Abstract
We have investigated anaerobic respiration of the archaeal model organism Halobacterium sp. strain NRC-1 by using phenotypic and genetic analysis, bioinformatics, and transcriptome analysis. NRC-1 was found to grow on either dimethyl sulfoxide (DMSO) or trimethylamine N-oxide (TMAO) as the sole terminal electron acceptor, with a doubling time of 1 day. An operon, dmsREABCD, encoding a putative regulatory protein, DmsR, a molybdopterin oxidoreductase of the DMSO reductase family (DmsEABC), and a molecular chaperone (DmsD) was identified by bioinformatics and confirmed as a transcriptional unit by reverse transcriptase PCR analysis. dmsR, dmsA, and dmsD in-frame deletion mutants were individually constructed. Phenotypic analysis demonstrated that dmsR, dmsA, and dmsD are required for anaerobic respiration on DMSO and TMAO. The requirement for dmsR, whose predicted product contains a DNA-binding domain similar to that of the Bat family of activators (COG3413), indicated that it functions as an activator. A cysteine-rich domain was found in the dmsR gene, which may be involved in oxygen sensing. Microarray analysis using a whole-genome 60-mer oligonucleotide array showed that the dms operon is induced during anaerobic respiration. Comparison of dmsR+ and ΔdmsR strains by use of microarrays showed that the induction of the dmsEABCD operon is dependent on a functional dmsR gene, consistent with its action as a transcriptional activator. Our results clearly establish the genes required for anaerobic respiration using DMSO and TMAO in an archaeon for the first time.
Extremely halophilic archaea (haloarchaea) generally grow heterotrophically under aerobic conditions in hypersaline environments, although they possess facultative anaerobic capabilities (9). These anaerobic capabilities are important since the high salt concentrations and elevated temperatures the organisms encounter, together with high cell densities promoted by aerobic growth and flotation, reduce the availability of molecular oxygen. Although haloarchaeal microorganisms frequently encounter microaerobic or even anoxic conditions, detailed knowledge regarding the extent of haloarchaeal anaerobic growth is still only beginning to emerge. Some species of haloarchaea can carry out primary energy conservation in the absence of molecular oxygen via photophosphorylation (13), substrate-level phosphorylation (13), and anaerobic respiration using nitrate (references 9 and 42 and references therein). In addition, two previous reports described the ability to use dimethyl sulfoxide (DMSO) and trimethylamine-N-oxide (TMAO) (23), and to some extent fumarate (22), as alternative electron acceptors for growth enhancement for some haloarchaea.
The molecular basis of the DMSO and TMAO respiratory systems has not been described for any member of the domain Archaea. By contrast, in various bacterial model organisms, DMSO reductases and the related TMAO reductases are well characterized as membrane-bound or periplasmic terminal reductases forming dimethyl sulfide or trimethylamine, respectively, as the final product (7, 17, 19, 29, 30, 32, 39). In Escherichia coli, the molecular components of the DMSO reductase are DmsA, the catalytically active subunit harboring molybdenum-molybdopterin (MPT) as a cofactor; DmsB, a subunit containing iron-sulfur clusters for electron transport; DmsC, a membrane anchor; and DmsD, a molecular chaperone. Other bacterial DMSO and TMAO reductases may contain a membrane-bound c-type cytochrome (DorC or TorC) instead of DmsB and DmsC (17, 31, 32). The expression of the corresponding dms and tor genes in bacteria may be regulated by a two-component regulatory system (e.g., TorR-TorS) (15, 33) or the global oxygen response regulator FNR (6). Additional regulation may be provided by the presence of the preferred terminal electron acceptor nitrate or the absence of molybdenum (1, 6).
In the present study, we describe the first functional genomic characterization of the DMSO/TMAO respiratory system in the haloarchaeal model organism Halobacterium sp. strain NRC-1. Previous annotation reported a single dmsA gene surrounded by additional, unspecified MPT oxidoreductase genes (20). Reexamination of the dmsA region, in this investigation, found a total of six genes, dmsREABCD, forming an operon that functions as the DMSO/TMAO respiration system (Fig. 1). Here, we report a combination of genetic and transcriptional data that establish the requirement for the dmsREABCD operon of Halobacterium sp. strain NRC-1 for anaerobic respiration, utilizing DMSO and TMAO, in an archaeon.
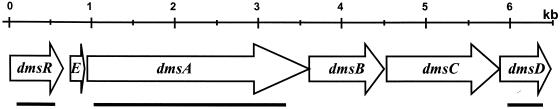
FIG. 1.
Physical map of the dms gene locus in Halobacterium sp. strain NRC-1. The locations and directions of open reading frames are indicated by arrows. Regions of dmsR, dmsA, and dmsD deleted in Halobacterium sp. strains JAM101, -102, and -103 are indicated by horizontal bars.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Halobacterium sp. strain NRC-1 was grown in complex medium containing peptone (10 g/liter) and 10 mM citrate (CM+ medium) (10) either under aerobic conditions or under anaerobic conditions in the presence of an alternative electron acceptor. For anaerobic growth, the medium was kept in an anaerobic glove box (Coy Laboratory Products, Ann Arbor, Mich.) with an N2-H2 (95:5 [vol/vol]) atmosphere for 1 day prior to inoculation. The medium contained the redox indicator resazurin to test for anoxic conditions. Even without added reducing agent, the pink color of oxidized resazurin disappeared in sterile, freshly prepared CM+ medium when kept overnight under an oxygen-free atmosphere. This was likely due to reduction of resazurin to the colorless form by redox-active components, e.g., cysteine, in the medium. The cultivation vessel containing the medium was sealed inside the anaerobic chamber with a butyl rubber stopper, and an electron acceptor (TMAO or DMSO) was added from a sterile stock solution. Cultivation vessels were shaken at 220 rpm at 42°C in the dark. Growth was monitored spectrophotometrically by determining the optical density at 600 nm. The concentration of dimethyl sulfide during growth on DMSO was quantified by gas chromatography (18). The presence of trimethylamine during growth on TMAO was detected by its characteristic odor. All cultivation experiments were performed at least in triplicates.
E. coli strain DH5α was used as a host for cloning vectors. The strain was grown in Luria-Bertani medium at 37°C, and ampicillin was added at 50 μg/ml when appropriate. The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source (reference) |
|---|---|---|
| Halobacterium sp. strains | ||
| NRC-1 | Wild type | Laboratory collection (28) |
| SK400 | NRC-1 Δura3 | Laboratory collection (31) |
| JAM101 | SK400 ΔdmsR | This study |
| JAM102 | SK400 ΔdmsA | This study |
| JAM103 | SK400 ΔdmsD | This study |
| E. coli strain DH5α | supE44 ΔlacU169 φ80ΔlacZM15 hsdR17 recA1 endA1gyrA96 thi-1 relA1 | Invitrogen |
| Plasmids | ||
| pUC19 | Cloning vector, lacZ bla | New England Biolabs |
| pJAM10 | pUC19 containing NRC-1 ura3 with flanking region | This study |
| pJAM101 | pJAM10 containing truncated dmsR with flanking region | This study |
| pJAM102 | pJAM10 containing truncated dmsA with flanking region | This study |
| pJAM103 | pJAM10 containing truncated dmsD with flanking region | This study |
DNA isolation and PCR amplification.
Standard protocols were used for DNA cloning and transformation into E. coli (27). Chromosomal DNA of strain NRC-1 was isolated as described previously (10). Purifications of PCR products, DNA fragments from restriction enzyme digests, and plasmids were performed with Qiaprep spin colums (Qiagen). PCR mixtures contained 100 ng of genomic DNA, a 200 nM concentration of each primer, 200 μM deoxynucleoside triphosphates, and 1 U of Taq polymerase (Promega) in PCR buffer with 1.5 mM MgCl2. PCR parameters were as follows: 3 min at 92°C; 30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C; and 5 min at 72°C.
Construction of deletion mutants.
A vector, plasmid pJAM10, was constructed for gene deletion in strain NRC-1 in combination with blue-white screening in E. coli. A 1.3-kb DNA fragment containing the Halobacterium sp. strain NRC-1 ura3 gene including 500 bp of the 5′ flanking DNA sequence regions was amplified with Pfu (Stratagene) and the primers ura3F (5′-AGGAGGCGTGCTGGAATGGTCGCT-3′) and ura3R (5′-GGGCCGCCGGCTGACTACCCG-3′). This PCR fragment was cloned into pUC19 digested with SspI.
For deletion of dmsR (VNG826), plasmid pJAM101 was generated. First, a 463-bp DNA fragment containing the 5′ region of dmsR, including 421 bp of 5′ flanking DNA sequence, and a 510-bp DNA fragment containing the 3′ region of dmsR, including 497 bp of 3′ flanking DNA sequence, were PCR amplified with primers dmsR5_F_Xba (5′-GCTCTAGACCACGACCGGCTCCCCAACC-3′) and dmsR5_R_Eco (5′-CGGAATTCACGAGCCCCACGAGCAGTAG-3′) and primers dmsR3_F_Eco (5′-CGGAATTCCAACACGCCGGACGCACACGAG-3′) and dmsR3_R_Xba (5′-CCTCTAGAGGCGAGCAGGCGGTTTTGACGAC-3′), respectively. Both fragments were digested with EcoRI, purified, and ligated. The ligated fragment was digested with XbaI, purified, and cloned into pJAM10 cut with XbaI. The correct incorporation of the 5′ and 3′ DNA sequence regions of dmsR into pJAM10 was verified by PCR analysis. Plasmid pJAM101 resulted in an in-frame deletion of codons 14 to 211 of dmsR (absolute positions 626492 to 627094). The plasmids pJAM102 for deletion of dmsA (with the primers dmsA5_F_Xba [5′-GCTCTAGACGACGGCGCTTGGCATCT-3′], dmsA5_R_Eco [5′-CGGAATTCCCGGGTGGCGTTGAGGTC-3′], dmsA3_F_Eco [5′-CGGAATTCCCCCGCCTGCGTCGTGAACTGTCC-3′], and dmsA5_R_Xba [5′-GCTCTAGAGGGGCGGCGTGCGGTCGTGTCG-3′]) and pJAM103 for deletion of dmsD (VNG832) (with the primers dmsD5_F_Xba [5′-GCTCTAGATCGTCGCGCCCGTGTTCAT-3′], dmsD5_R_Eco [5′-CGGAATTCCGCGCTGCAGGGGGAGATAG-3′], dmsD3_F_Eco [5′-CGGAATTCACGCCGTCTCCAGCAGTCA-3′], and dmsD3_R_Xba [5′-CGTCTAGACGCCATCCGGTCTTTGTA-3′]) were generated in an analogous manner. Plasmid pJAM102 resulted in an in-frame deletion spanning the region between codons 10 and 808 of dmsA (absolute positions 627475 to 629869), and plasmid pJAM103 resulted in an in-frame deletion between codons 9 and 183 of dmsD (absolute positions 632074 to 632597).
Transformation of Halobacterium sp. strain NRC-1 Δura3, a uracil auxotroph, with plasmid pJAM101, pJAM102, or pJAM103 was carried out essentially as described previously (24, 37). In brief, first-crossover integrants were obtained by using uracil-deficient medium. The integration of plasmid was confirmed by screening for the presence of the ura3 gene. Second-crossover recombinants were selected for on rich CM+ plates in the presence of 0.25 mg of 5-fluoro-orotic acid (Toronto Research Chemicals) per ml. Recombinants were screened by PCR to confirm loss of the ura3 gene and for the presence of either the deletion or wild-type target gene.
RT-PCR.
Total RNA was prepared from DMSO-grown strain NRC-1 cells in mid-log phase. Cells were harvested by centrifugation of the culture (8,000 × g, 5 min, 2°C) in chilled beakers. RNA was isolated immediately after cell harvest by using spin columns (Agilent, Palo Alto, Calif.). Lysis of cells was carried out directly in lysis buffer. Two treatments with DNase (Invitrogen) were carried out in order to remove contaminating DNA from the RNA preparations. Synthesis of cDNA was done from 500 ng of RNA and 2 pmol of specific primer with SuperScript II RNase H− reverse transcriptase as described by the supplier (Invitrogen). For amplification of parts of dmsR and intergenic regions of dmsR and dmsA, dmsE and dmsA, dmsA and dmsB, dmsB and dmsC, and dmsC and dmsD, six sets of primers were chosen for reverse transcriptase PCR (RT-PCR): dmsRf (5′-AGCGCACAGCCAGCCAGCCGAACA-3′) and dmsRr (5′-CGAGCCGCGTCAGGTGGGTCAGC-3′), i-dmsRf (5′-GATGCTCGCGGACTGTGGCTACG-3′) and i-dmsAr (5′-CGCGCTGATCGGGCTGTGG-3′), i-dmsEf (5′-CCAACACGCCGGACGCACACGAG-3′) and i-dmsAr (5′-GGCGAGCAGGCGGTTTTGACGAC-3′), i-dmsAf (5′-ACTTCGTCGACGGGCACCTCCAG-3′) and i-dmsBr (5′-CGTTCACCGGGCAGACCTTCACAC-3′), i-dmsBf (5′-CGGACGCCCCTGGAAGAGCAAGAA-3′) and i-dmsCr (5′-CAACAGCCCCGCGACGATGATG-3′), and i-dmsCf (5′-AGTACGCCGGCCTCACGAC-3′) and i-dmsDr (5′-TCCGGGCCCGACGAACAGT-3′). The PCR amplifications were done by using 1 U of Taq polymerase and standard conditions (30 cycles). Negative controls without reverse transcriptase to test for absence of genomic DNA in the assay were included.
Microarray design.
Oligomer (60-mer) probes were designed for 2,473 open reading frames (ORFs) of Halobacterium sp. strain NRC-1 by using the program OligoPicker (38). Up to three probes were designed per gene. Duplicate genes were considered only once for probe design. A BLAST search of all probes against the genome of Halobacterium sp. strain NRC-1 was carried out in order to ensure specificity. The melting temperature range of the probes was 3°C, with a mean melting temperature of 81°C. Microarray slides were fabricated by in situ oligonucleotide synthesis (Agilent) (14). The slides harbor 8,455 features per array, with two arrays per slide. The features include the gene probes (8,077), present randomly and often in multitude (the average occurrence of probes is 1.4 times), as well as negative and positive control spots to test hybridization conditions. Sequences of 364 control spots were designed by Agilent; sequences for additional 14 control spots were taken from the genomes of Mycobacterium tuberculosis (65.6% GC content) and Mycobacterium leprae (57.8% GC content), both of which have percent GC contents comparable to those of Halobacterium sp. strain NRC-1 genetic elements.
Microarray analysis.
Halobacterium sp. strain NRC-1 was grown under aerobic conditions and anaerobic conditions, using TMAO as an electron acceptor, to early exponential growth phase (optical density at 600 nm of 0.15 to 0.3). Cultivation of cells (50 ml) was carried out in 500-ml flasks to ensure proper mixing of culture fluid. Cells were harvested by chilling the incubation vessels in an ethanol-dry ice bath for 1 min followed by centrifugation of the culture (8,000 × g, 5 min, 2°C) in chilled beakers. Total RNA was isolated as described above. DNA was hydrolyzed in RNA preparations with DNase (Invitrogen). For cDNA synthesis, RNA preparations from three cultures grown under identical conditions were pooled to equal parts in order to minimize biological noise. Fluorescently labeled cDNA was prepared essentially as described by M. Laub et al. (http://caulobacter.stanford.edu/CellCycle/protocols.htm). In brief, 7 μg of total RNA was combined with 500 ng of random hexamers (Qiagen) and reverse transcribed to Cy3- or Cy5-dCTP (Amersham Pharmacia)-labeled cDNA by using SuperScript II RNase H− reverse transcriptase (Invitrogen). Subsequently, the cDNA preparations were purified after alkaline hydrolysis of RNA at 65°C with Qiagen spin columns. To control for labeling differences, duplicate reactions in which the Cy3 and Cy5 labels were switched during synthesis were carried out. The labeled cDNA targets were mixed with hybridization buffer (Agilent) and control targets (Agilent) and hybridized to microarray slides, assembled into a hybridization chamber (Agilent), for 17 h at 60°C in the dark. After hybridization, the slides were washed in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.005% Triton X-102 for 10 min at room temperature followed by 0.1× SSC-0.005% Triton X-102 for 5 min at 4°C. The microarray slides were scanned for the Cy3 and Cy5 fluorescent signals with an Agilent DNA microarray scanner (model no. G2565BA).
Image processing and statistical analysis were carried out with Agilent Feature Extraction software, version 7.1. Spot signal intensities were adjusted by subtracting local background, and a two-sided t test was performed to assess whether the signal was significantly different from the background signal. Fluorescence intensities were normalized by using the LOWESS method (5). Log ratios for each feature were calculated from the processed red and green signals. The significance of the log ratio was assessed by computing the most conservative log ratio error and significance value (P value), using a standard error propagation algorithm (Agilent) and a universal error model (Rosetta Biosoftware). After removal of outliers, the final log ratio, fold change of gene expression, log ratio error, and P value for a gene were calculated as arithmetic means of all probe values for that gene.
Computational analysis of DNA and amino acid sequences.
BLAST analysis and sequence manipulation were done with the bioinformatics tools available on our server and website (http://zdna2.umbi.umd.edu). Sequence data were obtained either from our website or from the National Center for Biotechnology Information. Nucleotide and amino acid sequence analyses were performed with the programs from the Wisconsin Genetics Computer Group software package (Accelrys, San Diego, Calif.), the DNAStar (Madison, Wis.) software package, Clustal_X (version 1.81), and Simple Modular Architecture Research Tool SMART (http://smart.embl-heidelberg.de). Phylogenetic analysis for Dms proteins was performed with Clustal_X for multiple-sequence alignment together with Treeview for neighbor-joining tree construction (34), as well as Treepuzzle (http://www.nsc.liu.se/software/biology/puzzle5).
RESULTS
Characterization of Halobacterium sp. strain NRC-1 anaerobic respiration using DMSO and TMAO.
Some enhancement of anaerobic growth of strain NRC-1 with DMSO and TMAO as terminal electron acceptors has been reported before (23). Since details were not published, we reinvestigated the use of both compounds for anaerobic respiration by strain NRC-1. NRC-1 grew in CM+ medium (containing peptone and citrate as carbon and electron sources) under strictly anaerobic conditions with either 40 mM DMSO (doubling time [td] = 1.2 days) (Fig. 2) or 40 mM TMAO (td = 1.1 days) as the sole electron acceptor. The respective respiration products dimethyl sulfide and trimethylamine accumulated concomitantly with increase in optical density. A lag in growth was observed after transfer of aerobically but not anaerobically grown cultures, indicating that components of anaerobic respiration are inducible. Microscopic examinations revealed the presence of abundant gas vesicles inside cells already during the early growth stage on DMSO or TMAO. Aerobically grown cells typically harbor abundant gas vesicles during late log and stationary phases.
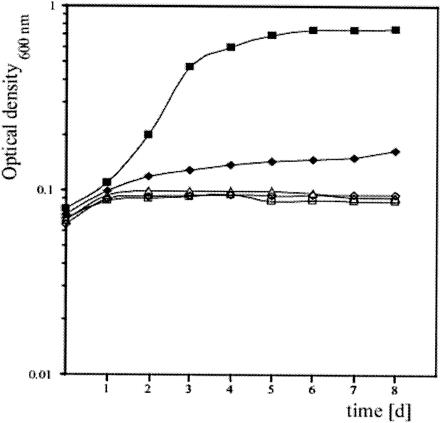
FIG. 2.
Test for anaerobic growth of Halobacterium sp. wild-type strain NRC-1 and dms deletion strains on DMSO (40 mM). Incubation was at 42°C with shaking at 200 rpm. Cultures were inoculated with cells grown on l-arginine (0.5%, wt/vol). The increase in optical density was measured at 600 nm. Closed squares, NRC-1; closed diamonds, JAM101 (ΔdmsR); closed triangles, JAM102 (ΔdmsA); open squares, JAM103 (ΔdmsD); open diamonds, NRC-1 without DMSO in the medium.
The MPT cofactor of DMSO and TMAO reductases contains the transition metal molybdenum. Addition of 3 μM molybdate to the medium did not change the growth profile, indicating the presence of sufficient molybdenum in the basal salt medium. In the presence of 10 mM tungstate, a molybdate antagonist, growth on DMSO and TMAO was severely impaired (td of >7 days), while aerobic growth and arginine fermentation were not affected.
Construction and characterization of dms deletion strains.
To confirm the involvement of the dmsREABCD operon in DMSO and TMAO respiration in Halobacterium sp. strain NRC-1, in-frame deletions of dmsR, dmsA, and dmsD were constructed. The resulting strains were designated JAM101 (ΔdmsR Δura3), JAM102 (ΔdmsA Δura3), and JAM103 (ΔdmsD Δura3) and tested for their ability to grow by anaerobic respiration. Strains JAM102 (ΔdmsA Δura3) and JAM103 (ΔdmsD Δura3) failed to grow on either DMSO (Fig. 2) or TMAO (not shown), while strain JAM101 (ΔdmsR Δura3) displayed a marked decrease in growth on DMSO and TMAO. During anaerobic fermentation of arginine and under aerobic conditions, the growth of the mutant strains was similar to that of the wild type (data not shown). These results clearly demonstrate that the dmsR, dmsA, and dmsD genes of Halobacterium sp. strain NRC-1 are essential for anaerobic growth with DMSO and TMAO as terminal electron acceptors. The finding of a DMSO- and TMAO-negative phenotype for the dmsR mutant was unexpected and indicated, together with bioinformatics analysis, that this gene encodes an activator regulating transcription of the dms genes (see below).
Transcriptional organization of the dms gene cluster.
To study the transcriptional organization of the dms gene cluster, RT-PCR analysis was performed with primer pairs that were designed to detect the dmsR transcript as well as cotranscription of adjacent genes. RT-PCR products were obtained for the intergenic regions of dmsE and dmsA, dmsA and dmsB, dmsB and dmsC, and dmsC and dmsD (Fig. 3). The RNA-specific recovery of these products demonstrates that the dmsEABCD genes are transcribed as a polycistronic unit and constitute an operon. No RT-PCR product was obtained for the intergenic region of dmsR and dmsA, indicating that dmsR, the putative activator, is transcribed separately.
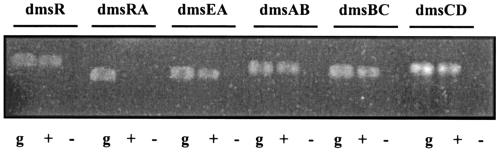
FIG. 3.
RT-PCR analysis of the dms operon in Halobacterium sp. strain NRC-1. Agarose gel electrophoresis of RT-PCR assays with primers targeting dmsR and intergenic regions of dmsR and dmsE, dmsE and dmsA, dmsA and dmsB, dmsB and dmsC, and dmsC and dmsD is shown. g, genomic DNA as template; +, RT-PCR assays with RNA as template and conducted with reverse transcriptase; −, RT-PCR assays with RNA as template and conducted without reverse transcriptase. Sizes of products were as predicted (for dmsR, 384 bp; for dmsRA, 544 bp; for dmsEA, 386 bp; for dmsAB, 415 bp; for dmsBC, 454 bp; and for dmsCD, 374 bp).
Microarray analysis.
To study dms gene expression and to obtain insight into the effect of anaerobic respiration on global gene expression, we compared the transcriptomes of TMAO-grown and aerobically grown cells of strain NRC-1 by using whole-genome oligonucleotide microarrays. In order to limit the effect of biological variability on the data, RNA preparations from three separately grown, comparably large quantities of cells were pooled to equal parts and fractions thereof were used for cDNA synthesis. The microarray data obtained was assessed to be of high quality based on the following criteria. Per array, only 10 to 15 features out of 8,455 total analyzed features had to be discarded as outliers due to background noise or nonuniform spot morphology. Features replicated within a single array showed low differences in absolute processed signal intensities (9% on average), and the spot-to-spot variation for replicate experiments was 11%. Green and red fluorescent signal intensities had a dynamic range in excess of 3 orders of magnitude, allowing simultaneous analysis of low- and high-intensity features. In the comparison of RNA samples derived from cultures grown under aerobic conditions, only one ORF (VNG725) displayed an expression change of greater than 1.8-fold (Fig. 4A). For the transcriptome comparison between anaerobic respiration and aerobic respiration, a gene was considered to be differentially expressed when the absolute value of the average log ratio was at least 0.3 (twofold expression change), the average log ratio error was <0.1, and the average P value of the log ratio was <10−5 (∼21% of all probes had a low P value of <10−5).
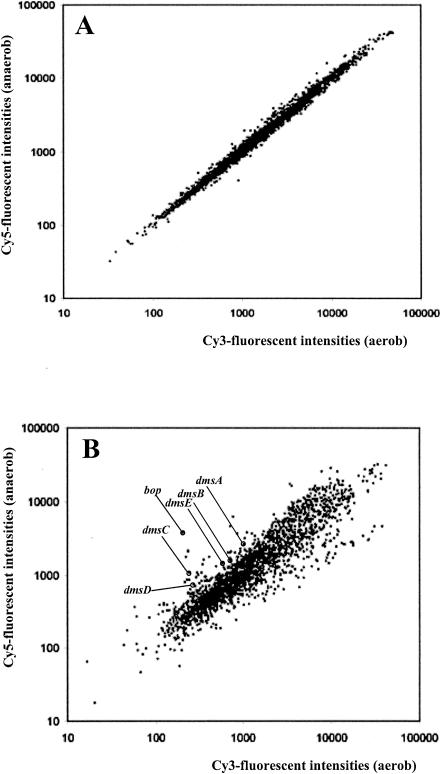
FIG. 4.
Scatter plots of fluorescence intensities of Cy3-labeled cDNA versus Cy5-labeled cDNA. (A) cDNA versus cDNA derived from two cultures grown under aerobic conditions in the dark. (B) cDNA versus cDNA from three cultures, each grown by aerobic and anaerobic respiration.
Of the 2,473 ORFs of Halobacterium sp. strain NRC-1 represented on the array, 104 were up-regulated and 137 were down-regulated at least twofold during anaerobic respiration. To obtain an overview, differentially expressed genes were sorted according to functional categories (Table 2) (for a list of the log ratios for all ORFs, see the supplemental material). Of the 109 genes of strain NRC-1 predicted to be involved in energy metabolism, 24 genes (22%) were differentially expressed during anaerobic respiration. Transcription of the dmsEABCD genes was induced approximately threefold during growth on TMAO (Fig. 4B). The dmsR gene was up-regulated 1.5-fold. The level of up-regulation of the dmsEABCD genes ranked among the highest of all differentially expressed ORFs, consistent with the requirement for these genes during anaerobic respiration.
TABLE 2.
Functional categories of genes differentially expressed during TMAO respiration
| Functional category | No. of genes | Anaerobic respiration vs aerobic respiration
|
|
|---|---|---|---|
| Genes up-regulated | Genes down-regulated | ||
| Energy metabolism | 109 | 11 | 13 |
| Amino acid metabolism | 123 | 2 | 6 |
| Carbohydrate and lipid metabolism | 89 | 0 | 6 |
| Cofactor and secondary metabolite metabolism | 128 | 0 | 8 |
| Nucleotide metabolism | 72 | 0 | 8 |
| DNA metabolism | 112 | 3 | 8 |
| Transcription and regulation | 167 | 6 | 7 |
| Translation | 152 | 0 | 47 |
| Cellular processes | 270 | 8 | 9 |
| Transport | 126 | 2 | 9 |
| General function prediction only | 51 | 0 | 1 |
| Unknown | 1,031 | 72 | 20 |
The bacterio-opsin gene bop displayed the highest up-regulation (15-fold) of all ORFs. The strong differential expression of the bop gene is consistent with its regulation by the oxygen- or redox-sensing activator Bat (2, 3, 40, 41). Genes involved in aerobic respiration in NRC-1 were not down-regulated in the absence of oxygen. NRC-1 contains genes for three different cytochrome oxidases: cbaABD (VNG2193, VNG2195, and VNG2196) (orthologous with the aberrant ba3-type cytochrome oxidases of Natronobacterium pharaonis and Thermus thermophilus), coxABC (aa3-type cytochrome oxidase [11]), and cydAB (orthologous with quinol bd oxidase of Azotobacter vinelandii). Interestingly, the cbaABD genes were markedly up-regulated (fivefold) under anaerobic conditions, suggesting an unusual requirement of this cytochrome oxidase under anoxic conditions (see Discussion). The cox and cyd genes as well as other genes encoding for respiratory enzymes (e.g., equivalents to mitochondrial complex I, II, and III) were not significantly differentially expressed, indicating constitutive regulation as in Pyrobaculum oguniense (21) and/or their requirement during TMAO respiration (e.g., complex I and II equivalents). The transcript levels of genes involved in the citrate cycle were also not significantly differentially expressed; however, the pyruvate:ferredoxin oxidoreductase-encoding porAB genes, channeling carbon and electrons into the citrate cycle, were down-regulated (2.3-fold) during TMAO respiration. The ATP synthase genes were also less expressed (on average 3.3-fold) during TMAO respiration, suggesting that the ATP/ADP ratio and/or the flux rates of the ATP-ADP pools are lower in Halobacterium sp. strain NRC-1 during anaerobic respiration.
Congruent with the microscopic observation of abundant gas vesicles inside anaerobically grown cells, several genes encoding regulatory and structural gas vesicle proteins (gvpED and gvpACNO) were induced (two- to fivefold) during anaerobic respiration. Likewise, transcript levels of the 34 ribosomal protein genes were lower (two- to sixfold) during anaerobic respiration, which most likely reflects the altered growth rate and presumably lower ribosome content during TMAO respiration. Genes needed for MPT biosynthesis (moaA to -E, moeAB, mobAB, modA, modC/cysU, and gdb) were not significantly induced during growth on TMAO; likewise, genes involved in biosynthesis of menaquinone (menA to -E), the typical physiological electron donor for DMSO and TMAO reductases, were not differentially expressed.
Transcriptome comparison of dmsR+ and ΔdmsR strains.
To verify activation of the dmsEABCD genes by DmsR and to potentially identify additional genes regulated by DmsR, we used microarrays to compare strain JAM101 (ΔdmsR Δura3) against its parent, strain NRC-1 Δura3. Both strains were grown by arginine fermentation, since strain JAM101 grows only slightly on DMSO and TMAO but preliminary microarray data had shown that the dms operon is highly induced during growth of strain NRC-1 by arginine fermentation. As expected, the signal intensities of dmsR spots were substantially lower (about 50-fold) for cDNA prepared from JAM101 than for cDNA prepared from its parent strain. The dmsE-D structural genes were five- to eightfold less expressed in the ΔdmsR strain than in its parent strain. This verifies that DmsR activates transcription of dmsEABCD. No indication that DmsR regulates additional genes was obtained.
Bioinformatics analysis of the dms operon.
The dmsEABCD genes (GC content, 67%) were found to be the only MPT oxidoreductase genes in the complete genome sequence of Halobacterium sp. strain NRC-1 (GC content, 68%). Identified domains and motifs of the Dms structural proteins were similar to those of other prokaryotic MPT oxidoreductases and are shown in Table 3. We also queried the unfinished genomes of Haloarcula marismortui and Haloferax volcanii for genes orthologous to the NRC-1 dms genes (sequence data are available at http://zdna2.umbi.umd.edu). Both genomes contain one identically organized dms gene cluster with 75 to 80% amino acid sequence identities of all of the deduced proteins. The reduction of DMSO and TMAO had been reported previously for both strains (23).
TABLE 3.
dms genes and Dms proteins in Halobacterium sp. strain NRC-1
| ORF | Gene | Protein domains and motifs | Predicted protein function | COG or Pfam description |
|---|---|---|---|---|
| VNG826 | dmsR | C-terminal DNA-binding domain | Regulator | COG3413, predicted DNA-binding protein |
| VNG828 | dmsE | Unknown | ||
| VNG829 | dmsA | Leader sequence (TAT motif), vestigial [4Fe-4S] cluster binding motif, molybdopterin domain, molybdopterin-binding domain | Catalytically active subunit | COG0243, anaerobic dehydrogenases (typically selenocysteine containing) |
| VNG830 | dmsB | Four iron-sulfur cluster binding motifs (ferredoxin type) | Electron transfer subunit | COG0437, Fe-S cluster-containing hydrogenase components |
| VNG831 | dmsC | Leader sequence (Sec pathway), 10 predicted transmembrane helices | Membrane anchor subunit | COG5557, polysulfide reductase |
| VNG832 | dmsD | Molecular chaperone | Pfam06192, cytoplasmic chaperone TorD |
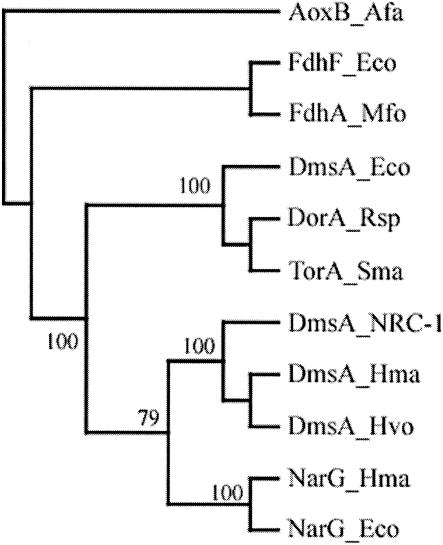
To examine the evolutionary relationships between haloarchaeal DmsA and members of the DMSO reductase family, an alignment was made from amino acid sequences of 90 MPT oxidoreductases and a neighbor-joining tree was constructed. A topologically representative tree containing 11 selected sequences was subsequently generated by the neighbor-joining method (Fig. 5) and by maximum-likelihood analysis. Amino acid sequence identity of DmsA from strain NRC-1 was highest with DmsA from E. coli, DorA from Rhodobacter spp., and NarG from E. coli and H. marismortui (all 21% ± 0.5% identity). Surprisingly, the topology of the phylogenetic tree suggests that haloarchael DmsA is more closely related to NarG-type nitrate reductases than to bacterial DMSO and TMAO reductases. Consistent with the phylogenetic analysis, the molybdenum atom-coordinating protein ligand of bacterial DMSO and TMAO reductases, a serine residue (7, 29, 30, 36), is not conserved; instead, a conserved aspartate residue, as in NarG of E. coli (4), was found in all haloarchaeal DmsA protein sequences. Based on the phylogenetic analysis, as well as the arrangement of cysteines in the N-terminal region, we classified haloarchaeal DmsA as a type II MPT oxidoreductase (35).
FIG. 5.
Neighbor-joining tree of representative molybdopterin oxidoreductases. DmsA_Eco, E. coli DmsA, NP_415414; DmsA_NRC-1, Halobacterium sp. strain NRC-1 DmsA, NP_279804; DmsA_Hma, putative DmsA from Haloarcula marismortui; DmsA_Hvo, putative DmsA from Haloferax volcanii; DorA_Rsp, DMSO reductase from R. sphaeroides, AAB94874; FdhA_Mfo, formate dehydrogenase from Methanobacterium formicicum, P06131; FdhF_Eco, formate dehydrogenase H from E. coli, NP_418503; NarG_Hma, NarG from H. marismortui, CAD22069; TorA_Sma, TMAO reductase from Shewanella massilia, O87948; AoxB_Afa, arsenite oxidase from Alcaligenes faecalis, Q7SIF4.
The closest homologs with known function to DmsB, DmsC, and DmsD of Halobacterium sp. strain NRC-1 were PsrB (40% identity) from Wolinella succinogenes and DmsC (16% identity) and DmsD (27% identity) from E. coli, respectively. Phylogenetic trees were constructed from amino acid alignments with DmsB, DmsC, DmsD, and all members of the respective cluster of orthologous genes (COG). The topology of the trees indicated that dmsB and dmsD are of archaeal evolutionary origin, while for dmsC, no clear correlation with the 16S rRNA phylogenetic tree was observed (data not shown). The small DmsE protein showed some identity (19%) with DorB from Rhodobacter capsulatus, which has been suggested to function in biogenesis of the DMSO reductase in that organism (32).
The regulator protein DmsR is predicted to harbor a helix-turn-helix DNA-binding domain in the C-terminal region, similar to that of the bop gene activator Bat (2) and of several predicted archaeal DNA-binding proteins (COG3413). The N-terminal region of DmsR is cysteine rich, comprising seven cysteines between Cys32 and Cys91. Three of these cysteines, arranged as C82-Xhydrophob-C-X7-C-(P/V), were conserved in two predicted regulators of dissimilatory nitrate reductase genes (narGH) from the related haloarchaea H. marismortui (accession number AJ429077) and H. volcanii.
DISCUSSION
The present study describes a detailed functional genomic analysis of anaerobic respiration, using DMSO and TMAO as terminal electron acceptors, in a model archaeon, Halobacterium sp. strain NRC-1. We employed in-frame gene deletion studies in combination with oligonucleotide microarray transcriptome analysis and physiological and phenotypic approaches to demonstrate that Halobacterium sp. strain NRC-1 carries a functional dms operon (dmsREABCD) as the sole system responsible for growth on either alternative electron acceptor. The dms structural genes are induced during anaerobic respiration. We further demonstrate that DMSO and TMAO respiration is under positive transcriptional control of a novel archaeal regulator, encoded by dmsR.
The DMSO reductase in strain NRC-1 is a novel member of the DMSO reductase family. Growth inhibition of DMSO or TMAO respiration by tungstate and bioinformatics analysis indicated that the catalytically active subunit, DmsA, contains molybdenum coordinated by MPT as a cofactor. Notably, DmsA of strain NRC-1 is more closely related to NarG-type dissimilatory nitrate reductases than to bacterial DMSO and TMAO reductases. Presumably, DmsA receives reducing equivalents from the iron-sulfur subunit DmsB, while both subunits are anchored to the membrane by interaction with DmsC. DmsD and possibly DmsE are involved in biogenesis in the DMSO reductase.
DmsR constitutes, along with Bat and various other archaeal proteins (e.g., a predicted regulator of narGH in H. marismortui and H. volcanii), a family of transcriptional regulators (COG3413). Members of this family harbor a Bat-like helix-turn-helix DNA-binding domain (Pfam HTH 10) in the C-terminal region of the protein. In general, transcriptional activators typically harbor their DNA-binding domain in the C-terminal region (25), rendering it likely that more or even all members of COG3413 constitute transcriptional activators. The environmental signal(s) governing expression of the dms genes is of interest. Preliminary data from transcriptome comparisons of arginine fermentation and aerobic respiration showed that the dms genes are also highly induced under anaerobic conditions in the absence of both DMSO and TMAO (J. A. Müller and S. DasSarma, unpublished data). This demonstrates that the absence of the terminal electron acceptor oxygen, and not the presence of either DMSO or TMAO, is essential for induction of the dms genes in strain NRC-1. DmsR may respond directly to changing oxygen concentrations. The protein contains a cysteine-rich region in the N-terminal region that may be involved in metal binding (e.g., of redox-active iron) or disulfide-bridge formation. Either possibility could lead to conformational changes of DmsR, affecting DNA-binding affinity and/or interactions with other transcriptional regulators, depending on the presence or absence of molecular oxygen in the cell.
The transcriptome comparison of anaerobic and aerobic respiration provides an insight into the survival strategy of Halobacterium sp. strain NRC-1 in the absence of molecular oxygen. Almost all genes required for the aerobic electron transport chain have essentially the same expression level under anaerobic conditions as under aerobic conditions. This suggests that strain NRC-1 stays primed for aerobic respiration under anaerobic conditions. The observed up-regulation of the cbaABD genes may fit this assumption. The homologous ba3-type cytochrome oxidase in T. thermophilus functions under low oxygen partial pressure (16). Assuming a similar function of the halobacterial homolog, expression of the cbaABD genes could enable the scavenging of molecular oxygen for use as an electron acceptor after a shift from anoxic to micro-oxic conditions. An alternative hypothesis is that the ba3-type oxidase of strain NRC-1 may be involved in denitrification. The T. thermophilus enzyme is bifunctional, as it can reduce both molecular oxygen and nitric oxide (NO) (12). Although strain NRC-1 appears to be unable to grow by denitrification (Müller and DasSarma, unpublished observations), the gene products of cbaABD may work together with that of VNG1187, a NirK-type nitrite reductase homolog, to facilitate auxiliary energy conservation under anaerobic conditions with nitrogen oxides as electron acceptors. This strategy would be similar to the contribution of phototrophy to the energy budget of strain NRC-1. Menaquinone is presumably used both as a mobile electron carrier during aerobic respiration (28) and as an electron donor for the DMSO/TMAO reductase. The flow of electrons (and carbon) through the citrate cycle is regulated at the level of pyruvate:ferredoxin oxidoreductase and not at each individual enzyme of the cycle. This regulatory scheme could result in a fast metabolic response to the availability of oxygen as electron acceptor. Finally, the induction of gvp genes, resulting in increased gas vesicle formation under anaerobic conditions, would enable Halobacterium sp. strain NRC-1 to float to aerobic zones in the water column (40).
Our results expand the understanding of the metabolic capabilities of Halobacterium sp. strain NRC-1 under anaerobic conditions. Previous work utilized a combination of genetic and genomic studies to establish the purple membrane regulon, which leads to coordinate synthesis of bacterio-opsin and its chromophore, retinal, to allow for light-driven proton pumping (2, 8, 40, 41). The previous work led to the identification of a novel positive regulator, Bat, which controls the expression of the bacterio-opsin (bop) gene, as well as the first and last steps in the retinal biosynthetic pathway. In addition, genes involved in arginine fermentation are up-regulated at low oxygen partial pressure (26). With those results together with our results on the dms operon, we now know of three metabolic systems which can be induced to allow energy conservation in Halobacterium sp. strain NRC-1 under conditions of limited oxygen supply in the hypersaline environment.
Supplementary Material
Acknowledgments
We thank Brian Berquist for helpful suggestions about the manuscript, as well as help with phylogenetic analysis, and Suraj Amonkar for help with microarray analysis. We also gratefully thank Kevin Sowers for providing access to an anaerobic glove box and Scott Vacha at Agilent Technologies for use of a microarray scanner.
This work was supported by NSF grant MCB0296017.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, L. A., E. McNairn, T. Luebke, R. N. Pau, and D. H. Boxer. 2000. ModE-dependent molybdate regulation of the molybdenum cofactor operon moa in Escherichia coli. J. Bacteriol. 182:7035-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baliga, N. S., S. P. Kennedy, W. V. Ng, L. Hood, and S. DasSarma. 2001. Genomic and genetic dissection of an archaeal regulon. Proc. Natl. Acad. Sci. USA 98:2521-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baliga, N. S., M. Pan, Y. A. Goo, E. C. Yi, D. R. Goodlett, K. Dimitrov, P. Shannon, R. Aebersold, W. V. Ng, and L. Hood. 2002. Coordinate regulation of energy transduction modules in Halobacterium sp. analyzed by a global systems approach. Proc. Natl. Acad. Sci. USA 99:14913-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertero, M. G., R. A. Rothery, M. Palak, C. Hou, D. Lim, F. Blasco, J. H. Weiner, and N. C. Strynadka. 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 10:681-687. [DOI] [PubMed] [Google Scholar]
- 5.Cleveland, W. S. 1979. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74:829-836. [Google Scholar]
- 6.Cotter, P. A., and R. P. Gunsalus. 1989. Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J. Bacteriol. 171:3817-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czjzek, M., J. P. Dos Santos, J. Pommier, G. Giordano, V. Méjean, and R. Haser. 1998. Crystal structure of oxidized trimethylamine N-oxide reductase from Shewanella massilia at 2.5 Å resolution. J. Mol. Biol. 284:435-447. [DOI] [PubMed] [Google Scholar]
- 8.DasSarma, S. 2004. Genome sequence of an extremely halophilic archaeon, p. 383-399. In C. M. Fraser, T. Read, and K. E. Nelson (ed.), Microbial genomes. Humana Press, Inc., Totowa, N.J.
- 9.DasSarma, S., and P. Arora. 2002. Halophiles, p. 458-466. In Encyclopedia of life sciences, vol. 8. Nature Publishing Group, London, United Kingdom.
- 10.DasSarma, S., and E. M. Fleischmann. 1995. Halophiles. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 11.Denda, K., T. Fujiwara, M. Seki, M. Yoshida, Y. Fukumori, and T. Yamanaka. 1991. Molecular cloning of the cytochrome aa3 gene from the archaeon (archaebacterium) Halobacterium halobium. Biochem. Biophys. Res. Commun. 181:316-322, [DOI] [PubMed] [Google Scholar]
- 12.Giuffre, A., G. Stubauer, P. Sarti, M. Brunori, W. G. Zumft, G. Buse, and T. Soulimane. 1999. The heme-copper oxidases of Thermus thermophilus catalyze the reduction of nitric oxide: evolutionary implications. Proc. Natl. Acad. Sci. USA 96:14718-14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartman, R., H.-D. Sickinger, and D. Oesterhelt. 1980. Anaerobic growth of halobacteria. Proc. Natl. Acad. Sci. USA 77:3821-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 15.Jourlin, C., A. Bengrine, M. Chippaux, and V. Méjean. 1996. An unorthodox sensor protein (TorS) mediates the induction of the tor structural genes in response to trimethylamine N-oxide in Escherichia coli. Mol. Microbiol. 20:1297-1306. [DOI] [PubMed] [Google Scholar]
- 16.Keightley, J. A., B. H. Zimmermann, M. W. Mather, P. Springer, A. Pastuszyn, D. M. Lawrence, and J. A. Fee. 1995. Molecular genetic and protein chemical characterization of the cytochrome ba3 from Thermus thermophilus HB8. J. Biol. Chem. 270:20345-20358. [DOI] [PubMed] [Google Scholar]
- 17.Méjean, V., C. Iobbi-Nivol, M. Lepelletier. G. Giorano, M. Chippaux, and M. C. Pascal. 1994. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol. Microbiol. 11:1169-1179. [DOI] [PubMed] [Google Scholar]
- 18.Miller, T. R., and R. Belas. 2004. Dimethylsulfoniopropionate (DMSP) metabolism by Pfiesteria-associated Roseobacter spp. Appl. Environ. Microbiol. 70:3383-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mouncey, N. J., and S. Kaplan. 1998. Redox-dependent gene regulation in Rhodobacter sphaeroides 2.4.1T: effects of dimethyl sulfoxide reductase (dor) gene expression. J. Bacteriol. 180:5612-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nunoura, T., Y. Sako, T. Wakagi, and A. Uchida. 2003. Regulation of the aerobic respiratory chain in the facultatively aerobic and hyperthermophilic archaeon Pyrobaculum oguniense. Microbiology 149:673-688. [DOI] [PubMed] [Google Scholar]
- 22.Oren, A. 1991. Anaerobic growth of halophilic archaeobacteria by reduction of fumarate. J. Gen. Microbiol. 137:1387-1390. [Google Scholar]
- 23.Oren, A., and H. G. Trüper. 1990. Anaerobic growth of halophilic archaeobacteria by reduction of dimethylsulfoxide and trimethylamine N-oxide. FEMS Microbiol. Lett. 70:33-36. [Google Scholar]
- 24.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Rueda, E., and J. Collado-Vides. 2000. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 28:1838-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruepp, A., and J. Soppa. 1996. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J. Bacteriol. 178:4942-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schäfer, G., M. Engelhard, and V. Müller. 1999. Bioenergetics of the archaea. Microbiol. Mol. Biol. Rev. 63:570-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindelin, H., C. Kisker, J. Hilton, K. V. Rajagopolan, and D. C. Rees. 1996. Crystal structure of DMSO reductase: redox-linked changes in molybdopterin coordination. Science 272:1615-1621. [DOI] [PubMed] [Google Scholar]
- 30.Schneider, F., J. Löwe, R. Huber, H. Schindelin, C. Kisker, and J. Knäblein. 1996. Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 Å resolution. J. Mol. Biol. 263:53-69. [DOI] [PubMed] [Google Scholar]
- 31.Shaw, A. L. A. Hochkoeppler, P. Bonora, D. Zannoni, G. R. Hanson, and A. G. McEwan. 1999. Characterization of DorC from Rhodobacter capsulatus, a c-type cytochrome involved in electron transfer to dimethyl sulfoxide reductase. J. Biol. Chem. 274:9911-9914. [DOI] [PubMed] [Google Scholar]
- 32.Shaw, A. L., S. Leimkuhler, W. Klipp, G. R. Hanson, and A. G. McEwan. 1999. Mutational analysis of the dimethylsulfoxide respiratory (dor) operon of Rhodobacter capsulatus. Microbiology 145:1409-1420. [DOI] [PubMed] [Google Scholar]
- 33.Simon, G., V. Méjean, C. Jourlin, M. Chippaux, and M. C. Pascal. 1994. The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of the trimethylamine N-oxide reductase genes. J. Bacteriol. 176:5601-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trieber, C. A., R. A. Rothery, and J. H. Weiner. 1996. Engineering a novel iron-sulfur cluster into the catalytic subunit of Escherichia coli dimethyl sulfoxide reductase. J. Biol. Chem. 271:4620-4626. [DOI] [PubMed] [Google Scholar]
- 36.Trieber, C. A., R. A. Rothery, and J. H. Weiner. 1996. Consequences of removal of a molybdenum ligand (DmsA-Ser-176) of Escherichia coli dimethyl sulfoxide reductase. J. Biol. Chem. 271:27339-27345. [DOI] [PubMed] [Google Scholar]
- 37.Wang, G., S. K. Kennedy, S. Fasiludeen, C. Rensing, and S. DasSarma. 2004. Arsenic resistance in Halobacterium sp. strain NRC-1 examined using an improved gene knockout system. J. Bacteriol. 186:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, X., and B. Seed. 2003. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics 7:796-802. [DOI] [PubMed] [Google Scholar]
- 39.Weiner, J. H., D. P. MacIsaac, R. E. Bishop, and T. Bilous. 1988. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdo enzyme with broad substrate specificity. J. Bacteriol. 170:1505-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, C.-F., and S. DasSarma. 1990. Transcriptional induction of purple membrane and gas vesicle synthesis in the archaebacterium Halobacterium halobium is blocked by a DNA gyrase inhibitor. J. Bacteriol. 172:4118-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, C.-F., J. M. Kim, E. Molinari, and S. DasSarma. 1996. Genetic and topological analyses of the bop promoter of Halobacterium halobium: stimulation by DNA supercoiling and non-B-DNA structure. J. Bacteriol. 178:84-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimatsu, K., T. Iwasaki, and T. Fujiwara. 2002. Sequence and electron paramagnetic resonance analyses of nitrate reductase NarGH from a denitrifying halophilic euryarchaeote Haloarcula marismortui. FEBS Lett. 516:145-150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.