Abstract
To examine the participation of P1 adhesin in gliding of Mycoplasma pneumoniae, we examined the effects of an anti-P1 monoclonal antibody on individual gliding mycoplasmas. The antibody reduced the gliding speed and removed the gliding cells from the glass over time in a concentration-dependent manner but had only a slight effect on nongliding cells, suggesting that the conformational changes of P1 adhesin and its displacement are involved in the gliding mechanism.
Mycoplasma gliding.
Mycoplasmas are parasitic, occasionally pathogenic, small-genome bacteria lacking a peptidoglycan layer (20). Several mycoplasma species, including Mycoplasma pneumoniae, M. genitalium, M. pulmonis, M. gallisepticum, and M. mobile, have distinct cell polarity and exhibit gliding motility in the direction of the tapered end (2, 10, 13). The mechanisms underlying gliding motility are intrinsically different from those of other motility systems and are not well understood (8, 12-15, 26).
M. pneumoniae and P1 adhesin.
M. pneumoniae, a human pathogen, forms a membrane protrusion, an attachment organelle, at a cell pole (11, 13). The cell surface of the attachment organelle exhibits clustering of a 170-kDa transmembrane protein, P1 adhesin, which is responsible for binding to animal cells and glass surfaces (4, 6, 19, 24). It shares structural similarities with the adhesion proteins of other mycoplasma species, such as MgPa of M. genitalium (7) and GapA of M. gallisepticum (5), but not with Gli349 of M. mobile, the fastest species (26). It is known that an antibody raised against P1 can block the binding of M. pneumoniae to animal cells and glass (4, 6, 19, 24), but the effects on glass binding or gliding have not been observed for individual cells (4, 6). Here we analyzed the effects of such an antibody on individual cells under conditions optimized for gliding.
Optimizing conditions for gliding.
In previous work, we found that a greater proportion of M. pneumoniae cells glided at higher speeds in a phosphate-buffered saline (PBS) solution containing serum than in the growth medium (9). Accordingly, in the present study we examined the effects of medium on gliding. M. pneumoniae M129 cells grown in Aluotto medium (1, 17) were suspended in fresh medium, dispersed as previously described (21, 22), put on a clean coverslip, and incubated at 37°C for 60 min to let the cells bind to the glass. The coverslip with mycoplasma cells was then assembled into a tunnel slide, as previously described (12, 26). After incubation of the cells on a microscope stage chamber at 37°C for 10 min, the growth medium was replaced by PBS containing 10% horse serum or by a fresh medium. The microscopic images were recorded and analyzed (15-17, 26). Since all cells are not always gliding (9), we examined both the proportion of gliding cells in relation to the total cells and the gliding speeds to evaluate the effects of the various conditions. The gliding activity presented by the two parameters did not change when the medium was replaced by fresh medium, but it increased in response to the replacement with PBS containing 10% serum. The proportion of gliding cells was 0 out of 406 cells at time zero but increased with time and reached 0.37 at 60 min, when the growth medium was replaced by PBS containing 10% serum. This proportion stayed at 0, however, when the growth medium was replaced with fresh medium. The gliding speed in PBS containing 10% serum also increased with time and plateaued at 0.93 μm/s at 15 min, although it did not change in the fresh medium. The average gliding speed of M. pneumoniae was originally reported to be as fast as 0.4 μm/s in a medium, comparable to the speed observed here in the PBS containing serum (3, 18). The content of the Aluotto medium used here was slightly different from that of the Hayflick medium used in the previous studies. We did try the Hayflick medium, but no difference in the gliding results was observed. These observations may suggest that the active gliding of M. pneumoniae is induced by starvation, which was unexpectedly achieved in the previous studies (3, 18).
We next examined the effects of serum concentrations, temperature, and gelatin. Once cells were bound to glass with 10% horse serum, gliding continued even in its absence but was better in concentrations ranging from 5 to 20%. The number of cells that glided was approximately the same over a temperature range of 27 to 42.5°C, but their speed increased linearly with temperature over this range from approximately 0.5 to 0.8 μm/s, as previously observed in the gliding of the fastest mycoplasma species, M. mobile (15). The addition of 1 to 5% gelatin did not prevent cells from leaving the glass during gliding (9, 18). Therefore, the effects of antibody were examined in PBS plus 10% horse serum without gelatin at 37°C.
Inhibition of gliding by anti-P1 adhesin antibody.
We made a monoclonal antibody by immunizing mice with a recombinant protein comprising 1,160 to 1,518 amino acids of a whole P1 molecule of 1,627 amino acids, which is known to have a site responsible for cell and glass binding (19). The specificity of antibody was confirmed by immunoblotting, immunofluorescence microscopy of fixed cells with and without permeabilization, and immunofluorescence microscopy of living cells (12, 22, 23, 26).
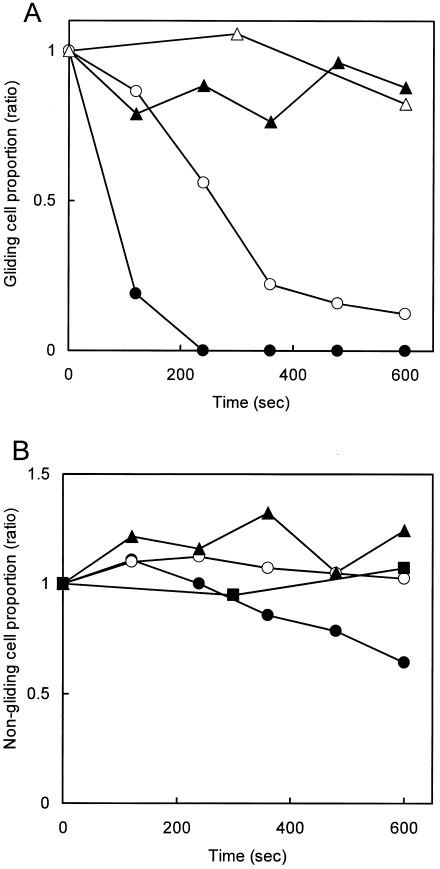
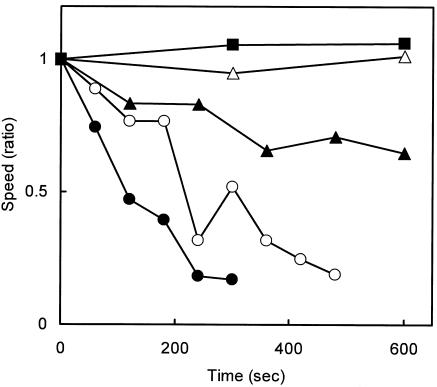
The effects of the antibody on gliding of individual cells were examined (Fig. 1 and 2). Cultured mycoplasma cells were resuspended in PBS containing 10% serum and bound to a clean coverslip at 37°C for 70 min. Then, PBS containing 10% serum was replaced by PBS containing 10% serum and various concentrations of the antibody, ranging from 0 to 300 μg/ml at time zero, and cells bound to glass with and without gliding motility were counted separately, as presented in Fig. 1A and B, respectively. The addition of antibody removed the gliding cells from the glass over time in a concentration-dependent manner (Fig. 1A). However, the antibody affected the glass binding of nongliding cells only slightly (Fig. 1B). These observations indicate that the displacement of a cell along a glass surface during gliding is essential to cell removal by the antibody. The effects of antibody on the gliding speed were examined (Fig. 2). The average speed of gliding cells was found to be reduced by the addition of antibody in a concentration-dependent manner, an effect similar to that for the inhibition of glass binding, indicating that the binding of antibody reduces the gliding speed.
FIG. 1.
Decrease in the number of bound cells after the addition of antibody. The number of bound cells relative to the initial number in a field of 9,600 μm2 is shown. (A) The ratio of gliding cells remaining on the glass is shown for each time point after the addition of antibody to 300 (closed circles), 30 (open circles), 3 (closed triangles), and 0 (open triangles) μg/ml relative to the number of cells gliding at time zero. (B) The number of nongliding cells remaining on the glass is shown relative to the number of nongliding cells at time zero. The same symbols as those in panel (A) are used. More than 100 cells were analyzed to determine the total number of gliding and nongliding cells at the zero time point.
FIG. 2.
Gliding speed after the addition of antibody. The gliding speeds normalized according to the initial speed are presented for each time point after the addition of antibody to 100 (closed circles), 30 (open circles), 10 (closed triangles), 3 (open triangles), and 0 (closed squares) μg/ml.
Involvement of P1 adhesin in gliding.
The dependence of the antibody's ability to inhibit glass binding during gliding on its concentration indicates that P1 is responsible not only for static binding but also for that during gliding (Fig. 1A).
The obvious difference in resistance to the antibody between gliding cells (Fig. 1A) and nongliding cells (Fig. 1B) suggests that P1 induces conformational changes in gliding. In other words, the P1 molecules should be in a state where the accessibility of the antibody is significantly reduced when the cell is not gliding on glass, compared to the states occurring in gliding. This observation can be explained by an assumption that the P1 molecule itself is involved in a “power stroke” that propels a cell like a leg.
The binding of antibody to a cell was found to decrease the gliding speed (Fig. 2), consistent with the observation of M. mobile with anti-Gli349 antibody (26). The results of the present study can be explained by one of the following three hypotheses, based on the assumption of a power stroke of the P1 molecule. The first hypothesis is that the binding of antibody reduces the rate of release of P1 molecules from the glass, resulting in generation of a drag force, and also blocks rebinding after the release, as discussed for M. mobile (refer to Fig. 7 of reference 26). The second hypothesis is that only a fraction of P1 molecules are in the propelling cycle, while others are in a state of static binding, keeping the cells on the glass and also causing a drag force in normal gliding. In this case, the binding of antibody causes a decrease in the number of P1 molecules in the cycle, resulting in a shortage of propelling force for a cell with the normal speed. In the third hypothesis, a fraction of P1 molecules are in the propelling cycle, as proposed in the second hypothesis, but the drag force is not large enough to balance the propelling force exerted through a P1 molecule in the cycle. However, the duration of a stroke is short, and the speed of a cell depends on the sum of stroke durations. In this case, the decrease in the number of P1 molecules in the cycle directly reduces the cell's gliding speed.
Conclusions.
We showed that P1 is involved in the gliding motility of M. pneumoniae. This finding may suggest that the gliding of other mycoplasma species sharing an adhesion protein structure with P1 (5, 7, 25) also depends on their adhesion molecules.
Acknowledgments
This work was supported in part by Grants-in-Aid for JSPS Fellows to S.S. and for Scientific Research (C) to M.M. from the Japan Society for the Promotion of Science and by grants for Science Research on Priority Areas (“Motor Proteins, ” “Genome Science, ” and “Infection and host response”) from the Ministry of Education, Science, Sports, Culture, and Technology to M.M.
REFERENCES
- 1.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 2.Bredt, W. 1979. Motility, p. 141-145. In M. F. Barile, S. Razin, J. G. Tully, and R. F. Whitcomb (ed.), The mycoplasmas, vol. 1. Academic Press, New York, N.Y.
- 3.Bredt, W. 1968. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol. Microbiol. (Basel) 32:321-326. [DOI] [PubMed] [Google Scholar]
- 4.Feldner, J., U. Gobel, and W. Bredt. 1982. Mycoplasma pneumoniae adhesin localized to tip structure by monoclonal antibody. Nature 298:765-767. [DOI] [PubMed] [Google Scholar]
- 5.Goh, M. S., T. S. Gorton, M. H. Forsyth, K. E. Troy, and S. J. Geary. 1998. Molecular and biochemical analysis of a 105 kDa Mycoplasma gallisepticum cytadhesin (GapA). Microbiology 144:2971-2978. [DOI] [PubMed] [Google Scholar]
- 6.Hu, P. C., R. M. Cole, Y. S. Huang, J. A. Graham, D. E. Gardner, A. M. Collier, and W. A. Clyde, Jr. 1982. Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science 216:313-315. [DOI] [PubMed] [Google Scholar]
- 7.Inamine, J. M., S. Loechel, A. M. Collier, M. F. Barile, and P. C. Hu. 1989. Nucleotide sequence of the MgPa (mgp) operon of Mycoplasma genitalium and comparison to the P1 (mpp) operon of Mycoplasma pneumoniae. Gene 82:259-267. [DOI] [PubMed] [Google Scholar]
- 8.Jaffe, J. D., M. Miyata, and H. C. Berg. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186:4254-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenri, T., S. Seto, A. Horino, Y. Sasaki, T. Sasaki, and M. Miyata. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 11.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 12.Kusumoto, A., S. Seto, J. D. Jaffe, and M. Miyata. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 150:4001-4008. [DOI] [PubMed] [Google Scholar]
- 13.Miyata, M. 2005. Gliding motility of mycoplasmas—the mechanism cannot be explained by current biology. .In A. Blanchard and G. Browning (ed.), Mycoplasmas: pathogenesis, molecular biology, and emerging strategies for control, in press. Horizon Scientific Press, Norwich, United Kingdom.
- 14.Miyata, M., and J. Petersen. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 186:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 17.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 18.Radestock, U., and W. Bredt. 1977. Motility of Mycoplasma pneumoniae. J. Bacteriol. 129:1495-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesion. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 20.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero-Arroyo, C. E., J. Jordan, S. J. Peacock, M. J. Willby, M. A. Farmer, and D. C. Krause. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence protein and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svenstrup, H. F., P. K. Nielsen, M. Drasbek, S. Birkelund, and G. Christiansen. 2002. Adhesion and inhibition assay of Mycoplasma genitalium and M. pneumoniae by immunofluorescence microscopy. J. Med. Microbiol. 51:361-373. [DOI] [PubMed] [Google Scholar]
- 25.Tham, T. N., S. Ferris, E. Bahraoui, S. Canarelli, L. Montagnier, and A. Blanchard. 1994. Molecular characterization of the P1-like adhesin gene from Mycoplasma pirum. J. Bacteriol. 176:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]