2C and D indicate that MvaT (at a ratio of one dimer per 160 bp) also has the ability to bridge DNA. In addition, higher-order complex formation occurs at high concentrations of MvaT (one MvaT dimer per 80 bp [Fig. 2E]). Rod-like complexes of various sizes are then observed. Interestingly, novel intermediate forms (different from sole lateral bridging) were also observed (Fig. 2D). In these complexes the DNA is bridged in a parallel rather than in an antiparallel fashion; DNA loops can be folded back inside other loops, which in turn can lead to more bridging between DNA tracts in these loops. In the most simple geometry, this will result in complexes half the length of complexes formed by only lateral bridging. Probably the different sizes of condensates directly reflect the pathway (involving different sizes and/or numbers of loops folded inward) towards condensation that was followed.
Similarities and differences.
Our observations indicate that the DNA binding properties of StpA are very similar to H-NS, as suggested by previous biochemical experiments (2, 3, 24, 30). The DNA binding properties of MvaT, as observed by SFM, also largely resemble those of H-NS and StpA. In fact, all three proteins have the ability to bridge DNA.
A clear difference between StpA and H-NS is that H-NS can locally induce higher-order compaction (in about 35% of the H-NS-DNA complexes) at intermediate protein concentrations (i.e., one H-NS dimer per 24 bp). This type of compaction by H-NS may be dependent on the ability of this protein to form large oligomeric structures consisting of dimers (14, 22). The oligomerization properties of StpA have not been biochemically characterized. It is, however, known that differences exist in the amino acid composition of the oligomerization domain. In particular, part of the linker region, which has been identified as being important for the formation of large oligomers (14), exhibits only low sequence identity between StpA and H-NS. This could explain the absence of higher-order compaction. However, it should be noted that, although DNA bridging is considered biologically relevant (12), the physiological relevance of the higher-order compaction remains unclear. The DNA bridging observed for StpA occurs at significantly lower protein/DNA ratios (one dimer per 200 bp instead of one dimer per 24 bp) than with H-NS (7). This probably reflects a higher binding affinity of StpA for DNA as suggested previously (24; R. Wagner and coworkers, unpublished observations). As a consequence the relatively low amounts of StpA (compared to H-NS) in the cell (15, 23, 30) can still have significant impact on DNA architecture. It thus seems likely not only that StpA can effectively replace (or assist) H-NS in its function as a repressor of transcription (16, 21, 23, 30) but also that it may be involved in the modulation of nucleoid compactness.
The binding properties of MvaT also exhibit differences from those of H-NS. MvaT does display the ability to form higher-order nucleoprotein structures. In doing so, an alternative pathway based on parallel bridging (not seen with H-NS or StpA), resulting in more compact structures, is observed. This alternative “mode” of bridging probably reflects subtle differences in protein-protein or protein-DNA interactions among the three proteins. The class of most compact complexes observed previously with H-NS (for which we had no structural explanation) could also result from following a similar type of condensation pathway, even though parallel bridged intermediate structures were not observed (7). The ability to compact DNA efficiently suggests that MvaT could be involved in the compaction of the Pseudomonas nucleoid. It has been proposed that the ability to bridge DNA allows recognition of regions containing strong DNA curvature or high local DNA flexibility (8). Therefore, apart from a possible role in the global organization of the nucleoid, StpA and MvaT are expected to exhibit preferential binding in vivo to such regions. Since these regions are often found close to promoters, this provides a direct means to repress transcription (12, 20). Moreover, nucleation of binding of these proteins at such sites may be a determinant in setting the boundary conditions for the global organization of the nucleoid.
Conclusions.
H-NS, StpA, and MvaT proteins each exhibit the ability to form bridges between adjacent DNA tracts. The ability to bridge DNA probably stems from the particular two-domain configuration of most H-NS-like proteins identified so far, resulting in dimers with two exposed independent DNA binding domains. DNA bridging may thus be a common property of H-NS-like proteins, and it is likely that they all use similar mechanisms to organize and compact DNA and/or to regulate transcription.
Acknowledgments
We acknowledge Christoph Schmidt for support.
This work was supported by a Nederlandse Organisatie voor Wetenschappelijk Onderzoek Vernieuwingsimpuls grant and a Nederlandse Organisatie voor Wetenschappelijk Onderzoek ALW open competition grant (to G.J.L.W. and R.T.D.).
REFERENCES
- 1.Ali, B. M., R. Amit, I. Braslavsky, A. B. Oppenheim, O. Gileadi, and J. Stavans. 2001. Compaction of single DNA molecules induced by binding of integration host factor (IHF). Proc. Natl. Acad. Sci. USA 98:10658-10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam, T. A., and A. Ishihama. 1999. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 274:33105-33113. [DOI] [PubMed] [Google Scholar]
- 3.Bertin, P., N. Benhabiles, E. Krin, C. Laurent-Winter, C. Tendeng, E. Turlin, A. Thomas, A. Danchin, and R. Brasseur. 1999. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol. Microbiol. 31:319-329. [DOI] [PubMed] [Google Scholar]
- 4.Bertin, P., F. Hommais, E. Krin, O. Soutourina, C. Tendeng, S. Derzelle, and A. Danchin. 2001. H-NS and H-NS-like proteins in Gram-negative bacteria and their multiple role in the regulation of bacterial metabolism. Biochimie 83:235-241. [DOI] [PubMed] [Google Scholar]
- 5.Bloch, V., Y. Yang, E. Margeat, A. Chavanieu, M. T. Auge, B. Robert, S. Arold, S. Rimsky, and M. Kochoyan. 2003. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 10:212-218. [DOI] [PubMed] [Google Scholar]
- 6.Ceschini, S., G. Lupidi, M. Coletta, C. L. Pon, E. Fioretti, and M. Angeletti. 2000. Multimeric self-assembly equilibria involving the histone-like protein H-NS. A thermodynamic study. J. Biol. Chem. 275:729-734. [DOI] [PubMed] [Google Scholar]
- 7.Dame, R. T., C. Wyman, and N. Goosen. 2000. H-NS mediated compaction of DNA visualized by atomic force microscopy. Nucleic Acids Res. 28:3504-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dame, R. T., C. Wyman, and N. Goosen. 2001. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 83:231-234. [DOI] [PubMed] [Google Scholar]
- 9.Dame, R. T., C. Wyman, R. Wurm, R. Wagner, and N. Goosen. 2002. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277:2146-2150. [DOI] [PubMed] [Google Scholar]
- 10.Deleage, G., C. Blanchet, and C. Geourjon. 1997. Protein structure prediction. Implications for the biologist. Biochimie 79:681-686. [DOI] [PubMed] [Google Scholar]
- 11.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Camara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 13.Dorman, C. J., J. C. Hinton, and A. Free. 1999. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 7:124-128. [DOI] [PubMed] [Google Scholar]
- 14.Esposito, D., A. Petrovic, R. Harris, S. Ono, J. F. Eccleston, A. Mbabaali, I. Haq, C. F. Higgins, J. C. Hinton, P. C. Driscoll, and J. E. Ladbury. 2002. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 324:841-850. [DOI] [PubMed] [Google Scholar]
- 15.Free, A., and C. J. Dorman. 1997. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J. Bacteriol. 179:909-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Free, A., M. E. Porter, P. Deighan, and C. J. Dorman. 2001. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol. Microbiol. 42:903-917. [DOI] [PubMed] [Google Scholar]
- 17.Goyard, S., and P. Bertin. 1997. Characterization of BpH3, an H-NS-like protein in Bordetella pertussis. Mol. Microbiol. 24:815-823. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, R. C., L. M. Johnson, J. Schmidt, and J. F. Gardner. 2005. Major nucleoid proteins in the structure and function of the Escherichia coli chromosome, p. 65-132. In N. P. Higgins (ed.), The bacterial chromosome. American Society for Microbiology, Washington, D.C.
- 18a.Munier, H., A. M. Gilles, P. Glaser, E. Krin, A. Danchin, R. Sarfati, and O. Barzu. 1991. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 196:469-474. [DOI] [PubMed] [Google Scholar]
- 19.Rescalli, E., S. Saini, C. Bartocci, L. Rychlewski, V. De Lorenzo, and G. Bertoni. 2004. Novel physiological modulation of the Pu promoter of TOL plasmid: negative regulatory role of the TurA protein of Pseudomonas putida in the response to suboptimal growth temperatures. J. Biol. Chem. 279:7777-7784. [DOI] [PubMed] [Google Scholar]
- 20.Schröder, O., and R. Wagner. 2002. The bacterial regulatory protein H-NS—a versatile modulator of nucleic acid structures. Biol. Chem. 383:945-960. [DOI] [PubMed] [Google Scholar]
- 21.Shi, X., and G. N. Bennett. 1994. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J. Bacteriol. 176:6769-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth, C. P., T. Lundback, D. Renzoni, G. Siligardi, R. Beavil, M. Layton, J. M. Sidebotham, J. C. Hinton, P. C. Driscoll, C. F. Higgins, and J. E. Ladbury. 2000. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 36:962-972. [DOI] [PubMed] [Google Scholar]
- 23.Sonden, B., and B. E. Uhlin. 1996. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 15:4970-4980. [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnenfield, J. M., C. M. Burns, C. F. Higgins, and J. C. Hinton. 2001. The nucleoid-associated protein StpA binds curved DNA, has a greater DNA-binding affinity than H-NS and is present in significant levels in hns mutants. Biochimie 83:243-249. [DOI] [PubMed] [Google Scholar]
- 25.Tendeng, C., and P. N. Bertin. 2003. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 11:511-518. [DOI] [PubMed] [Google Scholar]
- 26.Tendeng, C., O. A. Soutourina, A. Danchin, and P. N. Bertin. 2003. MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins. Microbiology 149:3047-3050. [DOI] [PubMed] [Google Scholar]
- 27.van Noort, J., S. Verbrugge, N. Goosen, C. Dekker, and R. T. Dame. 2004. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. USA 101:6969-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woldringh, C. L., and T. Odijk. 1999. Structure of DNA within the bacterial cell: physics and physiology, p. 171-187. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington, D.C.
- 29.Zhang, A., and M. Belfort. 1992. Nucleotide sequence of a newly-identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 20:6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, A., S. Rimsky, M. E. Reaban, H. Buc, and M. Belfort. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 15:1340-1349. [PMC free article] [PubMed] [Google Scholar]