Abstract
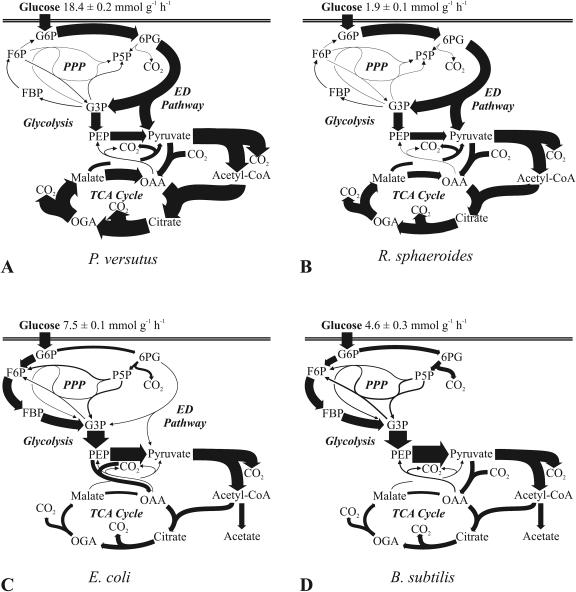
The structurally conserved and ubiquitous pathways of central carbon metabolism provide building blocks and cofactors for the biosynthesis of cellular macromolecules. The relative uses of pathways and reactions, however, vary widely among species and depend upon conditions, and some are not used at all. Here we identify the network topology of glucose metabolism and its in vivo operation by quantification of intracellular carbon fluxes from 13C tracer experiments. Specifically, we investigated Agrobacterium tumefaciens, two pseudomonads, Sinorhizobium meliloti, Rhodobacter sphaeroides, Zymomonas mobilis, and Paracoccus versutus, which grow on glucose as the sole carbon source, represent fundamentally different metabolic lifestyles (aerobic, anaerobic, photoheterotrophic, and chemoheterotrophic), and are phylogenetically distinct (firmicutes, γ-proteobacteria, and α-proteobacteria). Compared to those of the model bacteria Escherichia coli and Bacillus subtilis, metabolisms of the investigated species differed significantly in several respects: (i) the Entner-Doudoroff pathway was the almost exclusive catabolic route; (ii) the pentose phosphate pathway exhibited exclusively biosynthetic functions, in many cases also requiring flux through the nonoxidative branch; (iii) all aerobes exhibited fully respiratory metabolism without significant overflow metabolism; and (iv) all aerobes used the pyruvate bypass of the malate dehydrogenase reaction to a significant extent. Exclusively, Pseudomonas fluorescens converted most glucose extracellularly to gluconate and 2-ketogluconate. Overall, the results suggest that metabolic data from model species with extensive industrial and laboratory history are not representative of microbial metabolism, at least not quantitatively.
Based on 13C tracer experiments, metabolic-flux analysis emerged as a key methodology to identify the network topology of active reactions and to quantify the in vivo distribution of molecular fluxes throughout metabolism (38, 47). In contrast to global protein, mRNA, or metabolite concentration analyses that assess network composition, flux methods directly assess the operation of metabolic networks by quantifying in vivo reaction velocities. The general principle is based on mass spectrometry (MS) or nuclear magnetic resonance detection of 13C patterns in products of metabolism. Often, protein-bound amino acids that preserve the carbon backbone of eight metabolic key intermediates are used. The detected 13C isotope patterns then reflect the activity of intracellular pathways and reactions, whose fluxes can be quantified from the isotope data by using mathematical models with various levels of complexity. In the simplest approach, algebraic equations are used to determine strictly local ratios of converging fluxes analytically by so-called metabolic-flux ratio (METAFoR) analysis (3, 16, 41, 44). Absolute intracellular fluxes in millimoles per gram of biomass per hour may be estimated indirectly by combining such 13C data with quantitative physiological data on fluxes in and out of cells (47). In this case, the estimated fluxes are the best fit to the available data within the specified metabolic model. Beyond the quantification of flux through the well-known biochemical pathways, flux methods have recently demonstrated their value for the identification of novel (17) or unexpected (22, 34, 39, 40) metabolic pathways.
For obvious reasons, such flux methods were applied primarily to model microbes with industrial relevance, such as Escherichia coli (15, 24, 41), Bacillus subtilis (9, 40), Corynebacterium glutamicum (30, 48), and Saccharomyces cerevisiae (3, 19). While the accumulated biochemical and metabolic data on these species are also the basis of much of our textbook knowledge, it is clear that these model species are not representative for all, and perhaps not even for most, microbes. One example is the widely distributed Entner-Doudoroff (ED) pathway (6, 26), the genes for which are absent from B. subtilis, S. cerevisiae, and C. glutamicum and which is used by E. coli mainly during growth on gluconate (14). For glucose metabolism, all four model species rely primarily on the Emden-Meyerhof-Parnas (EMP) pathway and, in some cases, to a substantial extent also on the pentose phosphate (PP) pathway (9, 16, 19, 48, 53). These facts raise the general question of how representative the accumulated metabolic knowledge on these model species is. Here we attempt a quantitative comparison of the intracellular metabolisms of the two model microbes E. coli and B. subtilis with those of seven metabolically and phylogenetically distinct species that can grow on glucose as the sole carbon source.
In particular, we identify the network topology of active reactions by METAFoR analysis based on gas chromatography (GC)-MS analysis of proteinogenic amino acids from [U-13C]glucose and [1-13C]glucose batch experiments (16). Quantification of in vivo molecular fluxes is then achieved by 13C-constrained flux analysis (18, 40). In particular, we chose the anaerobic organism Zymomonas mobilis; members of the family Rhizobiaceae, i.e., Agrobacterium tumefaciens and Sinorhizobium meliloti; the metabolically versatile facultative phototroph Rhodobacter sphaeroides; the facultative autotroph Paracoccus versutus; and the versatile pseudomonads Pseudomonas fluorescens and Pseudomonas putida.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The following nine bacterial species or strains were analyzed: A. tumefaciens C58 (F. Narberhaus), P. fluorescens 52-1C (B. Witholt), P. putida KT2440 (B. Witholt), R. sphaeroides ATH 2.4.1 (German Collection of Microorganisms and Cell Cultures, DSMZ 158), Paracoccus versutus A2 (DSMZ 582), S. meliloti (DSMZ 1981), Z. mobilis NRRL B-806 (DSMZ 424), E. coli MG1655 (E. coli Genetic Stock Center, 6300), and B. subtilis 168 trpC2 (Bacillus Genetic Stock Center). Aerobic batch cultures were grown at 30°C in 500-ml baffled flasks with 50 ml of M9 minimal medium (Paracoccus versutus and Z. mobilis were grown in special minimal media) on a gyratory shaker at 225 rpm (250 rpm for P. fluorescens). Anaerobic cultures of Z. mobilis were grown at 30°C in 125-ml sealed glass flasks with 50 ml of minimal medium on magnetic stirrers at 225 rpm. The sterile medium was gassed with sterilely filtered N2 for 15 min.
The M9 minimal medium contained, per liter of deionized water, 7.52 g of Na2HPO4 · 2H2O, 3.0 g of KH2PO4, 0.5 g of NaCl, and 2.5 g of (NH4)2SO4. The following components (in indicated quantities per liter of final medium) were sterilized separately and then added: 1 ml of 0.1 M CaCl2, 1 ml of 1 M MgSO4, 0.6 ml of 100 mM FeCl3, 2 ml of vitamin solution (filter sterilized), and 10 ml of M9 trace salts solution. The vitamin solution contained (per 50 ml) 25 mg each of biotin, cyanocobalamin, niacin, calcium pantothenate, pyridoxine HCl, and thiamine HCl. The M9 trace salts solution contained (per liter) 0.18 g of ZnSO4 · 7H2O, 0.12 g of CuCl2 · 2H2O, 0.12 g of MnSO4 · H2O, and 0.18 g of CoCl2 · 6H2O. For R. sphaeroides, we used a special trace salts solution that contained (per liter) 1.5 g of nitrilotriacetic acid, 3.0 g of MgSO4 · 7H2O, 0.5 g of MnSO4 · H2O, 1.0 g of NaCl, 0.1 g of FeSO4 · 7H2O, 0.1 g of CoCl2 · 6H2O, 0.135 g of CaCl2 · 2H2O, 0.1 g of ZnSO4 · 7H2O, 0.01 g of CuSO4 · 5H2O, 0.01 g of H3BO3, 0.01 g of Na2MoO4 · 2H2O, 0.015 g of NiCl2, and 0.02 g of Na2SeO3; the pH was adjusted to 6.5 with KOH.
The Z. mobilis minimal medium contained (per liter) 0.18 g of KH2PO4, 0.082 g of MgSO4 · 7H2O, 0.002 g of FeSO4 · 7H2O, 0.87 g of NH4Cl, 0.142 g of trisodium citrate dihydrate, 10 g of potassium hydrogen phthalate, and 2 ml of the vitamin solution (filter sterilized) described above. The pH was adjusted to 5.8 with KOH. The Paracoccus versutus minimal medium was composed of two solutions that were mixed at a ratio of 1:10 after heat sterilization. Solution A contained (per 100 ml) 4.2 g of Na2HPO4 · 2 H2O, 1.5 g of KH2PO4, and 1.0 g of NH4Cl; the pH was adjusted to 9.0. Solution B contained (per liter) 0.1 g of MgSO4 · 7 H2O and 5.0 ml of trace metal solution; the pH was adjusted to 6.0 with KOH. The trace solution contained (per liter) 50.0 g of EDTA, 22.0 g of ZnSO4 · 7 H2O, 5.54 g of CaCl2 · 2 H2O, 5.06 g of MnCl2 · 4 H2O, 4.99 g of FeSO4 · 7 H2O, 1.10 g of MoNH4 · 4 H2O, 1.57 g of CuSO4 · 5 H2O, and 1.61 g of CoCl2 · 6 H2O.
In all experiments, sterile glucose was supplemented at a final concentration of 3 g of glucose/liter (4 g/liter for P. fluorescens and 10 g/liter for Z. mobilis). For 13C-labeling experiments, glucose was added either entirely in the form of the 1-13C-labeled isotope isomer (99% pure; Omicron Biochemicals, Inc., South Bend, Ind.) or in the form of a mixture of 20% (wt/wt) [U-13C]glucose (>99% pure; Martek Biosciences Corporation, Columbia, Md.) and 80% (wt/wt) natural glucose. To elucidate the influence of unlabeled CO2 on the 13C-labeling patterns of Z. mobilis, cultures were continuously flushed with filter-sterilized technical CO2 through an inlet-and-outlet needle.
Analytical procedures and physiological parameters.
Cell growth was monitored by measuring the optical density at 600 nm (OD600). Glucose and acetate concentrations were determined enzymatically with commercially available kits (Beckman, Enzytec, and Boehringer Mannheim). Ethanol concentrations were quantified by GC in a Hewlett Packard 5890 Series II Plus chromatograph with a Macherey-Nagel fused-silica capillary column (model CW20M-0.25, 25-m length, 0.25-mm inside diameter; Permabond) with butyrate as the internal standard. Organic acids in culture supernatants were detected by high-pressure liquid chromatography analysis (Perkin Elmer) at a wavelength of 210 nm, using a Supelcogel C8 column (4.6 by 250 mm) and 0.2% (vol/vol) sulfuric acid as the mobile phase at a flow rate of 0.3 ml/min at 30°C. The following physiological parameters were determined by regression analysis during the exponential growth phase in the batch culture, as described previously (41): maximum specific growth rate, biomass yield on glucose, specific glucose consumption, and by-product formation rates. Cellular dry weight (CDW) was inferred from OD600 measurements with a predetermined correlation factor, k. To determine k, CDW was determined from at least four parallel 2-ml cell suspensions that were harvested by centrifugation at 15,800 × g in an Eppendorf tabletop centrifuge using predried and weighed 2-ml Eppendorf cups. The pellets were washed with 0.9% NaCl and dried at 105°C for 24 h to a constant weight.
For P. fluorescens, the theoretical extracellular pool concentration of gluconate-2-ketogluconate, f, was calculated from the integrated product of the specific growth rate, μmax, and the gluconate-2-ketogluconate accumulation rate, c, as follows:
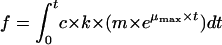 |
where m was a coefficient, c was the glucose decrease rate minus the total carbon uptake rate (see Results), and t was the time in hours for growth on glucose.
Sample preparation and GC-MS analysis.
Cell aliquots were harvested during mid-exponential growth by centrifuging 35 to 40 ml of culture broth at 1,200 × g and 4°C for 20 min. The pellet was washed twice with 1 ml of 0.9% NaCl, hydrolyzed in 1.5 ml of 6 M HCl for 24 h at 110°C in sealed 2-ml Eppendorf tubes, and desiccated overnight in a heating block at 85°C under a constant air stream. The hydrolyzate was dissolved in 50 μl of 99.8% pure dimethyl formamide and transferred into a new Eppendorf cup within a few seconds. For derivatization, 30 μl of N-methyl-N-(tert-butyldimethylsilyl)-trifluoroacetamide, which readily silylates hydroxyl groups, thiols, primary amines, amides, and carboxyl groups, (7) was added, and the mixture was incubated at 85°C with shaking at 550 rpm for 60 min. One microliter of the derivatized sample was injected into a series 8000 GC combined with a model MD 800 mass spectrometer (Fisons Instruments) and analyzed as described previously (7, 16).
Metabolic-flux ratio analysis.
For METAFoR analysis, mass spectra of the derivatized amino acids alanine, glycine, valine, leucine, isoleucine, proline, serine, threonine, phenylalanine, aspartate, glutamate, histidine, and tyrosine were corrected for the natural abundance of all stable isotopes and unlabeled biomass from the inoculum. Lysine and methionine are not required for the METAFoR analysis used in this study, whereas arginine, asparagine, cysteine, glutamine, and tryptophane are not detectable. The amino acids are synthesized from one or more metabolic intermediates, and the mass isotopomer distribution vectors (MDV) of these metabolites could easily be derived from the MDV of the amino acids (16). The metabolite MDV were then used to calculate the fractional contribution of a given pathway or reaction to a target metabolite pool (metabolic-flux ratios) by using sets of algebraic equations implemented in the MATLAB-based program Fiat Flux version 1.04 as described previously (16). The result is direct and quantitative evidence for strictly local ratios of two or more reactions and pathways to a metabolic intermediate.
Net-flux analysis and master reaction network.
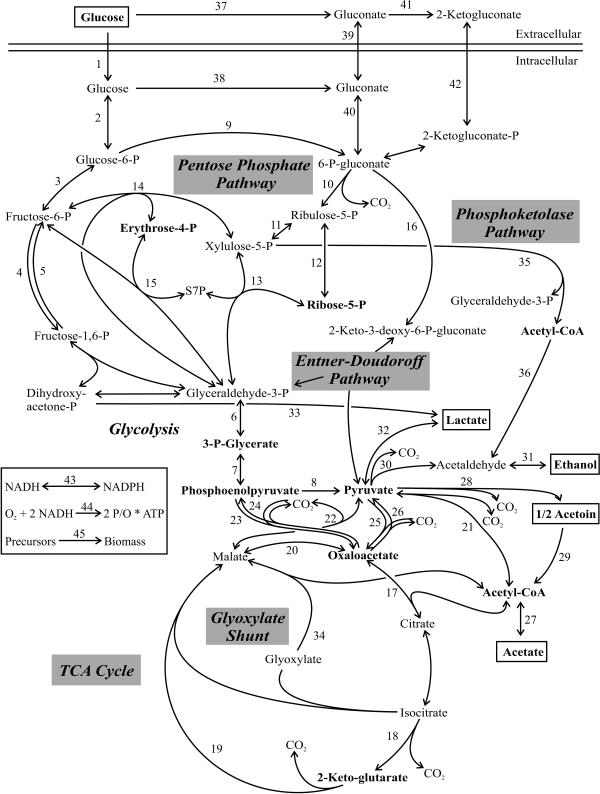
The metabolic models used for net-flux analysis were based on the master reaction network, which included 45 reactions and 33 metabolites and is shown in Fig. 1. Respiration, biomass formation, and a transhydrogenase reaction were included as additional reactions, and ATP and the cofactors NADH and NADPH were included as additional metabolites. Net fluxes were then calculated using (i) the stoichiometric reaction matrix, (ii) the METAFoR analysis-derived flux ratios, (iii) physiological data, and (iv) precursor requirements for biomass synthesis, as described previously (18). Specifically, the following flux ratios were used: serine derived through the EMP pathway, pyruvate derived through the ED pathway, oxaloacetate (OAA) originating from phosphoenolpyruvate (PEP) or pyruvate, PEP originating from OAA, the lower and upper bounds of pyruvate originating from malate, and the upper bound of PEP derived through the PP pathway. For the particular stoichiometric matrix of each organism, several reactions were omitted from the master network based on information from the 13C-labeling experiments and literature knowledge (see below). The stoichiometric matrix was then solved with the MATLAB-based program Netto version 1.04 (18) by minimizing the sum of the weighted square residuals of the constraints from both metabolite balances and flux ratios to obtain estimated net fluxes.
FIG. 1.
Master reaction network that was used as the basis for net-flux analysis. Metabolites in bold were precursors for amino acid biosynthesis, and metabolites in boxes were extracellular substrates or products. Doubled-headed arrows indicate reactions assumed to be reversible. Abbreviations: S7P, sedoheptulose-7-P; Acetyl-CoA, acetyl coenzyme A.
Biomass requirements and organism-specific networks.
E. coli and B. subtilis were routinely analyzed as control experiments using the previously established growth rate-dependent biomass requirements and network models (8, 15). For gram-negative P. fluorescens, P. putida, S. meliloti, A. tumefaciens, and R. sphaeroides, the biomass requirements were assumed to be similar to that of E. coli (15), and for the gram-positive organism Paracoccus versutus, the requirements were taken from the B. subtilis model (8). For Z. mobilis, the precursor requirements were based on the published biomass composition (10).
The metabolic network used for Z. mobilis lacks a functional EMP pathway due to the absence of the key enzyme phosphofructokinase. Furthermore, the tricarboxylic acid (TCA) cycle is incomplete and does not contain a transaldolase; thus, pentoses are synthesized from fructose-6-P and glyceraldehyde-3-P via the reversible transketolase (10, 32, 33). The resulting network of Z. mobilis included the following reactions in Fig. 1: 1 to 3, 6 to 14, 16 to 18, 21 to 23, 30, and 31. The network model for P. fluorescens and P. putida was adapted from previous literature (28, 46). Besides the direct uptake, glucose may also be converted extracellularly to gluconate and 2-ketogluconate, both of which can be taken up by the cell. Additionally, the EMP pathway is absent due to the lack of phosphofructokinase. Thus, the network of both pseudomonads included the following reactions: 1 to 3, 5 to 22, 24, 25, 37, and 39 to 42. Reaction 5 principally enables a cyclic flux through the ED pathway that could not be resolved by the present 13C-labeling experiments; nevertheless, a weak flux was obtained through reaction 5 in P. putida (see Table S1 in the supplemental material). The network models for A. tumefaciens and S. meliloti were basically the same as that for P. fluorescens (1, 12, 21, 25, 36). Since we did not detect gluconate or 2-ketogluconate accumulation, however, glucose uptake was assumed to be direct in both organisms. Thus, the networks consisted of the reactions 1 to 3, 5 to 22, 24, and 25. The network models for the metabolically versatile organisms R. sphaeroides (5, 6, 23, 52) and Paracoccus versutus (51) included the reactions 1 to 4, 6 to 22, 24, and 25.
RESULTS
Network identification and flux analysis.
To experimentally identify metabolic networks of active reactions, we used 13C-constrained metabolic-flux analysis that relies on the detection of mass isotopomer pattern in proteinogenic amino acids (16, 18). Since pathways of amino acid biosynthesis may differ among species, we first verified the biosynthetic routes. For each amino acid, the labeling pattern depended on that of the precursor from which it was synthesized; for example, the alanine carbon skeleton is derived from pyruvate. Since valine and leucine are typically synthesized also from pyruvate, the redundant information in all three amino acids must be the same if they are indeed synthesized from pyruvate. Generally, all detected labeling patterns were consistent with the amino acid biosynthesis pathways of E. coli (44), and no discrepancies were detected within the labeling patterns of the redundant amino acids.
Based on the established amino acid biosynthesis schemes, we then calculated intracellular-flux ratios from the labeling patterns of the amino acids by using the algebraic equations of METAFoR analysis (16). These flux ratios represent direct, local evidence for the in vivo activity of particular pathways and reactions. From the thus-elucidated network topology of active reactions and from literature data, organism-specific metabolic-reaction models were deduced from the master network model (Fig. 1). Absolute net fluxes were then calculated with these network models from the physiological data (Table 1) and the flux ratios (Table 2) (18, 40). Since the analytically determined flux ratios constrain the ratios of at least two fluxes within their margins of experimental error, this approach is referred to as 13C-constrained flux analysis.
TABLE 1.
Growth physiologya
| Organism | Maximum specific growth rate (h−1) | Biomass yield (CDW [g]/g of glucose) | Glucose uptake (mmol g−1 h−1) | By-product accumulation (mmol g−1 h−1)b |
|---|---|---|---|---|
| Z. mobilis | 0.34 ± 0.03 | 0.03 ± 0.01 | 61.5 ± 2.1 | 102.1 ± 5.0 |
| P. fluorescens | 0.49 ± 0.03 | 0.44 ± 0.01 | 4.5 ± 0.1c | |
| S. meliloti | 0.17 ± 0.01 | 0.41 ± 0.02 | 2.3 ± 0.1 | |
| A. tumefaciens | 0.3 ± 0.01 | 0.41 ± 0.04 | 4.1 ± 0.2 | |
| Paracoccus versutus | 0.70 ± 0.01d | 0.21 ± 0.03 | 18.9 ± 2.1 | |
| R. sphaeroides | 0.15 ± 0.02 | 0.41 ± 0.06 | 1.8 ± 0.1 | |
| E. coli | 0.39 ± 0.01 | 0.30 ± 0.03 | 7.8 ± 0.4 | 3.5 ± 0.1 |
| B. subtilis | 0.30 ± 0.03 | 0.35 ± 0.01 | 4.8 ± 0.3 | 2.1 ± 0.1 |
Values are given as means ± standard deviations.
The detected by-products were ethanol for Z. mobilis and acetate for E. coli and B. subtilis. The following metabolites were determined by high-pressure liquid chromatography to be present only in trace amounts: citrate, formate, fumarate, malonate, pyruvate, and succinate.
Total substrate uptake rate.
Values were determined from the culture that was fully adapted to growth on glucose (after two subcultivations).
TABLE 2.
Metabolic flux ratios obtained by METAFoR analysis of experiments with 100% [1-13C]glucose, 20% [U-13C]glucose, and 80% naturally labeled glucosea
| Organism | Mean relative split ratio (%) ± SD
|
||||||
|---|---|---|---|---|---|---|---|
| Ser through EMPb | Pyr through EDPb | PEP through PPP (UB) | OAA from PEP (or Pyr) | PEP from OAA | Pyr from Mal (LB) | Pyr from Mal (UB) | |
| Z. mobilis | —c | 100 ± 1 | — | 99 ± 4 | 1 ± 1 | — | — |
| P. fluorescens | — | 91 ± 1 | 2 ± 5 | 85 ± 4 | 17 ± 1 | 17 ± 2 | 55 ± 11 |
| S. meliloti | — | 95 ± 1 | 0 ± 5 | 34 ± 2 | 6 ± 7 | 13 ± 2 | 19 ± 3 |
| A. tumefaciens | — | 86 ± 1 | 0 ± 5 | 39 ± 2 | 10 ± 1 | 11 ± 1 | 18 ± 2 |
| Paracoccus versutus | 0 ± 1 | 100 ± 1 | 26 ± 5 | 44 ± 2 | 6 ± 1 | 11 ± 2 | 19 ± 3 |
| R. sphaeroides | 0 ± 1 | 100 ± 1 | 0 ± 5 | 46 ± 2 | 4 ± 1 | 13 ± 1 | 22 ± 3 |
| E. coli | 79 ± 1 | 7 ± 3 | 1 ± 5 | 63 ± 4 | 3 ± 1 | 0 ± 2 | 0 ± 4 |
| B. subtilis | 62 ± 1 | — | 27 ± 5 | 62 ± 3 | 5 ± 1 | 2 ± 2 | 5 ± 5 |
Abbreviations: Ser, serine; Pyr, pyruvate; EDP, ED pathway; PPP, PP pathway; Mal, malate; UB, upper bound; and LB, lower bound.
Split ratios obtained from experiments with 100% [1-13C]glucose.
—, this pathway was considered to be absent based on literature data.
Z. mobilis.
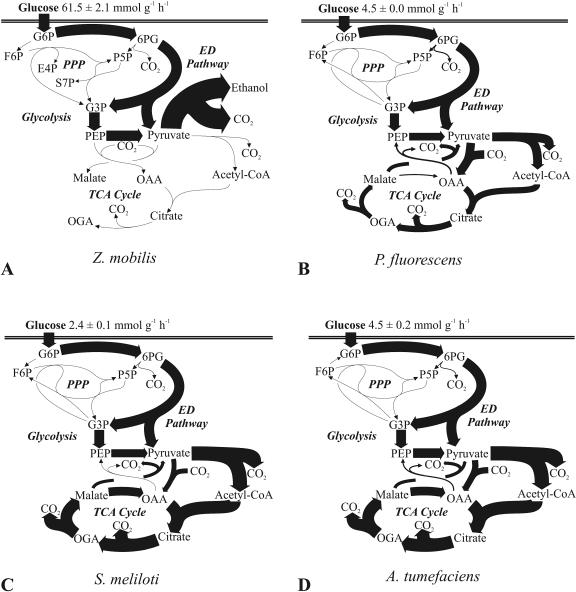
Without a respiratory chain, Z. mobilis is an obligate fermenter and produces a single ATP molecule per molecule of metabolized glucose. This energetically ineffective metabolism leads to very high fermentative fluxes with a very low biomass yield of 20 mg of cells per g of glucose (11). Among the species investigated here, the glucose uptake rate of Z. mobilis was at least an order of magnitude higher than those of most others (Fig. 2A; Table 1). This flux was exclusively catalyzed by the ED pathway (Fig. 2A; Table 2), the enzymes of which constitute up to 50% of a cell's total protein (42). The absence of the EMP pathway was confirmed by [1-13C]glucose experiments, where no 13C label was detected at the C-3 position of pyruvate, due to the missing phosphofructokinase (data not shown). The absence of transaldolase (10) was also verified from the pentose-labeling patterns found in the 20% [U-13C]glucose experiment, which were consistent with those expected when only transketolase B is active (Table 3).
FIG. 2.
In vivo carbon flux distribution in Z. mobilis (A), P. fluorescens (B), S. meliloti (C), and A. tumefaciens (D). All fluxes were normalized to the glucose uptake rate that is given at the top of each panel, and the widths of the arrows are scaled to the relative percentages of flux. Fluxes below 2.6% of the glucose uptake rate are represented by nonscaled hairlines. Possible cyclic fluxes through the ED pathways of P. fluorescens and S. meliloti are not resolved by the data and are not shown. Generally, the 95% confidence intervals were between 5 and 10% for the major fluxes. Larger confidence intervals were estimated for reactions with low flux. Abbreviations: G6P, glucose-6-P; 6PG, 6-P-gluconate; F6P, fructose-6-P; P5P, pentose-5-P; E4P, erythrose-4-P; S7P, sedoheptulose-7-P; G3P, glyceraldehyde-3-P; OGA, 2-ketoglutarate; PPP, PP pathway; Acetyl-CoA, acetyl coenzyme A.
TABLE 3.
Mass isotopomer distribution in pentose-5-P in Z. mobilisa
| Fractional label | m0b | m1 | m2 | m3 | m4 | m5 |
|---|---|---|---|---|---|---|
| Pentose-5-P 1-5 | 0.67 | 0.00 | 0.15 | 0.14 | 0.00 | 0.04 |
| Transketolase Bc | 0.64 | 0.00 | 0.16 | 0.16 | 0.00 | 0.04 |
| Transketolase A + transaldolased | 0.66 | 0.13 | 0.02 | 0.06 | 0.03 | 0.10 |
Mass isotopomer distribution in pentose-5-P was deduced from the mass isotopomer distribution in histidine. The data are from a 20% [U-13C]glucose experiment, and the theoretical patterns were calculated assuming only transketolase B or transaldolase and transketolase A to be active.
Values for m0 represent the fraction with the lowest mass, and those for mi>0 represent the abundances of molecules with higher masses.
Theoretical pattern expected from exclusive operation of transketolase B, which cleaves fructose-6-P into a C2 and a C4 fragment. The C2 fragment originating from fructose-6-P and the C3 fragment originating from glyceraldehyde-3-P combine to give the theoretical labeling pattern.
Theoretical pattern expected from the concerted operation of transaldolase (combines a C1 fragment from erythrose-4-P with fructose-6-P to give sedoheptulose-7-P) and transketolase A (transfers a C2 fragment from sedoheptulose-7-P to glyceraldehyde-3-P to give two pentoses).
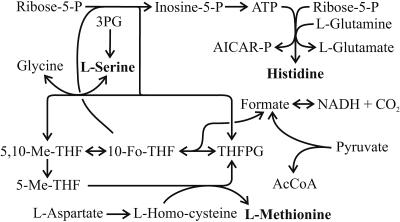
Although the EMP pathway was inactive, a 13C label was detected at the C-3 position of serine from 100% [1-13C]glucose experiments (Table 4). Thus, labeled carbon must have been introduced through C1 metabolism (Fig. 3). The tetrahydrofolyl (THF) cycle is replenished with C1 that originates from formate, which in turn is generated from pyruvate via pyruvate formate lyase or from CO2 via formate dehydrogenase. To differentiate between the two routes, we (i) added formate to strongly reduce the fraction of labeled formate in the medium and (ii) continuously gassed the culture with unlabeled CO2, which should have diluted the serine label if the formate dehydrogenase were active. Although noticeable, the influence of unlabeled CO2 on the m1 mass fraction signal of serine was far less significant than the effect of adding formate (Table 4). This finding indicates that pyruvate formate lyase rather than formate dehydrogenase generates the formate in Z. mobilis that leads to the labeling of C-3 of serine.
TABLE 4.
Mass isotopomer distribution of serine in Z. mobilis at different formate concentrations
| Condition | m0 | m1a | m2 | m3 |
|---|---|---|---|---|
| None (control) | 0.601 | 0.394 | 0.004 | 0.000 |
| 1 mM formate | 0.807 | 0.190 | 0.003 | 0.000 |
| 5 mM formate | 0.835 | 0.164 | 0.000 | 0.001 |
| CO2 by aeration | 0.665 | 0.334 | 0.001 | 0.001 |
The m1 mass fraction reveals the C-1 position of pyruvate if we assume that pyruvate-formate lyase is the predominant supply reaction for C-1 metabolism.
FIG. 3.
Possible reactions that introduce C1 fragments into serine, methionine, and histidine (31). Abbreviations: AICAR-P, 5-aminoimidazole-4-carboxamide-1-ribotide; THFPG, tetrahydrofolylpolyglutamate; 10-Fo-THF, 10-formyl-tetrahydrofolyl; 5,10-Me-THF, 5,10-methyl-tetrahydrofolyl; 3PG, 3-P-glycerate; AcCoA, acetyl coenzyme A.
Pseudomonads.
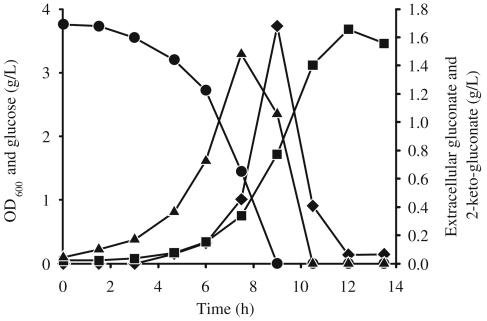
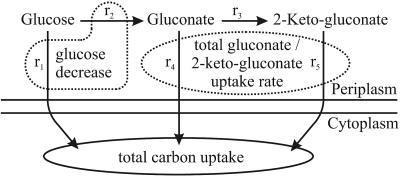
In P. fluorescens, but not in P. putida, extracellular glucose was converted to gluconate and 2-ketogluconate, which were subsequently consumed during the later growth phase (Fig. 4), as described previously (13, 28, 45). Considering the simplified glucose uptake scheme shown in Fig. 5, the total gluconate-2-ketogluconate accumulation rate, c, during growth on glucose was calculated as 15.21 mmol g−1 h−1 from the experimentally determined extracellular glucose decrease rate (a = 19.75 mmol g−1 h−1) minus the total carbon uptake rate (b = 4.54 mmol g−1 h−1). The experimentally determined extracellular concentration of gluconate and 2-ketogluconate was consistent with that calculated with the above equation, which strongly suggests that there was no cocatabolism of gluconate-2-ketogluconate during growth on glucose. Upon glucose depletion, the carbon uptake rate from both gluconate and 2-ketogluconate almost doubled to 9.42 mmol g−1 h−1. Based on the above conclusions, the total carbon uptake rate for net-flux analysis was assumed to be exclusively from glucose in the first phase and from gluconate-2-ketogluconate (g) in the second phase. The relative proportion of gluconate and 2-ketogluconate uptake could not be resolved, because the carbon skeleton remained unchanged, which led to identical labeling patterns.
FIG. 4.
Time courses of OD600 (squares) and concentrations of extracellular glucose (circles), gluconate (triangles), and 2-ketogluconate (diamonds) in P. fluorescens.
FIG. 5.
Glucose uptake in P. fluorescens. The following fluxes (shown by arrows) are involved in glucose uptake: direct glucose uptake (r1), membrane-bound glucose dehydrogenase (r2), gluconate dehydrogenase (r3), uptake of gluconate (r4), and 2-ketogluconate (r5). The experimentally determined extracellular glucose decrease rate, a, is defined as r1 plus r2; the total carbon uptake rate by the cell, b, is defined as r1 plus r4 plus r5; and the total gluconate-2-ketogluconate uptake rate, g, is defined as r4 plus r5.
During growth on glucose, P. fluorescens secreted very little in the way of metabolic by-products (Table 1), and the ED pathway and the TCA cycle were the almost exclusive catabolic pathways prior to (Fig. 2B; Table 2) and after (data not shown) glucose depletion. Unlike those in most other bacteria, however, the TCA cycle did not operate via malate dehydrogenase but mostly via the so-called pyruvate shunt, which is catalyzed by malic enzyme and pyruvate carboxylase (from malate to pyruvate to OAA). Moreover, a relatively high gluconeogenic flux of 17% through the PEP carboxykinase was detected. The higher carbon uptake rate of 9.42 mmol g−1 h−1 during the second growth phase resulted in a higher TCA cycle flux and a decreased biomass yield of 0.29, compared to a yield of 0.44 (CDW in grams per gram of glucose) produced during growth on glucose.
In P. putida, very little gluconate or 2-ketogluconate accumulated (below 2 mM); hence, carbon uptake was most likely via the direct glucose uptake route. Net-flux analysis with a lumped total carbon uptake resulted in a distribution that was very similar to that of P. fluorescens (data not shown).
Rhizobiaceae.
As a fast-growing rhizobium with a generation time of less than 6 h on complex medium, S. meliloti was reported to be able to convert glucose to gluconate, which then enters metabolism (36, 43). Since <1 mM gluconate accumulated and the labeling data did not allow the resolution of the different uptake routes, both were lumped for net-flux analysis. In S. meliloti, pyruvate carboxylase was assumed to be the anaplerotic reaction by analogy to A. tumefaciens (12). Both the PP and ED pathways were reported to be present, and glycolysis was reported to be absent, in both rhizobia (1, 43), which was confirmed by METAFoR analysis (Table 2). In both rhizobia, the ED pathway was basically the exclusive pathway of glucose degradation, while the pentose-5-P precursors for biomass were generated through the oxidative and nonoxidative branches of the PP pathway (Fig. 2C and D). With a flux well above 100% relative to the glucose uptake rate, both species exhibited higher TCA cycle fluxes than those of most species investigated here (Fig. 2C and D).
Paracoccus versutus.
As a member of the genus Paracoccus, Paracoccus versutus (synonyms, Thiobacillus versutus and Paracoccus strain A2) belongs to the group of α-proteobacteria and is closely related to the Rhodobacteraceae and distantly to pseudomonads (2). As a facultative autotroph, Paracoccus versutus is capable of heterotrophic growth on a wide rage of organic substrates that include mono-, di-, and trisaccharides (49). Extensive radiorespirometric and enzymatic analyses of the main carbon-degrading pathways indicated the presence of the EMP, ED, and PP pathways (49-51). Consistent with the reported adaptation to growth solely on glucose, the specific rate of growth increased from 0.29 to 0.70 h−1 (Table 1). In the rapidly growing culture, METAFoR analysis revealed an inactive EMP pathway, some PP pathway activity, and predominant catabolic flux through the ED pathway (Table 2). Since Paracoccus versutus exhibited the highest aerobic glucose uptake rate of all species investigated, it was surprising that its metabolism was fully respirative, with very high TCA cycle flux (Fig. 6A).
FIG. 6.
In vivo carbon flux distribution in Paracoccus versutus (A), R. sphaeroides (B), E. coli (C), and B. subtilis (D). All fluxes were normalized to the glucose uptake rates that are given at the top of each panel, and the widths of the arrows are scaled to the relative fluxes, expressed as percentages of the glucose uptake rates. Fluxes below 2.6% are represented by nonscaled hairlines. Generally, the 95% confidence intervals were between 5 and 10% for the major fluxes. Larger confidence intervals were estimated for reactions with low flux. Abbreviations: G6P, glucose-6-P; 6PG, 6-P-gluconate; F6P, fructose-6-P; FBP, fructose-1,6-bisphosphate; P5P, pentose-5-P; G3P, glyceraldehyde-3-P; OGA, 2-ketoglutarate; PPP, PP pathway; Acetyl-CoA, acetyl coenzyme A.
R. sphaeroides.
The purple, facultatively photosynthetic bacterium R. sphaeroides has an unusually versatile metabolism, capable of aerobic chemoheterotrophic and anaerobic photoheterotrophic growth (27). Aerobic growth on glucose, however, was rather slow (Table 1). While the EMP and ED pathways were reported to be present (4, 5), only the ED pathway was active during aerobic growth on glucose (Table 1; Fig. 6B). The m1 mass fractions of pyruvate fragments 1 to 3 and 2 to 3 (52 and 2%, respectively; detected in alanine, isoleucine, leucine, lysine, and valine) directly demonstrated exclusive use of the ED pathway, whereas the almost completely unlabeled PEP (detected in phenylalanine, tryptophane, and tyrosine) verified the absence of EMP pathway flux (data not shown). In a manner akin to that of most previously analyzed bacteria, glucose was catabolized almost exclusively via the ED pathway and the TCA cycle, with slight by-product formation but some contribution on the part of the pyruvate shunt via malic enzyme and pyruvate carboxylase (Fig. 6B). The PP pathway operated exclusively to provide precursors for biosynthesis, and the EMP pathway catalyzed even small gluconeogenic fluxes from glyceraldehyde-3-P to fructose-6-P.
E. coli and B. subtilis.
In sharp contrast to all other bacteria analyzed here, the gram-positive and gram-negative model bacteria E. coli and B. subtilis rely primarily on the EMP pathway for glucose catabolism (Fig. 6C and D). Moreover, the relative TCA cycle flux was much lower than that in the other species because secretion of the incompletely oxidized overflow product acetate was extensive. At 38%, the PP pathway flux in B. subtilis was the highest observed in all species (Table 5). Generally, the overall estimated flux distribution in both species was similar to that obtained previously from 37°C batch cultures, except for the elevated TCA cycle activity in E. coli at 30°C and the elevated anaplerosis (OAA from pyruvate) in B. subtilis at 30°C, which was due to lower relative acetate formation (39, 53). Apart from the about 30- to 50%-lower growth rates, there were only minor changes that are related to the lower overall rate of metabolism at 30°C, the temperature used here.
TABLE 5.
Relative molecular carbon flow into the PP pathway and catabolic PP pathway flux
| Organism | Mean ± SD
|
|
|---|---|---|
| Flux into PP pathway (%)a | Catabolic PP pathway flux (%)b | |
| Z. mobilis | 0 ± 0 | 0 ± 0 |
| P. fluorescens | 10 ± 1 | −2 ± 1c |
| S. melioti | 4 ± 1 | −2 ± 1 |
| A. tumefaciens | 6 ± 2 | −1 ± 2 |
| Paracoccus versutus | 0 ± 2 | −4 ± 2 |
| R. sphaeroides | 0 ± 5 | −7 ± 5 |
| E. coli | 21 ± 4 | 12 ± 4 |
| B. subtilis | 38 ± 1 | 27 ± 1 |
Carbon flux into the PP pathway was calculated as the flux through 6-P-gluconate dehydrogenase divided by the total carbon uptake rate.
For the catabolic PP pathway flux, the CO2 production and precursor flux of erythrose-4-P and ribulose-5-P into biomass were subtracted from those of 6-P-gluconate dehydrogenase.
Negative values indicate reverse flux into the PP pathway from fructose-6-P and/or glyceraldehyde-3-P.
DISCUSSION
In all seven newly investigated species, the ED pathway was the almost exclusive route of glucose catabolism, while the EMP pathway was mostly absent and the PP pathway served exclusively biosynthetic functions. Although the ED pathway is widely distributed (6, 26) and is probably the oldest catabolic pathway (37), its predominance was unexpected for species capable of all three routes of glucose catabolism, such as Paracoccus versutus (49) and R. sphaeroides (5). The generally held view that the EMP pathway is the major route of glucose catabolism may thus be a misconception, because most quantitative metabolic studies were done with the model microbes E. coli, B. subtilis, C. glutamicum, and S. cerevisiae, which exhibit unusually high glycolytic fluxes.
Beyond the ED pathway, the flux data presented here reveal a number of additional abnormalities in the glucose catabolism of the more frequently investigated model bacteria. First, all nonmodel species used the PP pathway exclusively for biosynthesis of biomass precursors, and in many cases the flux to the building blocks was through both the oxidative and the nonoxidative branches of the pathway (Table 5). In sharp contrast, the E. coli and B. subtilis PP pathway flux contributed substantially to catabolism (12 and 27%, respectively), which is consistent with the results of previous analyses (39, 53) and also true for C. glutamicum (48). Second, the E. coli and B. subtilis TCA cycle flux was relatively low, metabolism was not fully respiratory, and there was extensive overflow metabolism that is also referred to as aerobic fermentation. None of the aerobic species investigated here exhibited this phenomenon to a significant extent, which is particularly surprising for species such as Paracoccus versutus that exhibit much higher catabolic rates than do the model microbes. Thus, it is not unreasonable to speculate that overflow metabolism is not a typical microbial trait but rather an adaptation to industrial or laboratory conditions. The most prominent example is baker's yeast, which has been cultivated and selected over millennia by mankind and strongly increases its rate of aerobic fermentation at high growth rates (3). Third, all aerobic nonmodel species routed between 29 and 35% of their carbon flux through the so-called pyruvate shunt, which bypasses malate dehydrogenase. In the case of P. fluorescens, the pyruvate shunt was the predominant route of malate-to-OAA conversion. Although B. subtilis was reported to use the pyruvate bypass under glucose limitation (40), its activity is very low in glucose batch cultures (Fig. 6D) (53).
In addition to the linear ED pathway, which may be inducible in E. coli during growth on gluconate or constitutive, as it is in Z. mobilis, cyclic operation of the ED pathway was reported for organisms that lack phosphofructokinase, e.g., pseudomonads (28) and S. meliloti (25). In the absence of both phosphofructokinase and fructose-1,6-P aldolase, A. tumefaciens cannot operate a cyclic ED pathway but may alternatively use a cyclic PP pathway (1). Cyclic operation of either pathway is known from studies of members of the phylum Proteobacteria such as Acetobacter, Agrobacterium, Azotobacter, Pseudomonas, Rhizobium, Paracoccus, and Xanthomonas spp., which preferentially utilize organic acids rather than sugars and secrete exopolysaccharides (35). The cyclic pathway operation may facilitate the formation of the polysaccharide precursor fructose-6-P in cases where the EMP pathway is absent (21). The extent to which glyceraldehyde-3-P is recycled into the ED pathway is unclear from the present data and may vary between species. However, an increased demand for fructose-6-P and also glucose-6-P due to (exo)polysaccharide synthesis was assumed to be negligible under non-nitrogen-limited batch culture conditions (20, 35, 54).
Ideally, identification of reaction networks and quantification of metabolic fluxes is based on annotated genome data, a large body of biochemical data, and comprehensive tracer experiments. Here we demonstrate that important quantitative information can also be inferred from a much smaller data basis than is typically available for model microbes by using well-designed 13C experiments. Besides the above quantification of the different catabolic routes in each species, several new findings were made. Although the overall glucose flux was, of course, from hexoses to trioses, low but significant gluconeogenic flux occurred in R. sphaeroides and Paracoccus versutus from glyceraldehyde-3-P to glucose-6-P. These two were also the only species where pentose phosphates were mostly synthesized via the nonoxidative branch of the PP pathway, despite high catabolic fluxes passing through the other possible precursor, 6-P-gluconate. During the first growth phase, P. fluorescens converts a major fraction of glucose extracellularly to gluconate and 2-ketogluconate, which are metabolized during the later growth phases; this conversion is in contrast with the physiological data that suggested direct utilization of glucose (29). In Z. mobilis, C1 metabolism introduces a significant fraction of (13C-labeled) pyruvate formate lyase-derived formate into serine. Such experimental identification of metabolic networks and quantification of in vivo molecular fluxes by 13C tracer experiments has great potential for application in novel or not-yet-characterized species.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arthur, L. O., L. A. Bulla, Jr., G. St. Julian, and L. K. Nakamura. 1973. Carbohydrate metabolism in Agrobacterium tumefaciens. J. Bacteriol. 116:304-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. C., S. J. Ferguson, B. Ludwig, M. D. Page, O.-M. Richter, and R. J. M. van Spanning. 1998. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 62:1046-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blank, L. M., and U. Sauer. 2004. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology 150:1085-1093. [DOI] [PubMed] [Google Scholar]
- 4.Conrad, R., and H. G. Schlegel. 1977. Different degradation pathways for glucose and fructose in Rhodopseudomonas capsulata. Arch. Microbiol. 112:39-48. [DOI] [PubMed] [Google Scholar]
- 5.Conrad, R., and H. G. Schlegel. 1977. Influence of aerobic and phototrophic growth conditions on the distribution of glucose and fructose carbon into the Entner-Doudoroff and Embden-Meyerhof pathways in Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 101:277-290. [Google Scholar]
- 6.Conway, T. 1992. The Entner-Doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol. Rev. 9:1-27. [DOI] [PubMed] [Google Scholar]
- 7.Dauner, M., and U. Sauer. 2000. GC-MS analysis of amino acids rapidly provides rich information for isotopomer balancing. Biotechnol. Prog. 16:642-649. [DOI] [PubMed] [Google Scholar]
- 8.Dauner, M., and U. Sauer. 2001. Stoichiometric growth model for riboflavin-producing Bacillus subtilis. Biotechnol. Bioeng. 76:132-143. [DOI] [PubMed] [Google Scholar]
- 9.Dauner, M., T. Storni, and U. Sauer. 2001. Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J. Bacteriol. 183:7308-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Graaf, A. A., K. Striegel, R. M. Wittig, B. Laufer, G. Schmitz, W. Wiechert, G. A. Sprenger, and H. Sahm. 1999. Metabolic state of Zymomonas mobilis in glucose-, fructose-, and xylose-fed continuous cultures as analysed by 13C- and 31P-NMR spectroscopy. Arch. Microbiol. 171:371-385. [DOI] [PubMed] [Google Scholar]
- 11.Doelle, H. W., L. Kirk, R. Crittenden, H. Toh, and M. B. Doelle. 1993. Zymomonas mobilis—science and industrial application. Crit. Rev. Biotechnol. 13:57-98. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, M. F., G. Araiza, and T. M. Finan. 2001. Cloning and characterization of the pyruvate carboxylase from Sinorhizobium meliloti Rm1021. Arch. Microbiol. 176:355-363. [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg, R. C., S. J. Butters, S. C. Quay, and S. B. Friedman. 1974. Glucose uptake and phosphorylation in Pseudomonas fluorescens. J. Bacteriol. 120:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenberg, R. C., and W. J. Dobrogosz. 1967. Gluconate metabolism in Escherichia coli. J. Bacteriol. 93:941-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emmerling, M., M. Dauner, A. Ponti, J. Fiaux, M. Hochuli, T. Szyperski, K. Wüthrich, J. E. Bailey, and U. Sauer. 2002. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 184:152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, E., and U. Sauer. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270:880-891. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, E., and U. Sauer. 2003. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J. Biol. Chem. 278:46446-46451. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, E., N. Zamboni, and U. Sauer. 2004. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Anal. Biochem. 325:308-316. [DOI] [PubMed] [Google Scholar]
- 19.Gombert, A. K., M. Moreira dos Santos, B. Christensen, and J. Nielsen. 2001. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 183:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, J. E., G. M. York, and G. C. Walker. 1996. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene 179:141-146. [DOI] [PubMed] [Google Scholar]
- 21.Gosselin, I., O. Wattraint, D. Riboul, J. Barbotin, and J. Portais. 2001. A deeper investigation on carbohydrate cycling in Sinorhizobium meliloti. FEBS Lett. 499:45-49. [DOI] [PubMed] [Google Scholar]
- 22.Gunnarsson, N., U. H. Mortensen, M. Sosio, and J. Nielsen. 2004. Identification of the Entner-Doudoroff pathway in an antibiotic-producing actinomycete species. Mol. Microbiol. 52:895-902. [DOI] [PubMed] [Google Scholar]
- 23.Hickman, J. W., R. D. Barber, E. P. Skaar, and T. J. Donohue. 2002. Link between the membrane-bound pyridine nucleotide transhydrogenase and glutathione-dependent processes in Rhodobacter sphaeroides. J. Bacteriol. 184:400-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua, Q., C. Yang, T. Baba, H. Mori, and K. Shimizu. 2003. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J. Bacteriol. 185:7053-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irigoyen, J. J., M. Sanchez-Diaz, and D. W. Emerich. 1990. Carbon metabolism enzymes of Rhizobium meliloti cultures and bacteroids and their distribution within alfalfa nodules. Appl. Environ. Microbiol. 56:2587-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kersters, K., and J. De Ley. 1968. The occurrence of the Entner-Doudoroff pathway in bacteria. Antonie Leeuwenhoek 34:393-408. [DOI] [PubMed] [Google Scholar]
- 27.Klamt, S., S. Schuster, and E. D. Gilles. 2002. Calculability analysis in underdetermined metabolic networks illustrated by a model of the central metabolism in purple nonsulfur bacteria. Biotechnol. Bioeng. 77:734-751. [DOI] [PubMed] [Google Scholar]
- 28.Lessie, T. G., and P. V. Phibbs, Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38:359-388. [DOI] [PubMed] [Google Scholar]
- 29.Lynch, W. H., and M. Franklin. 1978. Effect of temperature on uptake of glucose, gluconate, and 2-ketogluconate by Pseudomonas fluorescens. Can. J. Microbiol. 24:56-62. [DOI] [PubMed] [Google Scholar]
- 30.Marx, A., A. A. deGraaf, W. Wiechert, L. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 49:111-129. [DOI] [PubMed] [Google Scholar]
- 31.Michal, G. 1999. Biochemical pathways. Spektrum Akademischer Verlag GmbH, Heidelberg, Germany.
- 32.Nipkow, A., B. Sonnleitner, and A. Fiechter. 1985. Effect of carbon-dioxide on growth of Zymomonas mobilis in continuous culture. Appl. Microbiol. Biotechnol. 21:287-291. [Google Scholar]
- 33.Osman, Y. A., T. Conway, S. J. Bonetti, and L. O. Ingram. 1987. Glycolytic flux in Zymomonas mobilis: enzyme and metabolite levels during batch fermentation. J. Bacteriol. 169:3726-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen, S., A. A. de Graaf, L. Eggeling, M. Mollney, W. Wiechert, and H. Sahm. 2000. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 275:35932-35941. [DOI] [PubMed] [Google Scholar]
- 35.Portais, J. C., and A. M. Delort. 2002. Carbohydrate cycling in micro-organisms: what can (13)C-NMR tell us? FEMS Microbiol. Rev. 26:375-402. [DOI] [PubMed] [Google Scholar]
- 36.Portais, J. C., P. Tavernier, I. Gosselin, and J. N. Barbotin. 1999. Cyclic organization of the carbohydrate metabolism in Sinorhizobium meliloti. Eur. J. Biochem. 265:473-480. [DOI] [PubMed] [Google Scholar]
- 37.Romano, A. H., and T. Conway. 1996. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 147:448-455. [DOI] [PubMed] [Google Scholar]
- 38.Sauer, U. 2004. High-throughput phenomics: experimental methods for mapping fluxomes. Curr. Opin. Biotechnol. 15:58-63. [DOI] [PubMed] [Google Scholar]
- 39.Sauer, U., F. Canonaco, S. Heri, A. Perrenoud, and E. Fischer. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279:6613-6619. [DOI] [PubMed] [Google Scholar]
- 40.Sauer, U., V. Hatzimanikatis, J. E. Bailey, M. Hochuli, T. Szyperski, and K. Wüthrich. 1997. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat. Biotechnol. 15:448-452. [DOI] [PubMed] [Google Scholar]
- 41.Sauer, U., D. R. Lasko, J. Fiaux, M. Hochuli, R. Glaser, T. Szyperski, K. Wüthrich, and J. E. Bailey. 1999. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 181:6679-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sprenger, G. A. 1996. Carbohydrate metabolism in Zymomonas mobilis: a catabolic highway with some scenic routes. FEMS Microbiol. Lett. 145:301-307. [Google Scholar]
- 43.Stowers, M. D. 1985. Carbon metabolism in Rhizobium species. Annu. Rev. Microbiol. 39:89-108. [DOI] [PubMed] [Google Scholar]
- 44.Szyperski, T. 1995. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids. An efficient analytical tool to investigate intermediary metabolism. Eur. J. Biochem. 232:433-448. [DOI] [PubMed] [Google Scholar]
- 45.Temple, L. M., A. E. Sage, H. P. Schweizer, and P. V. Phibbs, Jr. 1998. Carbohydrate metabolism in Pseudomonas aeruginosa. Biotechnol. Handb. 10:35-72. [Google Scholar]
- 46.Vicente, M., and J. L. Cánovas. 1973. Glucolysis in Pseudomonas putida: physiological role of alternative routes from the analysis of defective mutants. J. Bacteriol. 116:908-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiechert, W. 2001. 13C metabolic flux analysis. Metab. Eng. 3:195-206. [DOI] [PubMed] [Google Scholar]
- 48.Wittmann, C., and E. Heinzle. 2002. Genealogy profiling through strain improvement by using metabolic network analysis: metabolic flux genealogy of several generations of lysine-producing corynebacteria. Appl. Environ. Microbiol. 68:5843-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood, A. P., and D. P. Kelly. 1980. Carbohydrate degradation pathways in Thiobacillus A2 grown on various sugars. J. Gen. Microbiol. 120:333-345. [Google Scholar]
- 50.Wood, A. P., and D. P. Kelly. 1977. Heterotrophic growth of Thiobacillus A2 on sugars and organic acids. Arch. Microbiol. 113:257-264. [DOI] [PubMed] [Google Scholar]
- 51.Wood, A. P., D. P. Kelly, and C. F. Thurston. 1977. Simultaneous operation of three catabolic pathways in the metabolism of glucose by Thiobacillus A2. Arch. Microbiol. 113:265-274. [DOI] [PubMed] [Google Scholar]
- 52.Yakunin, A. F., and P. C. Hallenbeck. 1997. Regulation of synthesis of pyruvate carboxylase in the photosynthetic bacterium Rhodobacter capsulatus. J. Bacteriol. 179:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zamboni, N., and U. Sauer. 2003. Knockout of the high-coupling cytochrome aa3 oxidase reduces TCA cycle fluxes in Bacillus subtilis. FEMS Microbiol. Lett. 226:121-126. [DOI] [PubMed] [Google Scholar]
- 54.Zevenhuizen, L. P. 1981. Cellular glycogen, beta-1,2,-glucan, poly beta-hydroxybutyric acid and extracellular polysaccharides in fast-growing species of rhizobium. Antonie Leeuwenhoek 47:481-497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.