Abstract
Rebeccamycin, a member of the tryptophan-derived indolocarbazole family, is produced by Lechevalieria aerocolonigenes ATCC 39243. The biosynthetic pathway that specifies biosynthesis of this important metabolite is comprised of 11 genes spanning 18 kb of DNA. A presumed early enzyme involved in elaboration of the rebeccamycin aglycone is encoded by rebO, located at the left-hand region of the reb gene cluster. The deduced protein product, RebO (51.9 kDa), is an l-amino acid oxidase (l-AAO) that has 27% identity to an l-AAO from Scomber japonicus (animal, mackerel) and is a member of the family of FAD-dependent oxidase enzymes. In order to study the biochemical properties of this key enzyme, the rebO gene was overexpressed and purified from Escherichia coli. Biochemical characterization showed that RebO is dimeric, with a molecular mass of approximately 101 kDa. Further analysis revealed that the enzyme contains a noncovalently bound FAD cofactor and is reoxidized at the expense of molecular oxygen by producing one molecule of hydrogen peroxide. Based on kinetic studies, RebO shows significant preference for 7-chloro-l-tryptophan, suggesting its likely role as the natural early pathway substrate. Furthermore, the native RebO enzyme has evident, albeit limited, flexibility as shown by bioconversion studies with unnatural substrates. This work provides the first analysis of a structural enzyme involved in construction of this important class of indolocarbazole natural products.
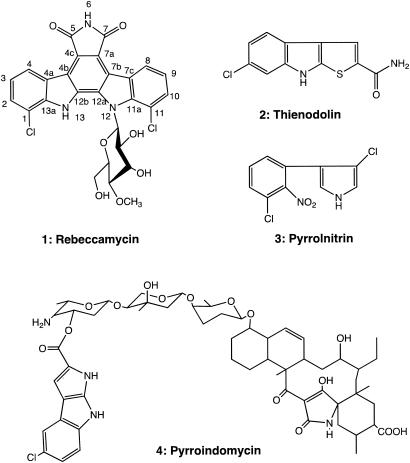
Over the past 50 years, many antibacterial, antifungal, and anticancer agents have been identified from secondary metabolites isolated from soil microorganisms. In particular, actinomycetes produce a wide variety of bioactive natural products. Based on phylogenetic analysis from 16S rRNA gene sequences (18), Lechevalieria aerocolonigenes ATCC 39243 is a member of the family Pseudonocardiaceae and is the producer of the indolocarbazole antitumor antibiotic rebeccamycin (Fig. 1) (3). The molecule contains a rare 7-chloro-tryptophan residue that is found in a small number of other natural products, including thienodolin, pyrrolnitrin, and pyrroindomycin (9, 35). Chloramphenicol is one of the best-known examples of a halogenated natural product that is also used clinically. However, its biosynthesis is unrelated to that of rebeccamycin, and there is no apparent similarity between the respective structural enzymes or halogenases (10).
FIG. 1.
Chemical structures of halogenated indole-derived natural products.
Previous studies with rebeccamycin and related analogs have shown their ability to inhibit topoisomerases I and II (22). Interest in the indolocarbazole-containing natural products, including rebeccamycin and related antibiotics K-252a and staurosporine (21), is due to their potent inhibition of protein kinase C and promising spectrum of antitumor activity (38). In some cases, a sugar moiety linked to one or both indole nitrogen atoms of the indolocarbazole core is required for biological activity (Fig. 1).
Due to the promising anticancer activity of rebeccamycin, studies were implemented to obtain a series of analogs with enhanced properties. These efforts included chemical synthesis of various tryptophan derivatives that were subsequently provided to L. aerocolonigenes by using a directed biosynthesis strategy (23). This approach provided rebeccamycin analogs with improved bioactivities (23) and allowed the compound to proceed into clinical trials to treat refractory neuroblastoma and advanced liver cancer (http://www.clinicaltrials.gov/ct/search?term = rebeccamycin).
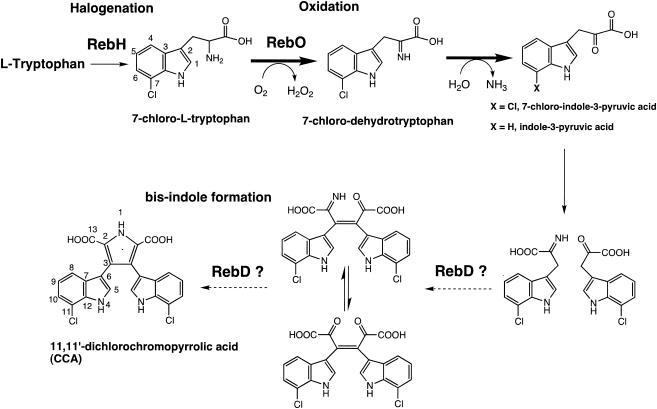
Recently, the rebeccamycin biosynthetic gene cluster has been cloned, sequenced, and characterized (32), and heterologous expression of part or the entire rebeccamycin biosynthetic gene cluster has been accomplished in Streptomyces albus and Escherichia coli (11, 32, 32a). Additionally, Onaka et al. reported disruption of several reb genes by using an E. coli-Streptomyces conjugation system, leading to accumulation of specific pathway intermediates from the resulting mutant strains (27). Furthermore, the gene cluster for staurosporine, whose core structure is very similar to rebeccamycin aglycone, has been cloned and sequenced (26). The combination of gene disruption and structural analysis of the accumulated pathway intermediates has begun to elucidate the biosynthetic order and mechanism of assembly of rebeccamycin. Thus, it is apparent that the indolocarbazole core proceeds through intermediate 11,11′-dichlorochromopyrrolic acid (CCA) (Fig. 2).
FIG. 2.
Proposed pathway for the early part of rebeccamycin biosynthesis leading to formation of CCA. The starter molecule is l-Trp. Dashed arrows indicated the presumed reaction sequence mediated by the RebD protein.
Although significant new information has been obtained from genetic studies of the reb cluster, there are no reports on the function, substrate specificity, or kinetic parameters of the enzymes that mediate rebeccamycin biosynthesis. A primary issue involves the specific identity of the aromatic amino acid residue that serves as a starter unit for the biosynthetic pathway. Moreover, there is no information about the timing of halogenation leading to modification of C-1 and C-11 on the indolocarbazole molecule. Understanding early steps of indolocarbazole biosynthesis will be important both for enhancing production of rebeccamycin by L. aerocolonigenes and for creation of analogs with improved therapeutic properties.
Herein we show that heterologous expression of RebO provides functional enzyme with l-amino acid oxidase (l-AAO) activity that requires FAD as cofactor. Based on kinetic substrate analysis of the purified protein, 7-chloro-l-tryptophan (7-chloro-l-Trp) is the preferred precursor for rebeccamycin biosynthesis. This included a detailed structure-activity analysis of RebO substrate specificity with a series of substituted tryptophan derivatives. The biochemical characterization of RebO represents an important step towards full understanding of the biosynthesis of indolocarbazole natural products.
MATERIALS AND METHODS
Amino acids and derivatives were purchased from Sigma-Aldrich. Restriction enzymes were obtained from New England Biolabs. Superdex 200 HR 10/30 was obtained from Amersham Biosciences, and Ni-nitrilotriacetic acid-agarose was from QIAGEN.
Media and culture conditions.
L. aerocolonigenes ATCC 39243 was grown on slants of yeast-malt extract agar for preproduction of rebeccamycin and for preparation of chromosomal DNA. This medium consisted of 4.0 g of glucose, 4.0 g of yeast extract, 10 g of malt extract, 1.5 g of CaCO3 and 15 g of agar in 1.0 liter of distilled water. The culture was incubated at 28 to 30°C and 250 rpm for 3 days (16, 29). E. coli DH5αMCR and JM109 were grown in liquid Luria-Bertani (LB) medium or on solid LB medium (1.5% agar) at 37°C (31). Ampicillin (100 μg/ml) and neomycin (25 μg/ml) were used for selection in E. coli cultures.
DNA isolation and manipulation.
Standard methods for DNA isolation and manipulation were performed as described by Sambrook et al. (31). DNA fragments were isolated from agarose gels by using a QIAEX II gel extraction kit (QIAGEN). Packaging was performed with Gigapack III Gold (Stratagene).
Amplification and cloning of the RebO protein corresponding to l-AAO.
The pET24b overexpression system (Novagen) was used to clone PCR products in which NdeI and HindIII sites were introduced by PCR with the following 5′-modified primers (restriction sites are underlined, and modified sequences are in italics): 5′-RebO (NdeI) (5′-TAA CAT ATG TCA CGC GGA CAC AAG AAG ATC-3′) and 3′-RebO (HindIII) (5′-TTC AAG CTT TCG TCC GTC GCC CGC CTC GAT CGC-3′). PCR using cosmid CLA14 as a template identified from an L. aerocolonigenes genomic library (constructed by partial Sau3AI digestion of chromosomal L. aerocolonigenes DNA and ligation into the BamHI sites of SuperCos I) was carried out with TaKaRa LA Taq (TaKaRa) as described by the manufacturer. Reactions were performed in a GeneAmp PCR 9700 system (Applied Biosystems). Typically, annealing was performed at 54°C for 45 s and was extended at 72°C for 1.5 min for 25 cycles. The PCR fragment containing rebO was digested with NdeI-HindIII and cloned into the NdeI-HindIII sites of pET24b, generating pDHS5514. The His6-tag fusion sites were confirmed directly by DNA sequencing. The T7 promoter primer and T7 terminator primer for sequencing were used according to the Novagen manual. Dideoxy chain termination with an Applied Biosystems automated sequencer (model 3100) was used to determine the nucleotide sequences of double-stranded template DNA fragments of the PCR products.
Overexpression and purification of the l-AAO protein.
The pDHS5514 plasmid containing PCR-amplified gene fragments of l-AAO was transformed in E. coli BL21(DE3) [pG-KEJ8] (TaKaRa) cells (25) and cultured in LB medium supplemented with kanamycin (20 μg/ml), chloramphenicol (12.5 μg/ml), and l-arabinose (0.5 to 1.0 mg/ml). Cells were grown at 28 to 30°C, and overproduction was induced with the addition of 5 mM isopropyl-β-d-thiogalactopyranoside and included 1 μg of FAD (flavin adenine dinucleotide)/ml at an A600 of 0.4 to 0.5. The culture was allowed to grow for an additional 3 to 4 h before being harvested. Purification of the His6-tagged proteins was carried out at 0 to 5°C. Cells were collected by centrifugation for 10 min at 3,000 × g and resuspended in sonication buffer (50 mM sodium phosphate [pH 7.2 at 25°C], 300 mM sodium chloride, and 10 mM imidazole [pH 8.0 at 25°C]) at 3 volumes/g of wet weight. Subsequently, lysozyme solution was added to the cell suspension (final concentration, 1 mg/ml), and freeze-thawing and sonication were carried out. After centrifugation at 13,500 × g for 25 min, the resulting supernatant was directly applied to a Ni2+-charged chelating column previously equilibrated with sonication buffer. The proteins were eluted by applying 250 mM imidazole in sonication buffer. After eluted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [4 to 20%]), the desired samples were pooled and dialyzed against assay buffer (100 mM Tris-HCl-100 mM NaH2PO4 [pH 7.6 at 25°C]-1 mM EDTA-50% glycerol) as described by Raibekas and Massey (30).
Protein determinations.
The protein concentrations were measured by using the Bradford assay kit with bovine serum albumin as a standard (Bio-Rad).
Molecular mass determination.
The relative molecular mass of the native enzyme was determined by gel filtration chromatography from its mobility relative to the mobility of protein standards under the following conditions: 50 mM NaH2PO4-150 mM NaCl (pH 7.2) buffer at a flow rate of 0.4 ml/min.
Spectroscopy.
Absorption spectra were recorded using a Beckman DU-640 spectrophotometer. The level of FAD cofactor in l-AAO was determined by absorbance changes observed following complete denaturation after heating for 5 min at 100°C in 50 mM potassium phosphate buffer, pH 7.5, containing 25 mM KCl and 3.0 M guanidine hydrochloride. To determine the flavin in denatured proteins, calculations were made using values determined from the extinction coefficient of free FAD in the presence of guanidine hydrochloride (Ε450 = 11,900 M−1 cm−1) (37).
l-AAO assay.
Activity of l-AAO was determined by the production of hydrogen peroxide, using an enzyme-coupled assay (1). Hydrogen peroxide generated by the RebO l-AAO was used by horseradish peroxidase to oxidize o-dianisidine to the radical cation, which was spectrophotometrically measured at 440-nm absorbance (A440) with a Beckman DU-640 spectrophotometer. o-Dianisidine was dissolved in 20% Triton X-100 at 5 mg/ml. Horseradish peroxidase was dissolved in 0.1 M sodium phosphate buffer, pH 7.8, at 1 mg/ml. One-milliliter assay mixtures contained 10 μl of the horseradish peroxidase solution, 100 μl of o-dianisidine solution, 20 μl of a 0.1 M l-amino acid solution, and 100 to 200 nM enzyme in 0.1 M sodium phosphate buffer, pH 7.8, and the incubation was performed at 28 to 30°C.
Chemical synthesis of 7-chloro-l-Trp.
Based on the method of Yokoyama et al. (39), an improved synthesis of 7-chloro-l-Trp was carried out as follows. 7-Chloroindole (488 mg; 3.20 mmol; 1.0 eq) and l-serine (672 mg; 6.39 mmol; 2.0 eq) suspended in 3:1 (vol/vol) glacial acetic acid-acetic anhydride were heated to 75°C. All solids dissolved within 30 min to afford a colorless solution, which gradually turned light yellow. After 2 h, the reaction mixture was cooled to room temperature and partitioned between ethyl acetate (30 ml) and saturated aqueous sodium chloride (30 ml). The organic layer was separated, and the aqueous layer was extracted with ethyl acetate (two times, 30 ml). The combined organic extracts were concentrated by rotary evaporation to provide a yellow oil. The remaining acetic acid was removed azeotropically with toluene (50 ml) to afford a yellow-white foam. Flash chromatography with 95:5:1 CHCl3-MeOH-acetic acid (AcOH) (1 liter) on silica gel (75 g) afforded 924 mg (102%) of the title compound as a yellow solid after azeotropic removal of the residual acetic acid with toluene (two times, 20 ml). The product was partially contaminated with α-acetamidoacrylic acid (thin-layer chromatography [TLC] Rf = 0.19; 95:5:1 CHCl3-MeOH-AcOH); thus, rechromatography under the same conditions afforded 745 mg (83%) of N-acetyl-7-chloro-dl-tryptophan as a white solid which was greater than 95% pure by high-performance liquid chromatography (HPLC; 210-nm detection). Recrystallization from ethyl acetate afforded 300 mg of white crystals from the first crop that was greater than 99.5% pure as assessed by HPLC. mp 201°C, lit (36). 200°C; TLC Rf = 0.10; 95:5:1 CHCl3-MeOH-AcOH; HPLC, tret = 28.7 min, Alltech C18 Econosil, 4.6 by 250 mm, 5 to 40% CH3CN-H2O plus 0.1% trifluoroacetic acid (TFA), flow rate 1 ml/min; 1H nuclear magnetic resonance (NMR; 500 MHz, acetone-d6) δ: 10.38 (br s, 1H), 7.58 (d, J = 7.9 Hz, 1H), 7.32 (ovlp br s, 1H), 7.30 (ovlp d, J = 2.2 Hz, 1H), 7.15 (d, J = 7.2 Hz, 1H), 7.03 (t, J = 7.8 Hz, 1H), 4.78 to 4.83 (m, 1H), 3.34 (dd, J = 14.8, 5.3 Hz, 1H), 3.20 (dd, J = 14.8, 7.2 Hz, 1H), 1.90 (s, 3H); 13C NMR (125 MHz, acetone-d6) δ: 173.4, 170.5, 134.3, 130.7, 125.6, 121.7, 120.6, 118.4, 117.1, 112.6, 53.8, 28.2, 22.7.
N-Acetyl-7-chloro-dl-tryptophan (505 mg; 1.80 mmol) was suspended in 0.1 M aqueous NaH2BO3 buffer (38 ml), and the pH was adjusted to 8.0 by the addition of 1.0 M aqueous NaOH. The suspension was vigorously stirred for 20 min until dissolution was complete. Then, 1.0 mM aqueous CoCl2 solution (2.0 ml) and Amano l-aminoacylase (5 mg; 150 U; 30,000 U/g) were added to afford a clear light brown solution, which was incubated for 24 h at 37°C. The solution was acidified to pH 4.0 by the addition of 1.0 M HCl. The protein was denatured by heating (100°C; 5 min) and then freezing the solution overnight at −20°C. After thawing, the precipitated protein was filtered and the supernatant was purified by ion-exchange chromatography (Sephadex strong cation-exchange resin SP-25; 10 g). The column was washed with water (400 ml), and the product was eluted with 1 M aqueous NH4OH (300 ml) to afford the (−)-7-chloro-l-Trp compound (143 mg; 89%) as a light tan solid that was greater than 99.5% pure by reverse-phase HPLC. HPLC tret = 27.6 min, Alltech C18 Econosil 4.6 by 250 mm, 5 to 40% CH3CN-H2O plus 0.1% TFA, 40 min, flow rate 1 ml/min; 1H NMR (300 MHz, CD3OD) δ: 7.55 (d, J = 7.9 Hz, 1H), 7.17 (s, 1H), 7.03 (d, J = 7.6 Hz, 1H), 6.92 (t, J = 7.7 Hz, 1H), 3.74 (dd, J = 9.2, 4.0 Hz, 1H), 3.38 (dd, J = 15.2, 4.0 Hz, 1H), 3.06 (dd, J = 15.2, 9.2 Hz, 1H); 13C NMR; ESI-(+): 239.1 (100), 240.1 (12%), 241 (28%), 242 (4%).
Synthesis of 7-chloro-indole-3-pyruvic acid (7-Cl-IPA).
Synthesized 7-chloro-l-Trp was dissolved in H2O at 100 mM (4.77g). l-AAO (from Crotalus atrox; Sigma-Aldrich) was dissolved in 0.05 M sodium phosphate buffer (NaH2PO4, pH 7.2, at 0.2 U/mg). One-milliliter reaction mixtures contained 100 μl of the 7-chloro-l-Trp solution, 10 U of the l-AAO solution, and 15 μl of the catalase solution (33,600 U/ml). The incubation was performed at 25 to 27°C for 3 h, during which the reaction mixture became pale yellow. To quench the oxidase reaction, the reaction sample was acidified to approximately pH 5 by using 1.0 N aqueous HCl, and then the solutions were filtered using Microcon centrifugal filter devices (Microcon YM-3000 MWCO; Amicon) in order to remove enzyme. 7-Chloro-indole pyruvic acid (0.9 mg; 38%) was isolated and purified from the filtrated products as a light brown solid that was >99.5% pure by reverse-phase HPLC. HPLC tret = 19.0 min, Alltech C18 Econosil 4.6 by 250 mm, 10 to 100% CH3CN-H2O plus 0.1% TFA, 30 min, flow rate 1 ml/min; 1H NMR (500 MHz, dimethyl sulfoxide-d6) δ: 11.24 (s, 1H), 7.90 (s, 1H), 7.71 (d, J = 7.9 Hz, 1H), 7.22 (d, J = 7.6 Hz, 1H), 7.08 (t, J = 7.8, 1H), 6.74 (s, 1H).
Kinetic calculations.
Km and kcat were determined using the Prism (version 4) software.
HPLC analysis of IPA.
Bioconversions were preformed from the reaction between l-Trp (final concentration, 0.2 mM) and RebO in 50 mM Tris-HCl buffer, pH 8.0, containing 100 U of catalase for 4 to 5 h at 28°C (500 μl of reaction buffer), and the reaction buffer was centrifuged using an Ultrafree-MC 10,000 centrifugal filter (Amicon) at 10,000 × g, 25°C, for 15 min. The production of indole-3-pyruvic acid (IPA) by enzymatic reaction was analyzed with HPLC in a reverse-phase column (μBondapak C18; 3.9 by 300 mm; Waters). The eluant system used was a 10-to-60% acetonitrile gradient in 0.1% TFA over 30 min, at a flow rate of 1.0 ml/min. Chromatographic identification was carried out using a Beckman Coulter System Gold 126 solvent module supplied with a System Gold 168 detector.
For the detection of 7-chloro-indole-3-pyruvic acid (7-Cl-IPA), the reaction with RebO protein was performed in 50 mM Tris-HCl buffer, pH 8.0, containing 100 U of catalase for 3 to 4 h at 28°C (1 ml of reaction buffer). The reaction buffer was converted to pH 3.0 to 4.0 with 1.0 N aqueous HCl and the reaction was stopped. The 7-Cl-IPA generated by enzymatic reaction was extracted with an equal volume of ethyl acetate and repeated three times. The organic layer was isolated and evaporated to dryness under reduced pressure. The residue was dissolved in 200 μl of methanol and analyzed with HPLC in a reversed-phase column [C18 (218TP); 4.6 by 250 mm; Vydac]. The eluant system used was a 0-to-90% acetonitrile gradient in 0.1% acetic acid-0.02% TFA over 30 min, at a flow rate of 1.0 ml/min for conversion of 7-Cl-IPA. The detector wavelengths were set at 275 and 310 nm. 7-Cl-IPA products were analyzed by LC-mass spectrometry (LC-MS) using Thermo Finnigan Surveyor and Finnigan MAT LCQ systems.
Nucleotide sequence accession number.
The nucleotide sequences of rebeccamycin biosynthetic genes have been submitted to DDBJ under accession number AB090952.
RESULTS
Homology analysis and characterization of the deduced amino acid sequence.
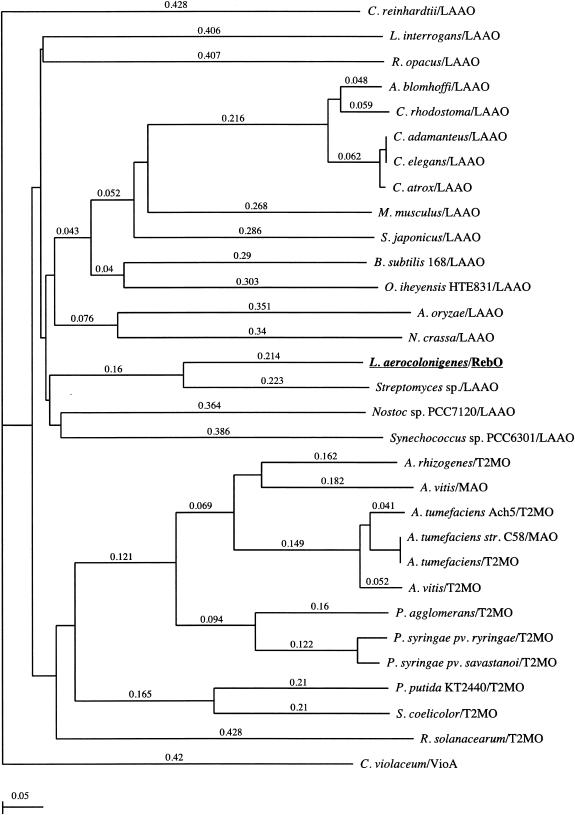
According to the deduced amino acid sequences and comparison of databases, RebO has 27% sequence identity to l-AAO of Scomber japonicus. l-AAO enzymes occur widely and have been studied from a number of different sources, such as bacteria, fungi, and algae (4, 6-8, 14, 24) as well as from mouse milk, snake venom, and snail (5, 19, 20, 28, 33). Based on an amino acid alignment analysis among several l-AAOs and monooxygenases, RebO showed a conserved N-terminal βαβ-dinucleotide-binding motif, G-X-G-X-X-G-X-X-X-[G/A], which is a typical FAD binding site for amine oxidases of this class, and a GG motif. This dinucleotide binding motif and the GG doublet appear in several families of flavoproteins (34). The phylogenetic relationship between RebO, related l-AAOs, and monooxygenases was analyzed by neighbor joining using ClustalW (version 1.4). The data showed that all l-AAOs are tightly clustered and that RebO resides firmly within bacterial l-AAOs in the l-AAO family (Fig. 3).
FIG. 3.
Phylogenetic tree representation of the apparent evolutionary distances of l-AAO (LAAO) proteins from several bacteria, fungi, and venomous snakes. Agkistroden blomhoffi LAAO, AB072392; Aspergillus oryzae LAAO, AB078782; Bacillus subtilis 168 LAAO, Z99114; Crotalus adamanteus LAAO, JE0266; Crotalus atrox LAAO, AF093248; Caenorhabditis elegans LAAO, NM_077599; Chlamydomonas reinhardtii LAAO, T08202; Calloselasma rhodostoma LAAO, P81382; Chromobacterium violaceum VioA, AF172851 and AB032799; Leptospira interrogans LAAO,AE011573; Mus musculus LAAO, BC017599 and NP_598653; Neurospora crassa LAAO, ABBX01000266; Nostoc sp. strain PCC7120 LAAO, NC_003276; Oceanobacillus iheyensis HTE831 LAAO, AP004597; Rhodococcus opacus LAAO, AY053450; S. japonicus LAAO, AJ400871; Streptomyces sp. LAAO, AB088119; Synechococcus sp. strain PCC6301 LAAO, Z48565; Agrobacterium tumefaciens strain C58 MAO, NC_003065; Agrobacterium vitis MAO, AF126447; Agrobacterium rhizogenes T2MO, Q09109; A. tumefaciens Ach5 T2MO, P04029; A. tumefaciens T2MO, NC_003308; A. vitis T2MO, P25017; Pantoea agglomerans T2MO, Q47861; Pseudomonas putida KT2440 T2MO, NC_002947; Pseudomonas syringae pv. syringae T2MO, A53376; P. syringae pv. savastanoi T2MO, A25493; Ralstonia solanacearum T2MO, NC_003295; S. coelicolor T2MO, AL939109. The tree was constructed by the neighbor-joining method using ClustalW (version 1.4). The scale for the tree indicates inferred evolutionary distances. MAO, monoamine oxidase; T2MO, tryptophan 2-monooxygenase.
Expression and purification of RebO protein in E. coli.
In order to obtain adequate amounts of protein for biochemical analysis, we sought to overproduce l-AAO of RebO in E. coli by using an expression vector (pDHS5514) containing rebO. A KLAAALEHHHHHH amino acid sequence was engineered onto the C terminus of RebO to facilitate subsequent purification by a Ni-nitrilotriacetic acid strategy. The plasmid pDHS5514 was transformed into E. coli BL21(DE3) and E. coli BL21(DE3)[pG-KJE8], respectively. The coexpressed plasmid pG-KJE8 was used to facilitate protein folding and to prevent aggregation and degradation of RebO in E. coli BL21(DE3) (25). The E. coli BL21(DE3)[pG-JKE8] system was found to be essential in order to obtain RebO as a soluble, functional enzyme. Recombinant RebO protein estimated to be >95% pure was observed by SDS-PAGE analysis (data not shown). By contrast, poor rebO overexpression was observed with the E. coli BL21(DE3) strain.
Purification of RebO from crude extracts of E. coli BL21(DE3)[pG-KJE8] cells yielded a protein with an apparent molecular mass on SDS-PAGE of approximately 55 to 56 kDa (data not shown). In the purification step, it was observed that overproduced RebO was slightly larger than the predicted molecular mass for the recombinant RebO protein (53,412.8 Da). The amino acid sequences of RebO showed an abundance of hydrophilic and nonpolar amino acid residues (42.6%, except His6 tag sequence). This phenomenon has been reported for other proteins, such as the principal sigma factors in E. coli and cyanobacteria (2, 12). To measure more precisely the molecular mass of the RebO protein, size exclusion chromatography was performed. Gel permeation chromatography on a Superdex 200 column of purified proteins indicated that RebO forms dimers with a molecular mass of ∼101 kDa (data not shown). The logarithm for molecular masses of standard proteins was plotted against a volume of elution with a linear profile (R2 = 0.96).
Determination of the RebO cofactor.
Since other l-AAOs are known to use FAD as a cofactor (20), we evaluated the RebO flavin content by UV spectroscopy. To characterize and identify the RebO cofactor, an absorption spectrum of the enzyme was obtained that displayed a characteristic flavoprotein pattern with maxima at 385 and 460 nm and a shoulder at ∼480 nm (data not shown). The absorbance of the denatured enzymes at 450 nm in 3 M guanidine hydrochloride was used to determine the noncovalently bound flavin, as detailed under Materials and Methods. On this basis, RebO was found to contain noncovalently bound flavin of 0.58 ± 0.05 mol per mol of protein. RebO protein that was overexpressed without supplementation of FAD in E. coli BL21(DE3) contained less than 0.5 mol of noncovalently bound flavin per mol of enzyme. To determine whether other cofactors are required for RebO function, NAD and pyridoxal phosphate were added to the reaction mixture with l-Trp. However, addition of these cofactors had no effect on enzyme activity (data not shown).
Substrate tolerance of the RebO protein.
Typically, l-AAOs (EC 1.4.3.2) are flavoenzymes that catalyze the oxidative deamination of l-amino acids to the corresponding α-imino acids, which are subsequently hydrolyzed to α-keto acids. The activity of RebO was characterized by analysis of various amino acid substrates. Moreover, as the marker of the l-AAO activity, the time-dependent production of H2O2 was measured. Remarkably, in a survey of the 20 natural l-amino acids, RebO showed high oxidase activity only against l-Trp (data not shown). Since d-Trp was not a substrate for RebO, as shown by lack of H2O2 production, the results showed clearly that RebO is specific for the l-amino acid enantiomer.
Enzymatic activity of RebO toward tryptophan derivatives.
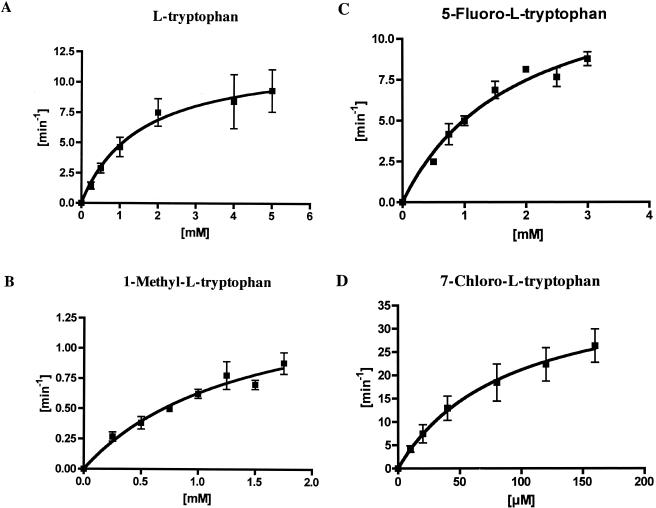
Substrate specificity was investigated by comparing the rate of conversion of selected tryptophan analogs under standard l-AAO reaction conditions (see Materials and Methods). Using recombinant RebO protein, oxidase activity was evaluated using a variety of tryptophan analogs. 1-Methyl-l-tryptophan (1-methyl-l-Trp), 5-methyl-dl-tryptophan (5-methyl-dl-Trp) and 5-fluoro-l-tryptophan (5-fluoro-l-Trp) were found to be substrates for RebO based on production of H2O2. Significantly, 7-chloro-l-Trp was found to be the preferred substrate based on the ability of RebO to convert O2 to H2O2 in the assay system. In contrast, two substrates with modified side chains, l-tryptophanamide and N-acetyl-tryptophan, were not accepted as substrates by RebO. These data indicate that both carboxyl and amino groups are essential for substrate recognition by the enzyme. The kinetic parameters of the reactions were obtained using l-Trp, 1-methyl-l-Trp, 5-fluoro-l-Trp, and 7-chloro-l-Trp (Fig. 4 and Table 1). The highest production of H2O2 was observed in the presence of 7-chloro-l-Trp as substrate. The specific activity profile of RebO showed that the kcat/Km of 7-chloro-l-Trp is 57-fold greater than that of l-Trp. To calculate Km and kcat values, the concentrations of l-Trp, 1-methyl-l-Trp, 5-fluoro-l-Trp, and 7-chloro-l-Trp were analyzed against the production of H2O2 per min according to Michaelis-Menten plots, with nonlinear best fit. The rate of production of H2O2 per minute was plotted against concentration, giving a linear plot (r > 0.9). Comparison of Km and kcat values for 7-chloro-l-Trp against l-Trp and the corresponding Trp analogs provided compelling evidence that 7-chloro-l-Trp is the key monomer species for rebeccamycin biosynthesis.
FIG. 4.
Determination of steady-state kinetic parameters of various substrates for RebO. Experimental conditions are given in Materials and Methods. The x axis is the substrate concentration, and the y axis is the initial velocity of hydrogen peroxide production. The total enzyme concentration was 100 nM in a reaction buffer. Error bars indicate standard deviations.
TABLE 1.
Substrate specificity of recombinant RebO enzyme
| Substrate | Km (mM) | kcat (min−1) | kcat/Km |
|---|---|---|---|
| l-Tryptophan | 1.53 ± 0.61 | 12.12 ± 0.61 | 7.94 |
| 1-Methyl-l-tryptophan | 1.43 ± 0.52 | 1.53 ± 0.30 | 1.07 |
| 5-Fluoro-l-tryptophan | 1.84 ± 0.42 | 14.32 ± 1.62 | 7.80 |
| 7-Chloro-l-tryptophan | 0.088 ± 0.036 | 39.82 ± 7.67 | 453 |
Identification of reaction products.
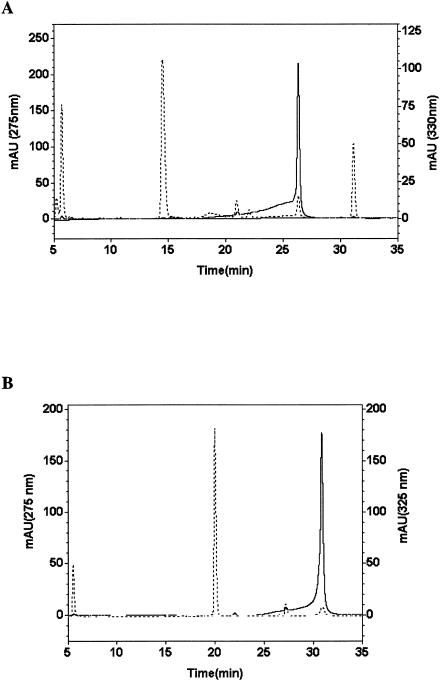
In an effort to determine the identity of early intermediates in rebeccamycin biosynthesis, we proceeded to identify RebO reaction products by using reverse-phase chromatography. l-Trp was used as substrate and for ease of detection and for correlation with IPA, the predicted reaction product of the RebO bioconversion. Following 4 h of incubation, conversion of l-Trp yielded two resolved peaks when using an absorbance from 200 to 400 nm (Fig. 5A). The first major peak represented nonconverted substrate (l-Trp; retention time, 14.51 min; λmax = 275 nm), whereas the second major peak (retention time, 26.33 min), characterized by an absorption band at 330 nm, was analyzed further (Fig. 5A). As judged from the HPLC analysis (retention time, 26.18 min; λmax = 330 nm [data not shown]), the second major peak was confirmed by coinjection with an authentic standard as IPA (Fig. 2). Moreover, UV spectrometry results were coincident with authentic IPA and the compound obtained from bioconversion (λmax = 232 and 330 nm). In order to determine the yield of 7-Cl-IPA in the reaction by RebO with 7-chloro-l-Trp, bioconversion of 7-chloro-l-Trp was carried out. 7-Cl-IPA was obtained as the product, as determined by LC-MS (retention time, 20.66 min; λ = 310 nm; electrospray ionization-MS (ESI-MS) [M+H]+, m/z 238.0) and the observed isotope pattern of the chlorine atom in 7-Cl-IPA (ESI-MS [M+H]+, m/z 238.0, 239.1, 240.0). The 7-Cl-IPA absorption band at λmax of 232 and 324 to ∼325 nm (retention time, 30.88 min; λmax = 325 nm) (Fig. 5B) and UV spectrometry profile were identical with those of authentic 7-Cl-IPA (data not shown). Furthermore, the major peak was confirmed by coinjection with a 7-Cl-IPA authentic standard (see Materials and Methods) (Fig. 2).
FIG. 5.
Chromatogram of HPLC for direct measurement of product with RebO protein. (A) Analysis of the reaction of L-Trp with RebO. The elution was detected at 275 nm (- - -) and 330 nm (—). (B) Analysis of the reaction of 7-chloro-L-Trp with RebO. The elution was followed at 275 nm (- - -) and 325 nm (—). Experimental conditions are given in Materials and Methods. In vitro reactions were performed with RebO enzyme (1.0 μM). The x axis is the retention time (in minutes), and the y axis is the relative absorbance level (photodiode array detector).
DISCUSSION
This study represents the first characterization of an enzyme from the rebeccamycin biosynthetic pathway. Previous work that involved in vivo incorporation of l-Trp derivatives demonstrated that there is inherent substrate tolerance in rebeccamycin structural enzymes (16). However, these studies provided no specific information on the key components involved in starter unit choice in the rebeccamycin system. Understanding control points for creation of new secondary metabolites can have a significant impact on developing effective strategies to expand chemical diversity through metabolic engineering. In this study, we focused on the RebO enzyme, due to its likely pivotal role in controlling product diversity of the indolocarbazole scaffold. The results demonstrated that 7-chloro-l-Trp is oxidized more efficiently than l-Trp by in vitro reaction of RebO in a coupled oxidase activity assay, and the results support the idea that 7-chloro-l-Trp serves as a preferred substrate over l-Trp. Accordingly, this finding implies that conversion of l-Trp to 7-chloro-l-Trp is the likely first step in rebeccamycin biosynthesis and serves to supply a unique monomer precursor pool. The creation of a specific precursor pool might be advantageous, since it provides a mechanism for the organism to avoid competition of carbon flux between primary and secondary metabolic processes.
Several previous studies involving cloning, analysis, and heterologous expression of the rebeccamycin gene cluster have provided important information on the specific genes and putative functional roles for individual enzymes involved in its biosynthesis (11, 27, 32). However, despite this progress, important details about enzyme function, specificity, and control have awaited detailed analysis. Due to the combination of precursor feeding studies that showed the ability to incorporate nonnatural substrates (17), along with ambiguous information on the specific identity of the starter unit monomer species (29), we decided to investigate the role of early enzymes in control of precursor formation in rebeccamycin biosynthesis.
Although heterologous expression of all or part of the rebeccamycin gene cluster had been accomplished for Streptomyces and E. coli (11, 32), and several gene disruption experiments had led to identification of key pathway intermediates (27), ambiguity remained regarding l-Trp or 7-chloro-l-Trp as the starting monomer unit. Sánchez et al. (32, 32a) proposed that halogenation of tryptophan occurs during an early step in rebeccamycin production before the oxidation of tryptophan. They reported isolation of deschloro-rebeccamycin and deschloro-4′-demethylrebeccamycin from heterologous expression studies in S. albus containing a plasmid bearing rebGODCPMRFU. Significantly, this plasmid did not include rebH, encoding the presumed halogenase, thus explaining production of the nonchlorinated derivatives of the natural product. Since this initial study did not include heterologous expression of the complete set of rebeccamycin pathway genes, it was not possible to compare production levels of rebeccamycin relative to those of 1,11-deschloro-rebeccamycin (32). These studies indicated that halogenation is not required to generate early or advanced intermediates in the rebeccamycin pathway, which is consistent with an overall substrate tolerance of the metabolic enzymes.
A subsequent study involving analysis of accumulated intermediates from mutant strains of L. aerocolonigenes provided further insight into substrate tolerance of rebeccamycin biosynthetic enzymes. Specifically, disruption of the rebH gene resulted in exclusive production of the 1,11-deschloro-4′-demethylrebeccamycin derivative (27). However, a key question that remained from these studies was the point at which halogenation occurs. Clearly, advanced intermediates can be generated in the absence of a chlorine atom at C-7 of tryptophan. However, the lack of direct comparisons of levels of chlorinated versus nonchlorinated compounds precluded further insight on the likely identity of the monomeric substrate. Currently, a comparative analysis of production levels regarding deschloro-rebeccamycin from an L. aerocolonigenes rebH mutant and rebeccamycin from the wild-type strain (27) has not been performed; however, such an analysis might provide some insight into metabolite production efficiency in the absence of the 7-chloro-l-Trp substrate. Furthermore, we presume that the preference for one monomeric species over another would lead to higher levels of the corresponding intermediate (e.g., a 7-chloro-l-Trp preference would provide higher levels of rebeccamycin than corresponding deschloro-rebeccamycin levels from l-Trp monomer) due to higher conversion rates of the corresponding biosynthetic enzymes.
The potential for broad application of the rebeccamycin gene cluster was demonstrated recently by heterologous expression in E. coli (11). In this work, isolation of rebeccamycin aglycone provided additional evidence that halogenation likely occurs prior to completion of aglycone ring closure or during the early stages of the first-ring synthesis.
Interestingly, early precursor incorporation experiments that included feeding 5-fluoro-dl-Trp and 6-fluoro-dl-Trp to the fermentation culture of the wild-type L. aerocolonigenes strain resulted in isolation of a novel indolocarbazole, fluoroindolocarbazole A, B, and C (17). This synthesis study demonstrated that no further halogenation (e.g., chlorination) occurred to 5- and 6-fluoro-dl-Trp in vivo. According to the RebO in vitro kinetic analysis described above, the unnatural substrate 5-fluoro-l-Trp undergoes oxidation at a level that is very similar to that of l-Trp. Taken together, these data suggest that RebH prefers l-Trp or IPA as a substrate during rebeccamycin biosynthesis.
Perhaps some of the most compelling information regarding the exact nature of the Trp monomer incorporation into rebeccamycin can be gleaned by analogy from reports on initial steps in pyrrolnitrin biosynthesis (9, 13, 15). In this case, a gene encoding a chloro-halogenase (prnA) in the pyrrolnitrin cluster (prn) from Pseudomonas fluorescens shows 52% amino acid sequence identity with RebH. Kirner et al. demonstrated that l-Trp is the substrate providing 7-chloro-l-Trp, which serves as the starter unit in the pyrrolnitrin pathway (15).
The objective of our present study was to isolate and obtain a detailed characterization of RebO, a key early pathway enzyme in rebeccamycin biosynthesis. Through analysis of its substrate specificity and direct comparison of 7-chloro-l-Trp and l-Trp, and related intermediates, a clear role for these monomers can be established in this system. Indeed, our results suggested that 7-chloro-l-Trp is the preferred monomer substrate for rebeccamycin biosynthesis, thus supporting the proposed pathway that halogenation occurs before oxidation of l-Trp (Fig. 2). l-Trp clearly plays an important part in primary metabolism of L. aerocolonigenes. Since RebO appears to prefer 7-chloro-l-Trp over l-Trp pools, it has created an effective branch in which to control the overall burden of secondary metabolic processes on primary metabolism.
Acknowledgments
We thank Leng Chee Chang and Eranda Jayamaha for helpful discussions and are grateful to Chizuko Nishizawa-Harada for technical assistance. David Ballou and Barrie Entsch are gratefully acknowledged for helpful comments on the manuscript.
This study was supported by National Institutes of Health grant U01 AI48521.
REFERENCES
- 1.Avrameas, S., and B. Guilbart. 1972. Enzyme-immunoassay for the measurement of antigens using peroxidase conjugates. Biochimie 54:837-842. [DOI] [PubMed] [Google Scholar]
- 2.Burgess, R. R., and J. J. Jendrisak. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634-4638. [DOI] [PubMed] [Google Scholar]
- 3.Bush, J. A., B. H. Long, J. J. Catino, W. T. Bradner, and K. Tomita. 1987. Production and biological activity of rebeccamycin, a novel antitumor agent. J. Antibiot. XL:668-678. [DOI] [PubMed] [Google Scholar]
- 4.Coudert, M. 1975. Characterization and physiological function of a soluble L-amino acid oxidase in Corynebacterium. Arch. Microbiol. 102:151-153. [DOI] [PubMed] [Google Scholar]
- 5.Ehara, T., S. Kitajima, M. Kanazawa, T. Tamiya, and T. Tsuchiya. 2002. Antimicrobial action of achacin is mediated by l-amino acid oxidase activity. FEBS Lett. 531:509-512. [DOI] [PubMed] [Google Scholar]
- 6.Furuya, Y., H. Sawada, T. Hirahara, K. Ito, T. Ohshiro, and Y. Izumi. 2000. A novel enzyme, l-tryptophan oxidase, from a basidiomycete Coprinus sp. SF-1: purification and characterization. Biosci. Biotechnol. Biochem. 64:1486-1493. [DOI] [PubMed] [Google Scholar]
- 7.Genet, R., C. Denoyelle, and A. Ménez. 1994. Purification and partial characterization of an amino acid α,β-dehydrogenase, l-tryptophan 2′,3′-oxdase from Chromobacterium violaceum. J. Biol. Chem. 269:18177-18184. [PubMed] [Google Scholar]
- 8.Geueke, B., and W. Hummel. 2003. Heterologous expression of Rhodococcus opacus L-amino acid oxidase in Streptomyces lividans. Protein Express. Purif. 28:303-309. [DOI] [PubMed] [Google Scholar]
- 9.Hammer, P. E., D. S. Hill, S. T. Lam, K.-H. van Pée, and J. M. Ligon. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Micobiol. 63:2147-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, J., N. Magarvey, M. Piraee, and L. C. Vining. 2001. The gene cluster for chloramphenicol biosynthesis in Streptomyces venezuelae ISP5230 includes novel shikimate pathway homologues and a monomodular non-ribosomal peptide synthetase gene. Microbiology 147:2817-2829. [DOI] [PubMed] [Google Scholar]
- 11.Hyun, C.-G., T. Bililign, J. Lia, and J. S. Thorson. 2003. The biosynthesis of indolocarbazoles in a heterologous E. coli host. ChemBioChem. 1:114-117. [DOI] [PubMed] [Google Scholar]
- 12.Imamura, S., S. Yoshihara, S. Nakano, N. Shiozaki, A. Yamada, K. Tanaka, H. Takahashi, M. Asayama, and M. Shirai. 2003. Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J. Mol. Biol. 325:857-872. [DOI] [PubMed] [Google Scholar]
- 13.Keller, S., T. Wage, K. Hohaus, M. Hölzer, E. Eichhorn, and K.-H. van Pée. 2000. Purification and partial characterization of tryptophan 7-halogenase (PrnA) from Pseudomonas fluorescens. Angew Chem. Int. 39:2300-2302. [DOI] [PubMed] [Google Scholar]
- 14.Khanna, P., and M. S. Jorns. 2001. Characterization of the FAD-containing N-methyltryptophan oxidase from Escherichia coli. Biochemistry 40:1441-1450. [DOI] [PubMed] [Google Scholar]
- 15.Kirner, S., P. E. Hammer, D. S. Hill, A. Altmann, L. Fischer, L. J. Weislo, M. Lanahan, K.-H. van Pée, and J. M. Ligon. 1998. Functions encoded by pyrrolinitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam, K. S., S. Forenza, T. W. Doyle, and C. J. Pearce. 1990. Identification of indolepyruvic acid as an intermediate of rebeccamycin biosynthesis. J. Ind. Microbiol. 6:291-294. [DOI] [PubMed] [Google Scholar]
- 17.Lam, K. S., D. R. Schroeder, J. M. Veitch, K. L. Colson, J. A. Matson, W. C. Rose, T. W. Doyle, and C. J. Pearce. 2001. Production, isolation and structure determination of novel fluoroindolocarbazoles from Saccharothrix aerocolonigenes ATCC 39243. J. Antibiot. 54:1-9. [DOI] [PubMed] [Google Scholar]
- 18.Lebeda, D. P., K. Hatano, R. M. Kroppenstedt, and T. Tamura. 2001. Revival of the genus Lentzea and proposal for Lechevalieria gen. nov. Int. J. Syst. Evol. Microbiol. 51:1045-1050. [DOI] [PubMed] [Google Scholar]
- 19.Macheroux, P., O. Seth, C. Bollschweiler, M. Schwaarz, M. Kurfürst, L.-C. Au, and S. Ghisla. 2001. l-Amino acid oxidase from the Malayan pit viper Calloselasma rhodostoma. Eur. J. Biochem. 268:1679-1686. [PubMed] [Google Scholar]
- 20.Massey, V., and B. Curti. 1967. On the reaction mechanism of Crotalus adamanteus l-amino acid oxidase. J. Biol. Chem. 242:1256-1264. [PubMed] [Google Scholar]
- 21.Meksuriyen, D., and G. A. Cordell. 1988. Biosynthesis of staurosporine. 2. Incorporation of tryptophan. J. Nat. Prod. 51:893-899. [DOI] [PubMed] [Google Scholar]
- 22.Moreau, P., F. Anizon, M. Sancelme, M. Prudhomme, D. Sevère, J.-F. Riou, J.-F. Goossens, J.-P. Héichart, C. Bailly, E. Labourier, J. Tazzi, D. Fabbro, T. Meyer, and A. M. Aubertin. 1999. Synthesis, mode of action, and biological activities of rebeccamycin bromo derivatives. J. Med. Chem. 42:1816-1822. [DOI] [PubMed] [Google Scholar]
- 23.Moreau, P., N. Gaillard, C. Marminon, F. Anizon, N. Dias, B. Baldeyron, C. Bailly, A. Pierreé, J. Hickman, B. Pfeiffer, P. Renard, and M. Prudhomme. 2003. Semi-synthesis, topoisomerase I and kinases inhibitory properties and antiproliferative activities of new rebeccamycin derivatives. Bioorg. Med. Chem. 11:4871-4879. [DOI] [PubMed] [Google Scholar]
- 24.Niedermann, D. M., and K. Lerch. 1990. Molecular cloning of the l-amino acid oxidase gene from Neurospora crassa. J. Biol. Chem. 265:17246-17251. [PubMed] [Google Scholar]
- 25.Nishihara, K., M. Kanemori, H. Yanagi, and T. Yura. 2000. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 66:884-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onaka, H., S. Taniguchi, Y. Igarashi, and T. Furumai. 2002. Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP-A0274 and its heterologous expression in Streptomyces lividans. J. Antibiot. 55:1063-1071. [DOI] [PubMed] [Google Scholar]
- 27.Onaka, H., S. Taniguchi, Y. Igarashi, and T. Furumai. 2003. Characterization of the biosynthetic gene cluster of rebeccamycin from Lechevalieria aerocolonigenes ATCC 39243. Biosci. Biotechnol. Biochem. 67:127-138. [DOI] [PubMed] [Google Scholar]
- 28.Pawelek, P. D., J. Cheah, R. Coulombe, P. Macheroux, S. Ghisla, and A. Vrielink. 2000. The structure of l-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 19:4204-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce, C. J., T. Doyle, S. Forenza, K. S. Lam, and D. R. Schroeder. 1988. The biosynthetic origins of rebeccamycin. J. Nat. Prod. 51:937-940. [DOI] [PubMed] [Google Scholar]
- 30.Raibekas, A. A., and V. Massey. 1996. Glycerol-induced development of catalytically active conformation of Crotalus adamanteus l-amino acid oxidase in vitro. Proc. Natl. Acad. Sci. USA 93:7546-7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sánchez, C., I. A. Butovich, A. F. Braña, J. Rohr, C. Méndez, and J. A. Salas. 2002. The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarazole deribatives. Chem. Biol. 9:519-531. [DOI] [PubMed] [Google Scholar]
- 32a.Sánchez, C., L. Zhu, A. F. Braña, A. P. Salas, J. Rohr, C. Méndez, and J. A. Salas. 2005. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc. Natl. Acad. Sci. USA 102:461-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, Y., E. Nonobe, Y. Kobayashi, T. Kuraishi, F. Aoki, K. Yamamoto, and S. Sakai. 2002. Characterization and expression of l-amino acid oxidase of mouse milk. J. Biol. Chem. 277:19080-19086. [DOI] [PubMed] [Google Scholar]
- 34.Vallon, O. 2000. New sequence motifs in flavoproteins: evidence for common ancestry and tools to predict structure. Proteins 38:95-114. [DOI] [PubMed] [Google Scholar]
- 35.van Pée, K.-H., and M. Hölzer. 1999. Specific enzymatic chlorination of tryptophan and tryptophan derivatives, p. 603-609. In G. Huether, W. Kochen, T. J. Simat, and H. Steinhart (ed.), Tryptophan, serotonin, and melatonin: basic aspects and applications. Kluwer Academic/Plenum Publishers, New York, N.Y. [DOI] [PubMed]
- 36.van Pée, K.-H., O. Salcher, and F. Lingens. 1981. Synthese von 7-chlor-l- and 7-chloro-d-tryptophan; Biosynthese von pyrrolnitrin. Liebigs Ann. Chem. 1981:233-239. [Google Scholar]
- 37.Wanger, M. A., P. Kahanna, and M. S. Jorns. 1999. Structure of the flavoenzyme of two homologous amine oxidases: monomeric sarcosine oxidase and N-methyltryptophan oxidase. Biochemistry 38:5588-5595. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita, Y., N. Fujii, C. Murakata, T. Ashizawa, M. Okabe, and H. Nakano. 1992. Induction of mammalian DNA topoisomerase I mediated DNA cleavage by antitumor indolocarbazole derivatives. Biochemistry 31:12069-12075. [DOI] [PubMed] [Google Scholar]
- 39.Yokoyama, Y., H. Hikawa, M. Mitsuhashi, A. Uyama, and Y. Murakami. 1999. Syntheses without protection: a three-step synthesis of optically active clavicipitic acid by utilizing biomimetic synthesis of 4-bromotryptophan. Tetrahedron Lett. 40:17803-17806. [Google Scholar]