Abstract
Proteins containing GGDEF domains are encoded in the majority of sequenced bacterial genomes. In several species, these proteins have been implicated in biosynthesis of exopolysaccharides, formation of biofilms, establishment of a sessile lifestyle, surface motility, and regulation of gene expression. However, biochemical activities of only a few GGDEF domain proteins have been tested. These proteins were shown to be involved in either synthesis or hydrolysis of cyclic-bis(3′→5′) dimeric GMP (c-di-GMP) or in hydrolysis of cyclic AMP. To investigate specificity of the GGDEF domains in Bacteria, six GGDEF domain-encoding genes from randomly chosen representatives of diverse branches of the bacterial phylogenetic tree, i.e., Thermotoga, Deinococcus-Thermus, Cyanobacteria, spirochetes, and α and γ divisions of the Proteobacteria, were cloned and overexpressed. All recombinant proteins were purified and found to possess diguanylate cyclase (DGC) activity involved in c-di-GMP synthesis. The individual GGDEF domains from two proteins were overexpressed, purified, and shown to possess a low level of DGC activity. The oligomeric states of full-length proteins and individual GGDEF domains were similar. This suggests that GGDEF domains are sufficient to encode DGC activity; however, enzymatic activity is highly regulated by the adjacent sensory protein domains. It is shown that DGC activity of the GGDEF domain protein Rrp1 from Borrelia burgdorferi is strictly dependent on phosphorylation status of its input receiver domain. This study establishes that majority of GGDEF domain proteins are c-di-GMP specific, that c-di-GMP synthesis is a wide-spread phenomenon in Bacteria, and that it is highly regulated.
The approximately 170-amino-acid-long protein domain GGDEF (http://www.sanger.ac.uk/Software/Pfam), also referred to as domain of unknown function 1 (designated DUF1) (http://smart.embl-heidelberg.de), is a conserved domain in Bacteria. However, its function has yet to be properly characterized. The domain name originates from the amino acid motif GGDEF (Gly-Gly-Asp-Glu-Phe). This domain is present in most sequenced genomes from all branches of the phylogenetic tree of Bacteria. This implies that GGDEF domain-containing proteins play important roles in Bacteria; however, such roles remain largely unknown. The presence of GGDEF domains in the genomes of representatives of the deepest branches of the bacterial tree, e.g., Aquifex and Thermotoga, suggests their ancient evolutionary origin (10, 11). It is evident that proper elucidation of structure-function relationships of the GGDEF domain and GGDEF domain-containing proteins will improve our understanding of the physiology, metabolism, and behavior of bacteria and will therefore enhance our ability to manipulate them.
The limited, yet rapidly accumulating, genetic and biochemical evidence suggests that the GGDEF domain is involved in the synthesis and hydrolysis of cyclic diguanylate, or cyclic-bis(3′→5′) dimeric GMP (c-di-GMP) (reviewed in references 8 and 13). This compound was originally identified as an allosteric activator of cellulose synthase in Gluconacetobacter xylinus, formerly known as Acetobacter xylinum (20, 22). Three paralogous diguanylate cyclases, (DGC) from G. xylinus involved in c-di-GMP synthesis were found to contain two conserved protein domains, GGDEF and EAL (http://www.sanger.ac.uk/Software/Pfam), where EAL is also known as DUF2 (http://smart.embl-heidelberg.de). A DGC was shown to catalyze the synthesis of c-di-GMP from two molecules of GTP via two distinct pyrophosphate-releasing steps with a linear dinucleotide, pppGpG, as an intermediate (20). Interestingly, enzymes involved in the original step in c-di-GMP hydrolysis, phosphodiesterases A (PDE-A), purified from G. xylinus also contain GGDEF and EAL domains. PDE-A cleave a single phosphodiester bond in the cyclic structure, yielding a linear dimer GpGp (l-di-GMP), which is further converted to 5′-GMP by different PDE (20, 21).
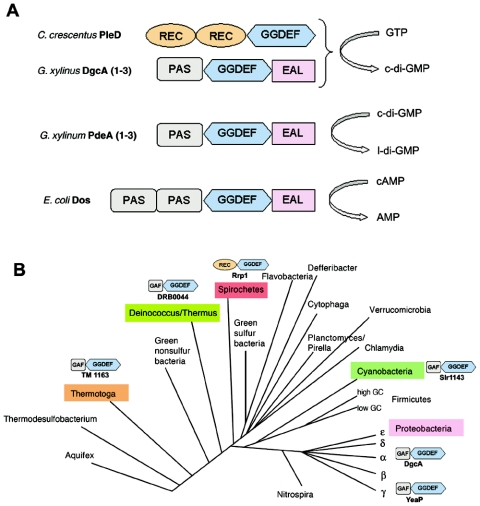
Pei and Grishin noticed low yet significant sequence similarity between GGDEF domains and catalytic domains of mammalian adenylyl cyclases. They predicted that the GGDEF domain may possess DGC activity (18). Up to date, biochemical activities of only a few GGDEF domain-containing proteins have been tested directly, all originating from the proteobacterial species. These proteins include PleD from Caulobacter crescentus (17) and Dos from Escherichia coli (23), as well as the G. xylinus proteins mentioned above. PleD functions as DGC (17), whereas Dos possesses cyclic AMP (cAMP)-dependent PDE activity (Fig. 1A) (23).
FIG. 1.
(A) Domain organization of the GGDEF domain proteins whose biochemical activities have been tested prior to this study. (B) Phylogenetic tree of Bacteria showing proteins investigated in this study. GAF, signal sensor domain often found in phytochromes and cyclic GMP-specific PDE; EAL, PDE domain probably specific to c-di-GMP; PAS, ubiquitous signal sensor domain; REC, receiver domain of response regulators from two-component systems (http://www.sanger.ac.uk/Software/Pfam; http://smart.embl-heidelberg.de).
There is a growing body of evidence linking additional GGDEF domain proteins to c-di-GMP synthesis. For example, when overexpressed in rhizobia, some proteobacterial GGDEF domain proteins were able to activate cellulose synthesis, which is anticipated to be c-di-GMP dependent (2). Overexpression of the GGDEF domain-containing proteins AdrA from Salmonella enterica serovar Typhimurium (24) and VCA0956 from Vibrio cholerae (29) resulted in increased intracellular levels of c-di-GMP, thus implying that these proteins possess DGC activity. However, in these instances, alternative explanations, i.e., indirect effects of the GGDEF proteins, cannot be excluded.
Our study addressed the following questions concerning GGDEF domain proteins. How specific are GGDEF domain proteins to synthesis or hydrolysis of c-di-GMP? Are other substrates, cAMP in particular, involved to an appreciable degree? How widespread is c-di-GMP synthesis in Bacteria? Is the GGDEF domain sufficient for DGC activity? What role do domains adjacent to GGDEF play in enzymatic activity? To answer these questions, we cloned several genes encoding GGDEF domain proteins from various bacteria and assayed for their enzymatic activities.
MATERIALS AND METHODS
Construction of plasmids for overexpression of the GGDEF domain-containing proteins.
Genomic DNA from Rhodobacter sphaeroides 2.4.1, Synechocystis sp. PCC6803, and E. coli DH5α was purified from bacterial cells using the Bactozol kit (Molecular Research Center, Cincinnati, Ohio). Genomic DNA from Thermotoga maritima DSM3109, Deinococcus radiodurans R1, and Borrelia burgdorferi B31 was purchased from the American Type Culture Collection. The genes encoding GGDEF domain proteins from these bacteria were PCR amplified with Pfu Hotstart DNA polymerase (Stratagene) and the following primers (shown in parentheses): R. sphaeroides RSP3513 (F3513-EcoRI, 5′-CGGGAATTCCAGGACTGCGAGAAACTTCTCG; and F3513-NsiI, 5′-CCAATGCATGACTGTCCTCACGCTCGCGAGG), E. coli yeaP (YeaP-XbaI, 5′-GCTCTAGATCAGATCAGATTATCGCCCGC; and YeaP-HindIII, 5′CAGAAGCTTGGAATGTAGCGCTGGATGCG), T. maritima TM1163 (TM1163- BamHI, 5′-CGGGATCCGTTGATGGACTTTCAAAACTCG; and TM1163-NsiI, 5′-CAATGCATCGCTCAGGACGATGCTCACGTC), D. radiodurans DRB0044 (DRB0044-XbaI, 5′-GCTCTAGAACCACAGCGGCGCCACAGCCTC; and DRB0044-HindIII, 5′-CAGAAGCTTTGTGATCCAGACCGACCACTTC), Synechocystis sp. slr1134 (SLR1143-XbaI, 5′-GCTCTAGAGAAGCTAAATTACCGCAAAATG; and SLR1134-HindIII, 5′-CAGAAGCTTTTATTCTGCCAGTTGAAAATTGCC), and B. burgdorferi BB0419 and rrp1 (BB0419-BamHI, 5′-CGGGATCCGCTTTTGAAGCAGAGAATCAGAAGC; and BB0419-NsiI, 5′-CCAATGCATCCACGATTCTATTGGAATGTCTATAAC). The DNA fragments encoding isolated GGDEF domains were amplified with the following primers: for RSP3513, DgcA-XbaI (5′-GCTCTAGAGAGCTGGAACTCCGGCAG) and F3513-NsiI; for slr1143, 1143_GG-XbaI (5′-GCTCTAGAGAACTTGAAAGGGTGGCCATGG) and SLR1143-HindIII. The DNA fragment encoding GGDEF and coiled-coil (CC) domains from slr1143 was amplified with 1143_CC-XbaI 5′-GCTCTAGAGAACTGGCGGCGATCGCCC) and SLR1143-HindIII. Restriction sites introduced for cloning purposes are present in primer names, and their recognition sequences are underlined in the primer sequences shown above.
PCR fragments were gel purified with the Gel Recovery kit (Zymo Research), digested with the indicated restriction enzymes, and cloned into vector pMAL-c2x (New England Biolabs) in strain E. coli DH5α or into vector pET23a (Invitrogen, Carlsbad, Calif.) in strain E. coli BL21(DE3) containing either pLysS or pLysE (Invitrogen).
Protein overexpression and purification.
The constructed plasmids were used for overexpression of proteins and protein domains as fusions to maltose binding protein (MBP). GGDEF domain protein overexpression was performed essentially according to the manufacturer's instructions (pMAL expression system; New England Biolabs). Briefly, IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration, 0.2 mM) was added to exponentially growing E. coli DH5α cells containing the appropriate plasmids (at an optical density [A600] of 0.6 to 0.8). After 2 h of induction, cells were collected by centrifugation. Cell pellets were resuspended in a buffer containing 200 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 20 mM Tris-HCl (pH 7.6), and 5% glycerol that also contained protease inhibitors (phenylmethylsulfonyl fluoride and P8465; Sigma, St. Louis, Mo.) at the concentrations specified by the manufacturer. Cell suspensions were passed through a French pressure minicell (Spectronic Instruments), followed by brief sonification (Sonifier 250; Branson). Crude cell extracts were centrifuged at 15,000 × g for 45 min. Soluble protein fractions were collected and mixed with the preequilibrated amylose resin (New England Biolabs) for 1 h at 4°C. MBP-protein fusions were eluted with maltose and dialyzed against the cyclase assay buffer in Slide-A-Lyzer dialysis cassettes (Pierce) according to the instructions of the manufacturer. Protein purity was assessed by capillary electrophoresis (Bioanalyzer; Agilent Technologies) or sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein concentration was measured with the BCA protein assay kit (Pierce). Proteins requiring further purification were subjected to gel filtration by fast protein liquid chromatography (FPLC).
Enzymatic assays.
The assay buffer and reaction conditions were essentially as described elsewhere (22). A standard reaction mixture (total volume, 0.6 ml) contained 5 μM enzyme in 50 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 0.5 mM EDTA, and 50 mM NaCl. The reaction was started by the addition of 4.5 μl of substrate (final concentration, 150 μM) to the prewarmed reaction mixture and was carried out for 0, 3, 5, 15, 30, or 60 min. Aliquots (each, 100 μl) were withdrawn at the indicated time points and immediately placed in a boiling water bath for 3 min, followed by centrifugation at 15,000 × g for 2 min. The supernatant was filtered through a 0.22-μm-pore-size filter and analyzed by high-pressure liquid chromatography (HPLC). Activities of RSP3513, Slr1143, and Rrp1 were assayed at 30°C; YeaP and DRB0044 were assayed at 37°C, and TM1163 was assayed at 37 and 50°C.
HPLC.
Samples (each, 20 μl) were injected into the 15- by 4.6-cm Supelcosil LC-18-T column (Supelco) and separated by reversed-phase HPLC (Summit HPLC system; Dionex). The following buffers were used in the gradient program: buffer A (100 mM KH2PO4, 4 mM tetrabutyl ammonium hydrogen sulfate [pH 5.9]) and buffer B (75% buffer A, 25% methanol). The following protocol was used for separation (the values are times in minutes and percentage of buffer B used): 0.0, 0; 2.5, 0; 5.0, 30; 10.0, 60; 14.0, 100; 21.0, 100; 22.0, 50; and 23.0, 0 at a flow rate of 0.7 ml min−1. Nucleotides were detected at a wavelength of 254 nm. Equal amounts of nicotine ADP (final concentration, 15 μM) were added to each sample prior to injection for quantification purposes.
FPLC.
Purification of proteins and determination of their oligomeric state was done with a Superdexj 200 10/300 GL gel filtration column (Amersham Biosciences) equilibrated with the cyclase assay buffer at a flow rate of 0.4 ml min−1. The dependence of log(Mr) on retention volume was linear (R2 = 0.9995) in the wide range of molecular masses (from 17 to 670 kDa), allowing accurate estimates of molecular masses of protein mono- and oligomers.
Mass spectroscopy.
Reaction products were diluted 1:10 in the solution containing 10 mg ml−1 α-cyano-4-hydroxycinnamic acid, 0.1% trifluoroacetic acid, and 50% acetonitrile. A total of 1 μl was spotted onto a matrix-assisted laser desorption ionization-time of flight plate. The samples were analyzed in reflectron and negative-ion modes with the Voyager DE PRO mass spectrometer (Applied Biosystems). The resulting spectra were calibrated against bradykinin fragment 1-7 (molecular weight, 757.3997) and des-arg-bradykinin (molecular weight, 904.4681) by close external calibration.
RESULTS AND DISCUSSION
Toxicity of c-di-GMP to E. coli BL21.
For initial characterization, we chose gene RSP3513 from the anoxygenic photosynthetic proteobacterium R. sphaeroides (15). This gene is constitutively expressed under a variety of growth conditions (7, 16), suggesting that it encodes a functional protein. In addition to GGDEF, the RSP3513 protein contains a ubiquitous protein domain, GAF (http://www.sanger.ac.uk/Software/Pfam), which often functions as a small ligand binding sensory domain (33).
We noticed that the efficiency of cloning of the RSP3513 gene into the pET23a vector using E. coli strain BL21(DE3)(pLysS) host was unexpectedly low. After several attempts, we could isolate only a few plasmids expressing the RSP3513::His6 fusion proteins. Sequence analysis of these plasmids revealed that all contained point mutations or deletions in the sequence of RSP3513 (unpublished data). We pursued cloning of the intact RSP3513 gene with the strain with a tighter control of expression in the absence of induction, i.e., BL21(DE3)(pLysE). However, these attempts have also failed. We concluded that either the RSP3513 protein or the product(s) of its activity is toxic to strain BL21. Attempts to clone intact genes encoding GGDEF domain proteins from other bacteria, i.e., Synechocystis sp. and E. coli, resulted in the same outcome. Therefore, toxicity was not RSP3513 protein specific but was specific to the product of reaction catalyzed by these GGDEF domain proteins, i.e., c-di-GMP (see below). Interestingly, toxicity turned out to be strain dependent. As is shown below, we successfully overexpressed several GGDEF domain proteins in strain E. coli DH5α. The reason for strain-specific toxicity is unknown at present. Our results must be considered as a warning against using BL21-derived strains for cloning the GGDEF domain-encoding genes.
R. sphaeroides RSP3513 encodes DGC.
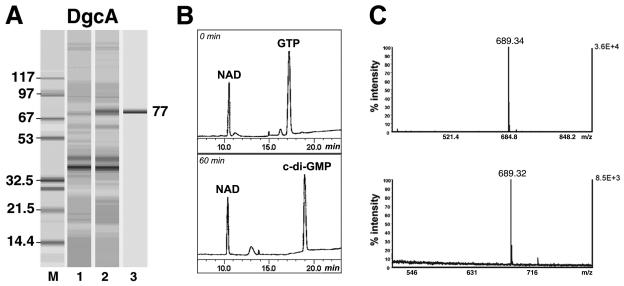
The intact RSP3513 gene was cloned into the pMAL-c2x vector with E. coli DH5α, overexpressed as an amino-terminal MBP fusion, and purified to homogeneity by affinity and gel filtration chromatography (Fig. 2A). Activity of the purified protein was tested with the following nucleotides: ATP, ADP, AMP, GTP, GDP, GMP, CTP, TTP, c-di-GMP, cAMP, and cGMP. GTP was found to be the sole substrate for the RSP3513 protein. GTP was quantitatively converted to c-di-GMP (Fig. 2B). Several lines of evidence show that the product of RPS3513 is c-di-GMP. (i) The retention time of the reaction product on the HPLC column is identical to that of the chemically synthesized c-di-GMP (data not shown). (ii) Treatment of the reaction product with the previously described c-di-GMP-specific PDE-A from G. xylinus, PdeA1 (26), resulted in l-di-GMP, as judged by the fact that its retention time is identical to that of the chemically synthesized c-di-GMP treated with G. xylinus PDE-A1 (not shown). Subsequent hydrolysis of l-di-GMP by the membrane fraction purified from G. xylinus yielded GMP, i.e., similar to what was described by Ross et al. (22). (iii) The molecular mass of the product of RSP3513 reaction measured by mass spectroscopy, 689.32 Da, was found to be identical to the molecular mass of the chemically synthesized c-di-GMP, 689.34 (Fig. 2C).
FIG. 2.
R. sphaeroides DgcA protein (RSP3513). (A) Protein overexpression and purification (protein chip, Bioanalyzer; Agilent Technologies). Lane M, molecular mass markers in kilodaltons; lane 1, crude extract of E. coli cells prior to induction of expression of the MBP::DgcA fusion protein; lane 2, crude extract of E. coli cells after 2-h induction with IPTG; lane 3, pure protein after affinity chromatography and gel filtration. (B) Enzymatic activity of DgcA. Substrate (top; 0 min) and products (bottom; 60 min) of the reaction, separated by reversed-phase HPLC. Nicotine ADP was used as an internal control for quantification purposes. (C) Mass spectroscopy analysis of the product synthesized by MBP::DgcA. The top panel corresponds to chemically synthesized c-di-GMP. The lower panel corresponds to the product of the HPLC fraction, with retention time of 19 min (see Fig. 2A, bottom).
This establishes RSP3513 as a DGC, which is hereby designated DgcA. This suggests that R. sphaeroides synthesizes c-di-GMP under a variety of growth conditions. The role of this compound in R. sphaeroides is currently under investigation.
DGC activity is ubiquitous in the bacterial world.
Based on the presence of GGDEF domain-encoding proteins in almost all sequenced bacterial genomes, it has been predicted that c-di-GMP is ubiquitous in bacteria (10, 11). However, it is unknown whether the GGDEF domain proteins from species outside of the proteobacterial branch are specific to c-di-GMP or cAMP, as both compounds have been associated with the GGDEF domain (Fig. 1A). It is possible that some GGDEF domain proteins have mixed specificities, which is characteristic of a group of mammalian PDE (31). It is also possible that a different type of nucleotide is used as a substrate for GGDEF domain proteins.
To investigate these possibilities, we tested proteins containing GGDEF domains from representatives of diverse branches of the phylogenetic tree of Bacteria. The following proteins were chosen: TM1163 from the hyperthermophilic marine bacterium T. maritima (branch Thermotoga), DRB0044 from the radiation- and desiccation-resistant soil bacterium D. radiodurans (Deinococcus/Thermus), Slr1143 from the oxygenic phototroph Synechocystis sp. (Cyanobacteria), Rrp1 (BB0419) from the intracellular parasite B. burgdorferi (spirochetes), and YeaP from the intestine inhabitant E. coli (γ subdivision of the Proteobacteria) (Fig. 1B). These proteins were chosen essentially at random. The anticipated ease of protein purification, i.e., relatively small size and lack of transmembrane domains, were used as sole criteria. The domain architecture of most chosen proteins turned out to be similar to that of R. sphaeroides DgcA, i.e., GAF plus GGDEF. The B. burgdorferi Rrp1 protein had a different domain structure, i.e., REC plus GGDEF (Fig. 1B), where REC is a receiver domain characteristic of response regulators of bacterial two-component systems (http://smart.embl-heidelberg.de). REC is also known as the CheY domain (http://www.sanger.ac.uk/Software/Pfam). None of the functions of these proteins was known prior to this study.
All selected genes were amplified from genomic DNA from their respective species and overexpressed in E. coli DH5α as fusions to the MBP protein. The recombinant proteins were purified by affinity chromatography and gel filtration, where needed, and assayed for biochemical activities with various nucleotides (Fig. 3A and data not shown). Four of five proteins (TM1163, DRB0044, Slr1143, and YeaP) used GTP as a substrate and showed DGC activity similar to that of the R. sphaeroides DgcA protein (Fig. 3B and data not shown).
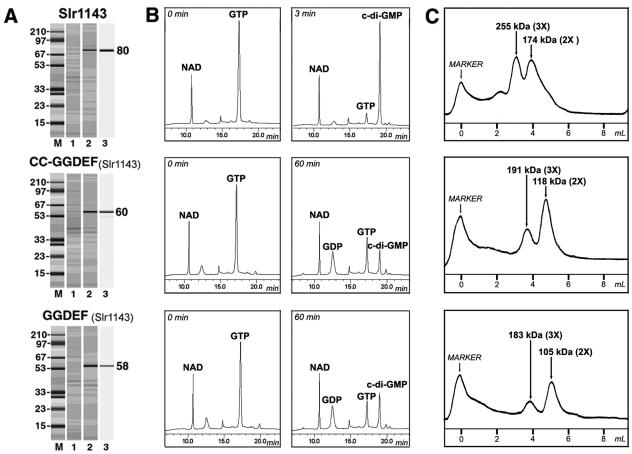
FIG. 3.
Synechocystis sp. Slr1143 protein. Top, full-length protein; middle, protein fragment corresponding to the CC-plus-GGDEF domains; bottom, protein fragment corresponding to the GGDEF domain. (A) Protein overexpression and purification. For details, see the legend to Fig. 2A. (B) Enzymatic activity. For details, see the legend to Fig. 2B. (C) Oligomeric state of MBP::Slr1143 and its derivatives assayed by FPLC. Marker, blue dextran.
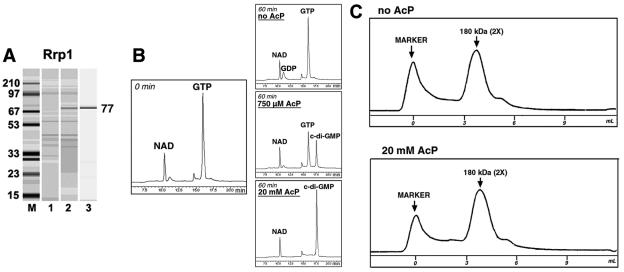
The purified recombinant Rrp1 protein (Fig. 4A) showed no activity, even on prolonged incubation (Fig. 4B, top right). This prompted us to investigate Rrp1 further. Considering the domain architecture of Rrp1, we predicted that its activity could depend on the phosphorylation status of its REC domain. To test this hypothesis, we incubated Rrp1 with acetyl phosphate, a small-molecule phosphate donor. Acetyl phosphate specifically phosphorylates the acceptor aspartyl residues of response regulators, which are phosphorylated in vivo by histidine protein kinases (14). It is worth noting that in the B. burgdorferi genome (GenBank accession number AE001146), a gene encoding histidine protein kinase, hpk1 (BB0420), is located immediately upstream of the rrp1 gene. Furthermore, hpk1 and rrp1 appear to be contranslated. This suggests that hpk1 is likely to encode the cognate kinase of Rrp1.
FIG. 4.
B. burgdorferi Rrp1 protein. (A) Protein overexpression and purification. For details, see the legend to Fig. 2A. (B) Enzymatic activity. The substrate (left; 0 min) and products (right; 60 min) of the reaction mixture separated by HPLC are shown. MBP::Rrp1 (7.5 μM) was incubated with the indicated concentrations of acetyl phosphate (AcP) for 30 min at room temperature, followed by removal of AcP by dialysis. (C) Oligomeric state of the MBP::Rrp1 protein assayed by FPLC.
The Rrp1 protein, which was incubated with acetyl phosphate and subsequently dialyzed, showed DGC activity. Preincubations with higher concentrations of acetyl phosphate resulted in higher levels of DGC activity (Fig. 4B and data not shown). These observations allow us to suggest that the DGC activity of Rrp1 depends on the phosphorylation status of its REC domain. Similar to other DGC proteins, Rrp1 was highly specific to GTP as a substrate.
The fact that six arbitrary chosen GGDEF domain proteins from diverse branches of Bacteria possess DGC activity establishes that synthesis of c-di-GMP is a widespread phenomenon and that c-di-GMP is ubiquitous in bacteria. The presence of DGC activity in T. maritima, which belongs to one of the deepest branches of the bacterial phylogenetic tree, suggests that c-di-GMP is an ancient molecule that apparently evolved at the dawn of bacterial evolution. This is supported by the presence of several GGDEF domain proteins in Aquifex aeolicus, a representative from yet another early diverging branch of bacteria (Fig. 1B). Our data strongly suggest that majority of GGDEF domain proteins encoded in bacterial genomes are involved in synthesis and/or hydrolysis of c-di-GMP, not cAMP.
No full-length GGDEF domains are encoded in the currently sequenced genomes of Archaea. We identified one protein, Methanopyrus kandleri MK0296, and one protein fragment, Haloferax volcanii Q9C4S7, that contain sequences of apparently truncated GGDEF domains, i.e., 144 and 84 amino acids, respectively. Whether or not these proteins are functional is unknown. It is possible that the archaean GGDEF-like domains have been acquired via interkingdom gene transfer from Bacteria. However, we could not predict this with certainty, based on the G+C content or the gene neighborhood of the MK0296 gene (http://www.tigr.org).
Six putative GGDEF domain-containing proteins are predicted in the unfinished genome of a eukaryote, mosquito Anopheles gambiae (http://smart.embl-heidelberg.de). Some of these genes, e.g., ENSANGG00000000049 and ENSANGP00000001831, are predicted to be composed of more than one exon, i.e., they have a eukaryotic gene structure (http://www.ensembl.org/Anopheles_gambiae). Interestingly, these proteins are very similar to the GGDEF domain proteins from the Proteobacteria, e.g., ENSANGG00000023215 from mosquito is 65% identical over the entire protein length to the putative Na+/Ca2+ antiporter from Desulfovibrio desulfuricans. Such level of similarity is unprecedented for an apparently nonessential protein. It is possible that the mosquito genes represent a recent case of interkingdom gene transfer from Bacteria. Alternatively, the GGDEF domain-encoding sequences may have originated from a contaminant genomic DNA of a bacterial symbiont or parasite of mosquito and genome sequence misassembly. In summary, functionality of the GGDEF domain proteins and the presence of c-di-GMP in Archaea and Eukarya are highly questionable.
Enzymatic activity of the GGDEF domains.
We investigated enzymatic activities of individual GGDEF domains. To this end, we cloned and overexpressed sequences corresponding to GGDEF domains from two proteins, R. sphaeroides DgcA and Synechocystis sp. Slr1143. The MBP::GGDEF fusion proteins were purified and assayed with various nucleotides. Low amounts of c-di-GMP were formed upon incubation of each of these proteins with GTP (Fig. 3 and data not shown). DGC activity of the GGDEF domains of DgcA and Slr1143 was significantly, i.e., approximately 2 orders of magnitude, lower than those of the corresponding full-length proteins. Both isolated GGDEF domains possessed somewhat higher phosphatase, i.e., GTPase, activity that converted GTP into GDP (Fig. 3B).
That DGC activity is an intrinsic property of the GGDEF domain is shown here for the first time. Why is such activity low compared to that of full-length proteins? We hypothesized that either conformation or the oligomeric state of individual GGDEF domains prevents expression of high-level DGC activity. The latter possibility seemed reasonable given that adenylyl cyclases, which share sequence similarity with the GGDEF domains (18), function as dimers (27). This prompted us to compare oligomeric states of the full-length proteins and individual GGDEF domains.
Oligomeric state of DGCs.
All six full-length DGC proteins analyzed in this study were found to exist primarily as dimers and/or trimers (Fig. 3C, top; Fig. 4C, top; and data not shown). The individual GGDEF domains from DgcA and Slr1143 also formed dimers and trimers (Fig. 3C, bottom, and data not shown). Therefore, GGDEF domains have intrinsic propensity to di- and trimerization. This suggests that low DGC activity of individual GGDEF domains did not result from an improper oligomeric state.
It is possible that, although dimers and trimers are formed by individual GGDEF domains, they are folded improperly. We explored this issue further by using the Slr1143 protein. Slr1143 contains a well-defined CC motif (http://smart.embl-heidelberg.de) between the GAF and GGDEF domains. Hydrophobic interactions between two CC motifs are known to promote protein dimerization. We reasoned that presence of the CC motifs would impose proper folding constraints on the CC-plus-GGDEF protein fragment, thus ensuring that the fold of the GGDEF domain resembles its fold in the full-length protein. We cloned and overexpressed the DNA fragment corresponding to the CC plus GGDEF portions of Slr1143 and purified the MBP::(CC + GGDEF) fusion (Fig. 3A, middle). The oligomerization state of the fusion protein was found to be identical to that of the individual GGDEF domain or full-length Slr1143 (Fig. 3C, middle). However, DGC activity of this construct remained low and similar to that of the single GGDEF domain (Fig. 3B, middle).
From these studies, we conclude that the ability of GGDEF domains to convert GTP into c-di-GMP, although intrinsic to the GGDEF domain, is strongly affected by neighboring protein domains or possibly by interacting proteins. In case of Rrp1, the phosphorylated REC domain is apparently required for high-level DGC activity. The nonphosphorylated REC domain either does not promote DGC activity or inhibits it. In the cases of five other proteins, the presence of the GAF domains is sufficient for expression of the DGC activity in vitro. We suggest that activities of these five proteins in vivo are further modulated via ligands anticipated to bind to the GAF domains. At present, the identities of these ligands, if any, are unknown. However, neither cAMP nor cGMP, both of which bind to some GAF domains (33), stimulated DGC activities of the tested proteins, and neither did c-di-GMP, thus excluding the possibility of positive autoregulation by the reaction product (data not shown).
Strong dependence on activating stimuli provides a possible answer to the question of how bacteria can maintain dozens of GGDEF domain proteins yet have very low levels of intracellular c-di-GMP (24, 29). Most DGC in cells are likely to be inactive or almost inactive, unless a specific environmental or intracellular signal activates them.
c-di-GMP as an underappreciated ubiquitous signaling molecule in Bacteria.
Architectures of GGDEF domain-containing proteins encoded by bacterial genomes indicate that they sense various environmental and intracellular signals, e.g., oxygen, light, small ligands, and membrane-derived signals (10, 11). Mutations in the GGDEF domain proteins or overexpression of such proteins affect exopolysaccharide synthesis in various proteobacterial species, including G. xylinus (20), V. cholerae (5, 19), Pseudomonas aeruginosa (9), Pseudomonas fluorescens (25), Agrobacterium tumefaciens (2), E. coli, and S. enterica (24, 32). In some of these species, this further affects exopolysaccharide-dependent formation of biofilms. In C. crescentus, flagellum ejection, which is required for the switch from motile to sessile lifestyle, is impaired in the GGDEF domain protein PleD mutation (1). In Synechocystis sp. PCC 6803 (30), P. aeruginosa (12), and Vibrio parahaemolyticus (4), mutations in the GGDEF domain proteins impair surface motility. In V. cholerae and Synechococcus elongatus, the proteins anticipated to be involved in c-di-GMP turnover affect gene expression (28, 29). Apparently, the GGDEF domain proteins integrate various environmental and intracellular stimuli into changes in c-di-GMP levels, which in turn control bacterial life, primarily life on surfaces. However, the full range of functions and targets of c-di-GMP has yet to be revealed. For example, in this work we observed strong toxicity of c-di-GMP to E. coli BL21. The target(s) of c-di-GMP action resulting in toxicity has yet to be identified.
Conclusions.
We provided biochemical evidence that one of the most ubiquitous protein domains encoded in bacterial genomes, GGDEF, is primarily associated with c-di-GMP, not cAMP. We reported that diverse species of Bacteria are capable of c-di-GMP synthesis. We showed that DGC activity of GGDEF domain proteins is apparently highly regulated by the adjacent sensory domains. When taken together with the physiological and behavioral data on the GGDEF proteins cited above, our work establishes c-di-GMP as a ubiquitous signaling molecule in Bacteria that apparently functions as a second messenger. Among the best characterized and widely distributed second messengers are cyclic mononucleotides, cAMP and cyclic GMP (3, 6). The time has come for c-di-GMP to join the ranks of these structurally related but currently much better understood second messengers.
Acknowledgments
This work was supported by NSF grant MCB-0316270.
We are indebted to the late Moshe Benziman and Haim Weinhoiz, Hebrew University of Israel, for G. xylinus and chemically synthesized c-di-GMP, as well as for generously sharing the assay protocols. We are thankful to Renata Green (supported by the Summer Research Fellowship from the Department of Molecular Biology, University of Wyoming) and Whitney Tarver (supported by the NSF EPSCoR Summer Research Apprenticeship Program for high school students at the University of Wyoming) for their contribution to cloning of the R. sphaeroides dgcA gene, to Mathew Chivvis for testing the effect of the pLysE plasmid, and to Justin Jones for help with mass spectrometry experiments performed at the Macromolecular Core Facility of the University of Wyoming.
Footnotes
This paper is dedicated to the memory of late Professor Moshe Benziman, Hebrew University, Israel, in whose laboratory c-di-GMP was discovered.
REFERENCES
- 1.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 2.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 3.Baker, D. A., and J. M. Kelly. 2004. Structure, function and evolution of microbial adenylyl and guanylyl cyclases. Mol. Microbiol. 52:1229-1242. [DOI] [PubMed] [Google Scholar]
- 4.Boles, B. R., and L. McCarter. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184:5946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botsford, J., and J. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45:827-836. [DOI] [PubMed] [Google Scholar]
- 8.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497-2502. [DOI] [PubMed] [Google Scholar]
- 9.D'Argenio, D., M. Calfee, P. Rainey, and E. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 12.Huang, B., C. Whitchurch, and J. S. Mattick. 2003. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 185:7068-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 14.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackenzie, C., M. Choudhary, F. W. Larimer, P. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. Donohue, J. Hosler, J. Newman, J. Shapleigh, E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2002. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 70:19-41. [DOI] [PubMed] [Google Scholar]
- 16.Pappas, C., J. Sram, O. Moskvin, P. Ivanov, R. Mackenzie, M. Choudhary, M. Land, F. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 186:4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei, J., and N. Grishin. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42:210-216. [DOI] [PubMed] [Google Scholar]
- 19.Rashid, M. H., C. Rajanna, A. Ali, and D. K. Karaolis. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227:113-119. [DOI] [PubMed] [Google Scholar]
- 20.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 21.Ross, P., R. Mayer, H. Weinhouse, D. Amikam, Y. Huggirat, M. Benziman, E. de Vroom, A. Fidder, P. de Paus, L. A. Sliedregt, G. A. van der Marel, and J. H. van Boom. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265:18933-18943. [PubMed] [Google Scholar]
- 22.Ross, P., Y. Aloni, H. Weinhouse, D. Michaeli, P. Weinberger-Ohana, R. Mayer, and M. Benziman. 1986. Control of cellulose synthesis in Acetobacter xylinum. A unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr. Res. 149:101-117. [Google Scholar]
- 23.Sasakura, Y., S. Hirata, S. Sugiyama, S. Suzuki, S. Taguchi, M. Watanabe, T. Matsui, I. Sagami, and T. Shimizu. 2002. Characterization of a direct oxygen sensor heme protein from Escherichia coli. Effects of the heme redox states and mutations at the heme-binding site on catalysis and structure. J. Biol. Chem. 277:23821-23827. [DOI] [PubMed] [Google Scholar]
- 24.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 25.Spiers, A., J. Bohannon, S. Gehrig, and P. Rainey. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50:15-27. [DOI] [PubMed] [Google Scholar]
- 26.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tesmer, J., and S. Sprang. 1998. The structure, catalytic mechanism and regulation of adenylyl cyclase. Curr. Opin. Struct. Biol. 8:713-719. [DOI] [PubMed] [Google Scholar]
- 28.Thomas, C., C. R. Andersson, S. R. Canales, and S. S. Golden. 2004. PsfR, a factor that stimulates psbAI expression in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 150:1031-1040. [DOI] [PubMed] [Google Scholar]
- 29.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilde, A., B. Fiedler, and T. Borner. 2002. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol. Microbiol. 44:981-988. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, K. Y., G. L. Card, Y. Suzuki, D. R. Artis, D. Fong, S. Gillette, D. Hsieh, J Neiman, B. L. West, C. Zhang, M. Milburn, S. H. Kim, J. Schlessinger, and G. Bollag. 2004. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol. Cell 15:279-286. [DOI] [PubMed] [Google Scholar]
- 32.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]
- 33.Zoraghi, R., J. Corbin, and S. Francis. 2004. Properties and functions of GAF domains in cyclic nucleotide phosphodiesterases and other proteins. Mol. Pharmacol. 65:267-278. [DOI] [PubMed] [Google Scholar]