Abstract
Carbamoyl phosphate is a precursor for both arginine and pyrimidine biosynthesis. In Lactobacillus plantarum, carbamoyl phosphate is synthesized from glutamine, ATP, and carbon dioxide by two sets of identified genes encoding carbamoyl phosphate synthase (CPS). The expression of the carAB operon (encoding CPS-A) responds to arginine availability, whereas pyrAaAb (encoding CPS-P) is part of the pyrR1BCAaAbDFE operon coding for the de novo pyrimidine pathway repressed by exogenous uracil. The pyr operon is regulated by transcription attenuation mediated by a trans-acting repressor that binds to the pyr mRNA attenuation site in response to intracellular UMP/phosphoribosyl pyrophosphate pools. Intracellular pyrimidine triphosphate nucleoside pools were lower in mutant FB335 (carAB deletion) harboring only CPS-P than in the wild-type strain harboring both CPS-A and CPS-P. Thus, CPS-P activity is the limiting step in pyrimidine synthesis. FB335 is unable to grow in the presence of uracil due to a lack of sufficient carbamoyl phosphate required for arginine biosynthesis. Forty independent spontaneous FB335-derived mutants that have lost regulation of the pyr operon were readily obtained by their ability to grow in the presence of uracil and absence of arginine; 26 harbored mutations in the pyrR1-pyrB loci. One was a prototroph with a deletion of both pyrR1 and the transcription attenuation site that resulted in large amounts of excreted pyrimidine nucleotides and increased intracellular UTP and CTP pools compared to wild-type levels. Low pyrimidine-independent expression of the pyr operon was obtained by antiterminator site-directed mutagenesis. The resulting AE1023 strain had reduced UTP and CTP pools and had the phenotype of a high-CO2-requiring auxotroph, since it was able to synthesize sufficient arginine and pyrimidines only in CO2-enriched air. Therefore, growth inhibition without CO2 enrichment may be due to low carbamoyl phosphate pools from lack of CPS activity.
In all organisms, pyrimidine nucleotides may be synthesized either de novo from bicarbonate and intermediates of central metabolism or via salvage of preformed pyrimidine bases and nucleosides present in the surrounding medium (17). When no exogenous pyrimidines are present in the culture, most organisms can perform de novo pyrimidine synthesis and produce UMP (Fig. 1). Bacteria living on rich medium that provide nucleotide precursors may lose their ability to synthesize them. Lactic acid bacteria are fastidious, and at least one third of natural isolates of Lactobacillus plantarum are high-CO2-requiring auxotrophs for pyrimidine nucleotides and arginine (5).
FIG. 1.
Simplified pyrimidine biosynthesis and link with arginine biosynthesis in an L. plantarum ΔcarAB mutant. Carbamoyl phosphate (CP) is a common precursor in arginine and pyrimidine biosynthesis. The wild-type strain CCM 1904 has two functional carbamoyl phosphate synthases (CPSs) (19). The carAB operon codes for the two subunits of the arginine-repressed CPS, CPS-A. The pyrAaAb genes present on the pyr operon (6) code for the pyrimidine-regulated CPS, CPS-P (19). Exogenous uracil enters the cell and is metabolized to UMP, which negatively controls the pyr operon, including the pyrAaAb genes. The carAB genes have been deleted in strain FB335, so that CPS-P is the only source of carbamoyl phosphate for pyrimidine and arginine biosynthesis (19). In the presence of added uracil, FB335 is unable to grow for lack of carbamoyl phosphate for arginine synthesis. For this reason, FB335 is sensitive to uracil (Uras).
Carbon dioxide hydrates into bicarbonate, which is a substrate for carbamoyl phosphate synthesis catalyzed by the carbamoyl phosphate synthase (CPS). Carbamoyl phosphate is a precursor of both pyrimidine nucleotide and arginine synthesis. In L. plantarum, carbamoyl phosphate is synthesized by two CPSs, CPS-P and CPS-A, encoded by pyrAaAb and carAB, respectively. The expression of pyrAaAb and carAB is subject to transcriptional regulation. Whereas pyrAaAb is repressed by pyrimidines, carAB responds to the arginine level. Carbamoyl phosphate is produced mainly from the pyrimidine-regulated CPS-P and not from the arginine-regulated CPS-A in wild-type L. plantarum when cultivated in ordinary air (without CO2 enrichment) as demonstrated with pyrAaAb and carAB mutants (19). High-CO2-requiring auxotrophs may have limited carbamoyl phosphate availability resulting from altered or deregulated pyrimidine-related genes (5). As a first step to identifying the genetic lesions or regulation alterations that occur in natural high-CO2-requiring auxotrophs, the pyrimidine-dependent regulation of nucleotide pools in L. plantarum was investigated in this work.
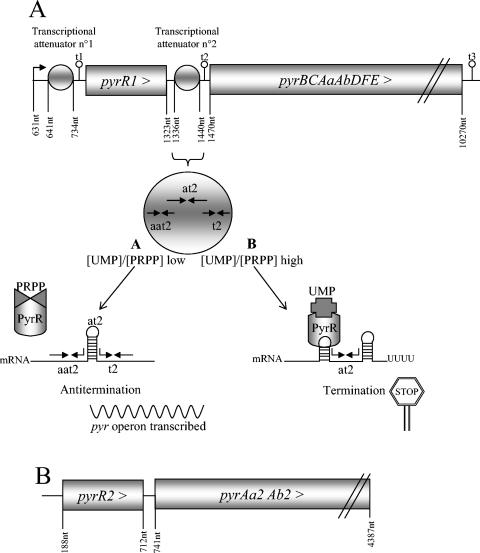
In L. plantarum, the enzymes catalyzing the de novo pyrimidine pathway are encoded by the pyr operon (3, 15, 19). The first gene of the operon is pyrR1, a Bacillus subtilis pyrR-like gene (Fig. 2A). In B. subtilis, PyrR regulates the expression of the downstream genes of the pyr operon by modulating the attenuation of transcription at three points in response to exogenous pyrimidines (11, 24). In the presence of UMP, PyrR binds to the target RNA that folds into a stem-loop structure called the anti-antiterminator, allowing the formation of the downstream termination structure, resulting in transcription termination. The phosphoribosyl pyrophosphate (PRPP) antagonizes the action of UMP on termination. PyrR also has catalytic activity as a uracil phosphoribosyltransferase, although this activity has no physiological importance (12). Consequently, the coregulators PRPP and UMP are proposed to bind to the active site of PyrR, like the substrate and product molecules. When the UMP/PRPP ratio is low, PyrR does not bind its RNA target so that the antitermination loop is formed and transcription proceeds beyond the terminator.
FIG. 2.
Gene organization of the pyr genes. A, Scheme of the transcription attenuation mechanism of the biosynthetic pyrimidine operon pyrR1BCAaAbDFE (EMBL database accession no. Z54240). Two transcription attenuators are found in the 5′ leader sequence of the pyr operon (6) which consists of overlapping repeated sequences that form exclusive RNA loops such as the anti-antiterminator loop (aat), the antiterminator loop (at), and terminator loop (t). Terminators t1, t2, and t3 are indicated. B, pyrR2pyrAa2Ab2 cluster (EMBL database accession no. AJ617795) with the pseudogene pyrAb2.
In vitro analysis of the three known B. subtilis pyr attenuation sites suggests that the RNA polymerase pauses after synthesis of the leading anti-antiterminator sequence so that the more stable antiterminator loop cannot be formed, allowing PyrR to bind to the anti-antiterminator loop (25). B. subtilis PyrR acts probably as a dimer (2), and the crystal structure of the ligand-free dimeric form has recently been resolved (accession no. 1A4X). Several regions involved in mRNA binding (8, 20), coregulator binding, and the dimerization process were characterized, and the structure of the domain homologous to other phosphoribosyltransferases was studied (23). A second pyrR-like gene was found in L. plantarum upstream of pyrAa2 and pyrAb2 (Fig. 2B). The resulting CPS is predicted to be nonfunctional, since frameshift mutations are found in the pseudogene pyrAb2 (10).
Pyrimidine-regulated transcription attenuation may be common in bacteria, as previously suggested (22). Sequence analysis of the L. plantarum pyr operon 5′ leader region suggested that in this organism the pyr operon may be regulated by transcription attenuation at two possible attenuation sites (Fig. 2A). The first attenuation site is located between the promoter and pyrR1. The second attenuator is located between pyrR1 and pyrB (6). In this work, the occurrence of pyrimidine-dependent transcription attenuation of the pyr operon was assessed on RNA extracted from L. plantarum grown in the presence and absence of exogenous uracil. Moreover, use of a classical genetic screening for deregulated mutants, analysis of independent mutants allowed us to study in vivo pyrimidine regulation of the pyr operon in L. plantarum. The pyr operon expression is modulated by PyrR1 and the sequence of the attenuation site located between pyrR1 and pyrB. The inability of some deregulated mutants to grow in air without additional carbon dioxide was linked to reduced nucleotide pool sizes.
MATERIALS AND METHODS
L. plantarum strains and growth conditions.
All strains derive from the pyrimidine prototrophic strain CCM 1904, as documented in Table 1. Routine Lactobacillus propagation was done in MRS (Difco Laboratories) at 30°C without agitation. The defined Lactobacillus medium DLA (4) was used when the concentration of pyrimidines needed to be controlled. When required, uracil was added at 50 μg/ml. For nucleotide pool determination, DLP medium was used. Compared to DLA, the DLP medium has less phosphates (2 mM instead of 10 mM), no cystine, and Tween 80 (1 g) replaced oleic acid and Tween 40.
TABLE 1.
Characteristics of the strains studieda
| Strain | Relevant genotype | Mutant name(s) | Growth | Pyrimidine excretion (μg/ml) | Comments or reference |
|---|---|---|---|---|---|
| Wild-type CCM 1904 | Prototroph | 0 | |||
| CCM 1904 mutants | ΔpyrD | PH1021 | Ura− | Not tested | 9 |
| aat2 (G1356A) | AE1021 | Prototroph | 5.7 | aat2 hexaloop mutated | |
| at2b | AE1023 | HCR | 0 | Unstable RNA at2 loop | |
| pyrR1 (G1090T) | AE1026 | Prototroph | 4.3 | PyrR1 D104Y | |
| ΔcarAB | FB335 | Uras prototroph | 0 | 19 | |
| Pyrimidine-deregulated isolates of FB335c | |||||
| RNA loops | ΔcarAB aat2 (Δ1332-1349) | U1, U8, U28 | Prototroph | 7.3 | aat2 loop absent |
| ΔcarAB aat2 (G1358A) | U4 | HCR | 2.5 | aat2 hexaloop mutated | |
| ΔcarAB aat2-at2 (G1360A) | U23 | Prototroph | 6.7 | aat2 loop mutated | |
| ΔcarAB ΔpyrR1 aat2 (Δ886-1340) | U25 | Prototroph | 14.7 | G36 to E180 PyrR1 deletion and aat2 loss | |
| ΔcarAB aat2 (G1361T) | U26 | Prototroph | 3.1 | aat2 loop mutated | |
| ΔcarAB at2 (T1401G) | U33 | HCR | 0 | at2 loop mutated | |
| PyrR1 | ΔcarAB pyrR1 (C1204T) | U3 | Prototroph | 4.6 | PyrR1 P124S |
| ΔcarAB pyrR1 (G901A) | U5 | Prototroph | 4.0 | PyrR1 G41R | |
| ΔcarAB pyrR1 (G1140) | U6 | Prototroph | 0.4 | PyrR1 M120I | |
| ΔcarAB pyrR1 (C991T) | U9 | Prototroph | 1.8 | PyrR1 R71W | |
| ΔcarAB ΔpyrR1 (1228-1296) | U12 | Prototroph | 4.6 | H170-G172 PyrR1 deletion | |
| ΔcarAB pyrR1 (A1193G) | U13 | Prototroph | 2.3 | PyrR1 H138R | |
| ΔcarAB pyrR1 (G1282T) | U15 | Prototroph | 2.4 | PyrR1 D168N | |
| ΔcarAB pyrR1 (C1195T) | U16 | Prototroph | 1.9 | PyrR1 R139C | |
| ΔcarAB ΔpyrR1 (769-1041) | U17 | Prototroph | 8.5 | No PyrR1 translated | |
| ΔcarAB pyrR1 (1141 insG) | U18 | Prototroph | 8.6 | D121-E180 PyrR1 deletion | |
| ΔcarAB pyrR1 (911insT) | U19 | Prototroph | 7.6 | F44-E180 PyrR1 deletion | |
| ΔcarAB pyrR1 (G997A) | U22, U29 | Prototroph | 1.2 | PyrR1 D73N | |
| ΔcarAB pyrR1 (924insA) | U24 | Prototroph | 7.7 | A49-E180 PyrR1 deletion | |
| ΔcarAB ΔpyrR1 (795-950) | U31 | HCR | 6.2 | V6-G57 PyrR1 deletion | |
| ΔcarAB pyrR1 (ins801)d | U32 | Prototroph | 7.6 | Q17-E180 PyrR1 deletion | |
| ΔcarAB pyrR1 (G783A) | U40 | HCR | 7.2 | PyrR1 truncated due to a point mutation in the initiation codon | |
| ΔcarAB pyrR1 (C1117T) | U41 | Prototroph | 5.4 | PyrR1 R113W |
Sequence data refer to EMBL accession number Z54240. Prototrophs grew without arginine and uracil in defined medium DLA. The growth of uracil-sensitive (Uras) prototrophs was inhibited by uracil in the absence of arginine. The high-carbon-dioxide-requiring (HCR) auxotrophs require exogenous arginine and uracil at low carbon dioxide levels (see Table 3).
TgTgaggaacTTaacggAtACCAGT; mutated bases in uppercase (nucleotides 1355 to 1379 in database accession no. Z54240).
Uracil-resistant (Urar) derivatives of strain FB335 that acquired the ability to grow in the presence of uracil when arginine was omitted from DLA plates incubated in CO2-enriched air.
The inserted sequence is 5′-GCAATGACCATGCGCCGCGCGCTGAC-3′.
Physiological tests.
Nutritional requirements were tested at 30°C on DLA agar plates supplemented or not with 50 μg of uracil (DLAU) or arginine (DLAA) per ml, in aerobiosis or CO2-enriched air. Aerobiosis was obtained by incubation in ordinary air. To calibrate the air at 4% pCO2, a water-jacketed CH/P incubator (Forma Scientific) was used. To test whether the different mutants excreted pyrimidines, the amount of pyrimidines present in the culture supernatant was measured with a bioassay. The indicator strain HN217 is a stable L. plantarum Ura− Arg− mutant because of the pyrAaAb carAB gene deletions (19). HN217 only grows in DLAA when pyrimidines are supplied.
Strains to be tested were cultivated without agitation for 3 days at 30°C in 2 ml of DLAA broth. Cells were eliminated by centrifugation (3,000 × g for 6 min). The supernatant was filter sterilized with a 0.45-μm-pore-size Millipore filter and mixed to new DLAA broth before inoculation with the indicator strain. To correlate the indicator strain growth yield with uracil concentrations and to compare the mutants with the parental FB335 strain, some used DLAA medium (after growth of FB335) was mixed with some fresh medium supplied with increasing amounts of uracil and inoculated with the indicator strain. After 4 days of incubation at 30°C without agitation, the growth yield was established by measuring the optical density at 660 nm. The cell densities in the culture with known amounts of uracil (0 to 7.5 μg/ml) were used to quantify the uracil excretion of the different mutants.
Nucleic acid techniques.
Three genetic loci were PCR amplified with the primer sets described in Table 2. Overlapping PCR products were amplified when the genetic loci exceeded 3 kb such as for pyrAaAb and the pyrR2 loci. PCR amplifications were done on a Peltier thermal cycler PTC-200 (MJ Research) with Taq polymerase (Sigma). PCR amplifications started with denaturation at 95°C for 1 min. Then, 35 cycles of successive denaturation (20 s at 94°C), hybridization (20 s at 56°C), and elongation (5 min at 72°C) were performed. The PCR was completed by a 10-min postelongation treatment at 72°C. Prior to sequencing of the PCR products, the nonincorporated primers and deoxynucleoside triphosphates were eliminated by passage on a Microspin S-400 HR column (Pharmacia Biotech). RNA extraction, primer extension, and Northern hybridization were performed according to Nicoloff et al. (19).
TABLE 2.
Primer list
| Genetic locus | PCR product
|
Primer name and sequence (5′→3′)a | EMBL accession no. | |
|---|---|---|---|---|
| Use | Size (kb) | |||
| pyrR1 and attenuators | Amplification and sequencing | 1.2 | N40, GTACTAATATTAGGCGACCG (F) N29, TGTGTGCGGGTGCTATTCTC (R) | Z54240 |
| pyrR1 mutagenesis | 1.1 | N44, GTTAGGGCGTTGGATGCC (F) | ||
| aem11, AATAAGACATAATCGACCAAG (R) | ||||
| 0.5 | N29, see above | |||
| aem10, CTTGGTCGATTATGTCTTATT (F) | ||||
| aat2 mutagenesis | 0.7 | aem1, TTGTTTCCAAAGAGGAACG (F) | ||
| 2056, CCATCGTCTGATATTCGCCG (R) | ||||
| 0.7 | aem2, CGTTCCTCTTTGGAAACAA (R) | |||
| N46, CCCCGTGGATTAAGGTGA (F) | ||||
| 1.1 | 2072, CTGACAAACTAATGGCACG (F) | |||
| 2042, TGTGACGGATGTTGCCCAC (R) | ||||
| at2 mutagenesis | 0.7 | aem5, AATTCTTTCCTGTGAGGAACTTAACGGATACC AGTTGCGTGACCACTTAATCGGAG (F) | ||
| 2056, see above | ||||
| 0.7 | aem6, CGCAACTGGT ATCCGTTAA GTTCCTCACAGGAAAGAATTAAATGGTTTTATTCAA (R) | |||
| N46, see above | ||||
| Primer extension | 2071, CGTGCCATTAGTTTGTCAG | |||
| pyrR1 probe | 0.5 | 2072, CTGACAAACTAATGGCACG (F) | ||
| N25, TGTTCATCCAAGGCCGTTGG (R) | ||||
| pyrB probe | 1.3 | 2052, GGTTCGGCATCCACGCGATC (F) | ||
| 2059, CCAACATGACATCACGAG (R) | ||||
| pyrAaAb | 5′-End amplification and sequencing | 2.5 | 2041, TAGTTGCGGGATTGTTGGATG (F) 2029, TGGGTGAACTTATCAAACGG (R) | Z54240 |
| 3′-End amplification and sequencing | 2.6 | 2380, CGGCCAGCACATTGTCA (F) 2002, CTCCAAAGCGACTGGTTACC (R) | ||
| pyrR2 | 5′-End amplification and sequencing | 1.9 | lp1782F, CGACAACCTTGGCTAAGCTTG (F) lp1783r, CTCGGCCAATGTCATTATGC (R) | AL935263 |
| 3′-End amplification and sequencing | 3.0 | lp1783f3, CAGATGGCGCTCCTGGTCCGAC (F) l904lp1784r2, TGATACTAGTGACTATCAGTG (R) | AJ617795 | |
F, forward primer; R, reverse primer. Nucleotides in bold differ from the wild-type sequence, and complementary sequences between aem5 and aem6 are underlined. Primers aem11 and aem10 as well as aem1 and aem2 are complementary over their whole sequence.
Spontaneous mutant selection and site-specific mutagenesis.
Independent spontaneous uracil-resistant (Urar) mutants of strain FB335 were selected on DLAU plates incubated 4 days at 30°C in 4% CO2-enriched air as previously described (18). Mutagenesis by allelic replacement has been successfully used in L. plantarum CCM 1904 with pGID023-derived plasmids (19). Such plasmids were constructed to mutate antiterminator site 2 (at2) in AE1023 with plasmid pAE1017, anti-antiterminator site 2 (aat2) in AE1021 with plasmid pAE1015, and pyrR1 in AE1026 with plasmid pAE1020.
To construct plasmid pAE1017, the PCR products of two amplifications with L. plantarum CCM 1904 genomic DNA as the template and primer set aem5 and 2056 and primer set aem6 and N46 were merged by a second PCR with primers N46 and 2056. Primers aem5 (55-mers) and aem6 (54-mers) have an overlapping 40-nucleotide-long sequence and 12 different nucleotides compared to the wild-type at2 sequence (Table 2). The resulting 1.340-bp PCR product was BclI restricted and cloned into the linearized BamHI pGID023 vector. To construct pAE1015, the PCR products of two amplifications with primer set aem1 and 2056 and primer set aem2 and N46 were merged by a second PCR with primers 2072 and 2042. Primers aem1 and aem2 overlapped and harbored a single nucleotide mutation compared to the wild-type aat2 (Table 2). The resulting 1.100-bp PCR product was BclI restricted and cloned into the linearized BamHI pGID023 vector. To construct pAE1020, the PCR products of two amplifications with primer sets N44 and aem11 and N29 and aem10 were merged by a second PCR with primers N44 and N29. Primers aem10 and aem11 are overlapping and harbor a single nucleotide mutation compared to the wild-type pyrR1 coding sequence (Table 2). The 893-bp PCR product was digested with the restriction enzymes Cac8I and RsaI and cloned into the linearized SmaI vector. Selection of recombinants harboring the mutated alleles was performed as previously described (19), and the mutations were confirmed by direct sequencing on the chromosome (6).
Intracellular nucleotide pool quantification.
The intracellular nucleoside triphosphate pool measurement in L. plantarum was optimized from the method described for Lactococcus lactis (14). Cells were grown exponentially for at least two generations in DLP medium in the presence of 33Pi until an optical density at 660 nm of 0.8 was reached. The 33P-labeled nucleotides were extracted from L. plantarum by transferring 190 μl of cell culture to 20 μl of 10 M formic acid at room temperature. After three freeze-thaw cycles followed by 30 min of incubation in an ice-water bath, the cell debris were removed by centrifugation.
For analysis, 20 μl of extract was mixed with 2 μl of marker solution (nucleoside triphosphates, 5 mM each) and applied to polyethyleneimine-cellulose plates. The plates were dried under cold air, washed for 10 min in ethanol, and dried. The first dimension of chromatography was performed at room temperature in a 0.85 M KH2PO4 solution adjusted to pH 3.42 by addition of an equimolar H3PO4 solution. Following chromatography, the plates were washed in 10% citrate (wt/vol) and two times in distilled water. The plates were dried and chromatographed in the second dimension with 0.75 M LiCl supplemented with 7.5% H3BO3 as the running buffer. The pH was adjusted to 6.8 by addition of solid LiOH. Individual nucleoside triphosphates were identified according to the spots of the unlabeled nucleoside triphosphate marker solutions visible in UV light. Finally, the plates were evaluated and enumerated in an Instant Imager. With the specific activity measured in the assays, the concentration of the individual nucleotides was then determined as a function of the radioactivity in their individual spots. In order to report nucleoside triphosphate pools as nanomoles per milligram of dry weight, we used the correlation factor 0.2 mg [dry weight] ml−1 corresponded to an optical density at 450 nm equal to 1. Two independent biological replicates were performed for each condition tested.
Nucleotide sequence accession number.
The nucleotide sequence of the pyrR2-pyrAa2-Ab2 locus in L. plantarum CCM 1904 reported in this paper has been submitted to the EMBL database and assigned accession number AJ617795.
RESULTS
L. plantarum pyr operon is regulated by transcription attenuation.
Two putative attenuation sites followed by terminators t1 and t2, respectively, were found in the pyr operon 5′ leader sequence (Fig. 2A). These proposed RNA hairpin structures suggested that the pyr operon is regulated by transcription attenuation (6). In this case, transcription initiation of the pyr operon is predicted to occur whether or not uracil is added to the medium. To test this, the pyr operon transcription start site was determined by primer extension with avian myeloblastosis virus reverse transcriptase. With primer 2071, located 11 nucleotides upstream of the pyrR1 gene, the transcription start site was determined at nucleotide C (position 631 of EMBL sequence accession no. Z54240), which is positioned 149 nucleotides before the pyrR1 initiation codon. A putative −10 box (TACACT) was found 6 nucleotides upstream of the transcription start site. This start site was detected with RNA extracted from cells cultivated in the absence and presence of uracil (Fig. 3A).
FIG. 3.
Pyrimidine-regulated transcription of the L. plantarum pyr operon. RNA was extracted from cells cultivated without agitation in defined medium without (−U) and with added uracil at a concentration of 200 μg/ml (+U). A, Primer extension by reverse transcription with primer 2071. The same primer was used for the sequencing reaction (A, C, G, and T tracks). The transcription start site is indicated (+1). B, Northern hybridization on 13 μg of RNA probed with two DNA fragments hybridizing to pyrR1 and to pyrB. The sizes of the bands detected were deduced from molecular size markers (lane M), and relevant bands are marked: a, 0.8 kb; b, 9.7 kb.
Once transcription is started, RNA polymerase may stop elongation at the putative t1 terminator located before pyrR1, at terminator t2 located in the intergenic pyrR1-pyrB region, and at terminator t3 located at the end of the pyr operon, leading to mRNAs with predicted sizes of 0.1, 0.8, and 9.7 kb, respectively. To test this model, the sizes of the pyr operon mRNAs were characterized by Northern hybridization with two different probes. These probes had no sequence similarities with the smaller 0.1-kb transcript so that the activity of terminator t1 was not assessed. With a pyrR1-specific probe, two mRNA bands of 0.8 kb and 9.7 kb were identified. With the pyrB-specific probe, only the pyrimidine-repressed 9.7-kb mRNA was detected (Fig. 3B; the band around 2.5 kb obtained with the pyrR1 and pyrB probes may correspond to a degradation product of the larger 9.7-kb transcript). These data show that pyr operon transcription initiation occurs in the presence and absence of uracil, that the pyrR1BCAaAbDFE transcript (9.7 kb) is detected only in cells grown without uracil, and that the pyrR1 transcript (0.8 kb) is detected in the presence and absence of uracil. These in vivo data demonstrate that the pyr operon in L. plantarum is pyrimidine-dependently regulated by transcription attenuation with termination between the pyrR1 and pyrB genes, possibly at the terminator t2 hairpin (Fig. 4A).
FIG. 4.
L. plantarum mutations in the pyr operon 5′ leader. Mutations are indicated with arrows. Mutant names are indicated in parentheses except for AE1023, whose mutations are marked with an asterisk. The deleted nucleotides are surrounded in U1 and highlighted with a gray background in mutant U25. Sequence coordinates refer to EMBL accession no. Z54240. A, Proposed hairpin structures of the anti-antitermination (aat2) and termination attenuator (t2) that favor transcription termination. Underlined, nucleotides (CAGAGA) in the repressor RNA binding aat2 hexaloop. B, Proposed hairpin structure of the antitermination attenuator at2 that allows transcription of the pyr operon.
Positive screening test for spontaneous mutants.
The positive screening for pyrimidine-deregulated mutants was based on the ability to supply the arginine biosynthesis pathway with sufficient carbamoyl phosphate produced by the pyrimidine-regulated CPS-P. Strain FB335 carries a deletion of the carAB genes, encoding the arginine-regulated CPS-A. Consequently, CPS-P is responsible for carbamoyl phosphate production for both arginine and pyrimidine biosynthesis. The presence of uracil (DLAU medium) represses transcription of the pyr operon, which harbors the structural genes for CPS-P, so that only a limited amount of CPS-P is present. The amount of carbamoyl phosphate produced under these conditions is not sufficient to support adequate biosynthesis of arginine. This explains why FB335 cannot grow in DLAU, a phenotype called uracil sensitivity (Uras). Mutants derived from FB335 that lost pyrimidine regulation of the pyr operon were expected to grow in DLAU, a phenotype called uracil resistance (Urar). The mutants were selected on DLAU plates incubated in 4% CO2-enriched air since CO2 is a substrate for carbamoyl phosphate synthesis and CO2 concentrations higher than those found in normal air are required for the growth of some L. plantarum CPS mutants (19).
Forty spontaneous mutants were isolated from independent cultures, restreaked on MRS plates, and tested again for the Urar phenotype on DLAU plates. All conserved their ability to grow in the presence of uracil, suggesting that they harbored stable mutations.
Phenotypic characterization of 40 independent Urar mutants.
The physiological abilities of the Urar mutants were compared to the parental Uras strain FB335 (ΔcarAB), the prototroph CCM 1904, and the uracil auxotroph PH1021 (ΔpyrD) (9). Two physiological traits were studied: requirements for uracil and arginine for growth (Table 3) and pyrimidine excretion (Table 1). When the growth requirement for uracil and arginine was assessed with the 40 Urar mutants, three main phenotypes were found (Table 3 and data not shown). Twenty-four mutants were prototrophic for uracil and arginine, as found with the wild-type strain CCM 1904. Fifteen mutants had a high-CO2-requiring phenotype (5), which means that they were only able to grow in the absence of arginine and uracil in CO2-enriched air. One mutant, U7, was auxotrophic for uracil and, unlike the pyrD strain, grew in the presence of uracil without CO2 enrichment. The prototroph U25 mutant also acquired the ability to grow in the presence of uracil without CO2 enrichment.
TABLE 3.
Arginine and pyrimidine nutritionnal needs of the strains studieda
| Phenotypeb | Carbon dioxide enrichment | Growth
|
Mutants | |||
|---|---|---|---|---|---|---|
| No addition | Ura | Arg | Ura and Arg | |||
| Prototroph (like wild-type CCM 1904) | No | + | − | + | + | U1, U3, U5, U6, U8, U9, U12, U13, U15-U19, U22-U24, U26, U28, U29, U32, U41, AE1021, AE1026 |
| Yes | + | + | + | + | ||
| Prototroph | No | + | + | + | + | U25 |
| Yes | + | + | + | + | ||
| Prototroph Uras | No | + | − | + | + | FB335 |
| Yes | + | − | + | + | ||
| HCR auxotroph | No | − | − | − | + | U4, U31, U33, U40, AE1023 |
| Yes | + | + | + | + | ||
| Ura auxotroph | No | − | − | − | + | PH1021 |
| Yes | − | + | − | + | ||
Growth was assessed on DLA agar plates supplemented with arginine and uracil and incubated in air or in air enriched with 4% CO2. + and −, growth and no growth after 3 days of incubation, respectively.
Uras (sensitive to Ura) refers to the growth inhibition observed when only Ura is provided in the medium. High-CO2-requiring (HCR) strains require air-enriched with CO2 to grow in the absence of both Arg and Ura.
Compared to the parental strain, some mutants may have lost the ability to control pyrimidine synthesis, which would result in increased levels of pyrimidine synthesis and consequently pyrimidine excretion. Pyrimidine excretion was evaluated in the Urar mutant collection and compared to that of the parental strain FB335. Excretion of pyrimidines was observed with most mutants (Table 1). The largest amount of pyrimidine nucleotides excreted was 15 μg/ml in strain U25. Phenotypic characterization of the 40 spontaneous Urar mutants revealed two major types of mutants, the prototrophs and the high-CO2-requiring auxotrophs, which can be further subdivided according to their ability to excrete pyrimidines.
Genetic localization of the mutation conferring Urar in strain FB335.
Genetic loci potentially implicated in the Urar phenotype were sequenced and compared to the wild-type genes. Sequencing of the two putative pyrimidine regulators pyrR1 and pyrR2 and the pyr operon 5′ end revealed mutations that may impair pyrimidine-dependent transcription attenuation of the pyr operon. Sequencing of the pyrAa-Ab genes searched for mutations in the structural genes of the CPS that may affect their activity or their allosteric regulation. Analysis of these loci in the 40 independent mutants allowed us to genetically characterize 26 mutants. All the Urar strains tested harbored wild-type pyrR2-pyrAa2-pyrAb2 and pyrAa1-pyrAb1 genes. The mutations were found in the 5′ leader sequence of the pyr operon including pyrR1 (between nucleotides 424 and 1620 in Fig. 2A).
The 26 genetic lesions identified were point mutations in 14 strains; 17- to 454-nucleotide-long deletions in seven strains; and 1- to 26-nucleotide-long insertions in four strains (Table 1). All the mutations were different except for a 17- nucleotide-long deletion in the 5′ leader region of the pyr operon (strains U1, U8, and U28) and a missense point mutation in pyrR1 (strains U22 and U29).
Site-specific mutagenesis of the wild-type strain CCM 1904.
Three mutants were constructed by site-specific mutagenesis of strain CCM 1904 for two reasons. First, the spontaneous Urar mutants were obtained in a ΔCPS-A derivative of the wild-type strain CCM 1904, and we wanted to see whether the same phenotype was obtained in the wild-type strain. Second, multiple mutational events may have occurred in the spontaneous mutants even though this is unlikely, since the frequency of spontaneous mutations was less than 10−7 mutants per plated cell and that most of the mutants studied harbored different mutations. To ensure that strains with the Urar phenotype can indeed result from a single-locus mutation in the wild-type strain, mutants AE1021, AE1023, and AE1026 were constructed by homologous recombination. One mutation invalidated the repressor PyrR1 (AE1026), and the other two mutations targeted the transcriptional attenuators (AE1021 and AE1023).
In strain AE1026, the D106Y mutation, known to impair B. subtilis PyrR binding to the mRNA loop of the pyr operon leader sequence (mutants BGH4, BGH14, and BGH19) (8), was introduced into the L. plantarum pyrR1 locus. In mutant AE1021, a point mutation (G1356A) alters the aat2 hexaloop, which is expected to prevent PyrR binding, as found in B. subtilis (2). This mutation slightly increases the stability of the at2 loop (ΔG of −21.7 instead of the wild-type −20.9 kcal mol−1) but has no effect on the stability of the proposed aat2 and t2 RNA loops (see Fig. 4). Thus, constitutive high expression of the pyr operon is predicted in AE1021. In strain AE1023, 11 bases were changed (Fig. 4) in order to destabilize the antiterminator at2 loop with a calculated ΔG of −9.2 kcal mol−1 instead of −20.9 kcal mol−1 in the wild-type strain. The terminator t2 was unchanged. The anti-antiterminator aat2 loop was slightly less stable (ΔG of −4.4 instead of −6.8 kcal mol−1 in the wild type) but harbored two mutations in the putative PyrR binding hexaloop (CUGUGA instead of CAGAGA in the wild type; see Fig. 4A). Thus, pyrimidine-independent high termination of transcription and low expression of the pyr operon were predicted in strain AE1023.
When assessed for arginine and pyrimidine requirements (Table 3), the constructed mutants AE1021, AE1025, and AE1026 were resistant to uracil (Urar) when grown in CO2-enriched air. AE1021 and AE1026 were prototrophs that excreted at least four times more pyrimidines than the wild-type prototroph CCM 1904. AE1023 had the high-CO2-requiring phenotype, and no excretion of pyrimidines could be detected. Thus, the constructed mutants AE1021, AE1023, and AE1026 have growth characteristics found in the equivalent Urar spontaneous mutants (Table 1).
cis-acting spontaneous mutations found to alter L. plantarum pyr operon expression.
Seven spontaneous Urar mutants (U4, U23, U26, U33, U1, U8, and U28) harbored mutations in the second transcription attenuator in the intergenic region between the pyrR1 and pyrB genes proposed to be involved in transcription attenuation of the pyr operon (Fig. 2A). No aat2 loop was found in the deletion mutants U25 and U1 (U8 and U28 harbored the same deletion as U1) (Fig. 4A). Mutant U25 in addition had a PyrR1 deletion. Point mutations were found at the top of the aat2 loop, either directly within the hexaloop (mutant U4) or at its stem (mutants U23 and U26). These mutations also concomitantly destabilized the at2 loop in mutants U1, U4, U23, and U26 (ΔG of about −16 instead of −20.9 kcal mol−1 calculated in the wild type). These Urar mutants excreted increased pyrimidines (Table 1), suggesting that mutations in the RNA aat2 loop conferred deregulated expression of the pyr operon.
Among the seven mutants, only U33 did not excrete significant quantities of pyrimidines (Table 1). The T→G point mutation in U33 slightly altered both the t2 loop (ΔG of −13.2 instead of −17.6 kcal mol−1 as found in the wild type) and the at2 loop (ΔG of −19.1 instead of −20.9 kcal mol−1 in the wild type).
Different mutations of PyrR1 modulate the levels of deregulation of pyrimidine synthesis.
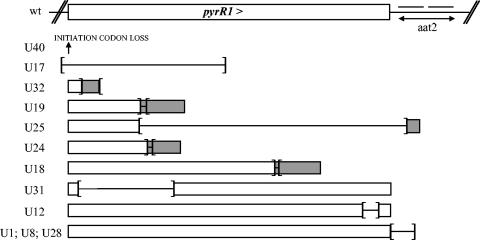
Eighteen out of 20 pyrR1 mutants excreted pyrimidine nucleotides in a range between 1 and 15 μg/ml (Table 1; 0 and 0.4 μg/ml of excreted pyrimidine nucleotides in mutants U33 and U6, respectively). The impact of deletion and insertion events on protein PyrR1 composition is summarized in Fig. 5. In mutants U40 and U17, complete absence of PyrR1 was the result of a lost translation initiation codon. Strain U32 also lost all PyrR1 functional domains with PyrR1 reduced to its 16 first residues. In mutant U31, loss of residues V6 to G57 deleted PyrR1 of two conserved regions (the first mRNA binding region and the PPi loop). Three mutants harbor repressors with less than 25% of the N-terminal part of wild-type PyrR1 due to mutations that introduced translation frameshift (1-nucleotide insert in mutants U19 and U24; 454-nucleotide deletion in mutant U25). The PyrR1 C-terminal domain has been lost completely in strain U18 (residues D121 to E180) and partially in strain U12 (loss of three residues, H170 to G172).
FIG. 5.
Deletions and insertions found in the spontaneous Urar mutants. Rectangles schematize the open reading frame that encodes wild-type PyrR1 protein (white) or chimeric peptides (gray). Insertion and deletion events are delimited by brackets and are oriented outwards and inwards, respectively. The point mutation in mutant U40 is localized with an arrow. The RNA loop binding the PyrR repressor is indicated as anti-antiterminator (aat2) and is mutated in mutants U25, U1, U8, and U28.
The 10 different missense mutations found in PyrR1 were located in highly conserved residues, except for residue M120 in mutant U6 (Fig. 6). The strains carrying pyrR1 point mutations had only partially derepressed phenotypes since excretion was increased to a lesser extent in these mutants (≤5 μg/ml in AE1026, U3, U5, U6, U9, U13, U15, U16, U22/29, and U41) compared to the deleted PyrR1 strains (≥5 μg/ml in U12, U17, U18, U19, U24, U25, U31, U32, and U40) (Table 1).
FIG. 6.
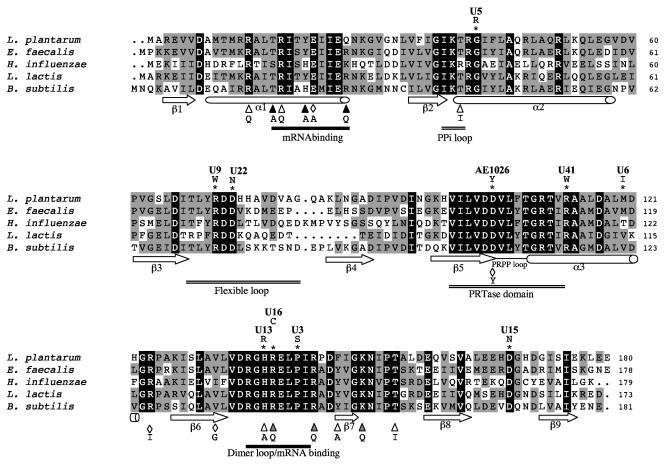
Sequence alignment of PyrR homologs and localization of L. plantarum PyrR1 mutations. Four functional gram-positive bacteria repressors were aligned, L. plantarum (accession no. Z54240), Bacillus subtilis (accession no. M59757), Enterococcus faecalis V583 (accession no. AF044978), and Lactococcus lactis subsp. lactis (accession no. Q9L4N8), and compared to the gram-negative Haemophilus influenzae Rd KW20 (accession no. U32728) homolog. Identical and similar amino acids are highlighted with black and gray backgrounds, respectively. The secondary structure of the B. subtilis protein is indicated; arrows and cylinders represent β strands and α helices, respectively (23). A basic concave surface at the dimer interface was proposed to be required for B. subtilis RNA binding and comprised two distinct regions (black lines) (20). The conserved phosphoribosyltransferase (PRTase) structural elements of substrate and product binding involve three regions marked with double lines: the phosphoribosyltransferase domain, the flexible loop which is disordered in the dimer structure, and the PPi loop (23). Dimerization of B. subtilis PyrR involves the α3 helix, the β6 strand, and the dimer loop (20). Amino acid substitutions in L. plantarum PyrR1 (*) and B. subtilis (diamonds [8] and triangles [20]) that lead to defects in pyrimidine regulation are marked. Black triangles indicate B. subtilis residues that were mutated and impaired mRNA binding without changing protein folding. The names of the L. plantarum mutants are indicated above the mutated residues.
Optimization of a method for measuring nucleoside triphosphate pools in L. plantarum.
The incorporation of radioactively labeled phosphate has previously been used successfully to determine the intracellular nucleotide pool of gram-positive bacteria (14, 21) but has not previously been assessed in L. plantarum. Accordingly, the internal nucleoside triphosphate pool of L. plantarum was determined by the incorporation of [33P]orthophosphate (50 μCi) in exponentially growing cells for two generations. In order to obtain a high specific activity, defined medium DLA was modified by lowering the phosphate concentration. A fivefold reduction of phosphate (from 10 to 2 mM) was shown not to affect growth (data not shown). This medium (DLP) was used for all nucleotide pool determinations. Cells were extracted by transferring aliquots of the culture to formic acid and treating these samples as described in Materials and Methods.
In order to optimize the method, we focused on the concentration of the formic acid used in the extraction. The reported concentrations of formic acid for the extraction of nucleotides from Salmonella enterica serovar Typhimurium (1) and Streptococcus equisimilis (16) were very different, 0.3 M and 13 M, respectively. We decided to test for the lowest concentration giving quantitative extraction. Final concentrations above 0.95 M formic acid gave quantitative extraction of nucleotides from L. plantarum. Accordingly, 0.95 M formic acid was used in all of the following experiments.
Intracellular nucleotide pool quantification.
The nucleotide pool sizes observed in L. plantarum are similar to those found in other organisms, whereas its ATP pool is the largest; the others' are two- to sixfold smaller (Table 4). Deletion of the carAB operon conferred a slight reduction in pyrimidine pool sizes. This could be explained by a reduced availability of carbamoyl phosphate (Table 4). In the absence of the pyrimidine regulatory protein PyrR1, the pyr operon would be overexpressed, resulting in increased amounts of the pyrimidine biosynthetic enzymes. Intuitively, one would expect an increase in pyrimidine nucleotide pool sizes. Surprisingly, this was not observed; strain AE1026 (pyrR1) had nucleotide pool sizes similar to that of the wild-type strain (Table 4). On the other hand, in the isolated mutant U25, with the pyrR1 gene and a part of the attenuation site deleted, the UTP and CTP pool sizes were increased two- to threefold (Table 4). In strain AE1023, the second attenuator in the pyr operon was mutated so that the formation of the antiterminator was affected, favoring the formation of the terminator structure (Fig. 4). Consequently, a reduction in the level of the biosynthetic enzymes is expected, resulting in reduced pyrimidine nucleotide pools. The UTP and CTP pool sizes were reduced three- and twofold, respectively, in this mutant (Table 4).
TABLE 4.
Nucleotide pools in wild-type strain CCM 1904 and isogenic derivatives impaired in pyrimidine metabolism
| Strain | Genotype | Generation time (min)a | Nucleotide pool size (nmol [mg (dry wt)]−1)
|
|||
|---|---|---|---|---|---|---|
| UTP | CTP | GTP | ATP | |||
| CCM 1904 | Wild type | 103 ± 6 | 2.7 ± 0.4 | 0.9 ± 0.2 | 2.2 ± 0.4 | 6.2 ± 0.7 |
| FB335 | ΔcarAB | 113 ± 4 | 2.0 ± 0.2 | 0.6 ± 0.0 | 2.0 ± 0.3 | 5.9 ± 1.4 |
| AE1026 | pyrR1 | 107 ± 5 | 2.9 ± 0.5 | 0.9 ± 0.1 | 2.0 ± 0.4 | 5.6 ± 0.2 |
| AE1023 | at2 mutated | 106 ± 11 | 0.6 ± 0.2 | 0.4 ± 0.1 | 3.3 ± 0.4 | 7.3 ± 1.4 |
| U25 | ΔcarAB pyrR1 | 120 ± 10 | 5.9 ± 1.7 | 1.3 ± 0.5 | 1.6 ± 0.6 | 5.7 ± 0.7 |
| U33 | ΔcarAB at2 | 115 ± 5 | 2.9 ± 0.0 | 0.9 ± 0.1 | 2.7 ± 0.3 | 7.5 ± 0.0 |
In defined DLP medium in the presence of purines (adenine guanine, hypoxanthine, and xanthine) at final concentrations of 83, 50, 33, and 5 μg/ml, respectively.
DISCUSSION
Based on Northern hybridization studies and the characterization of pyrimidine-dependent deregulated mutants, we proposed that pyr operon expression is regulated in response to pyrimidine by transcriptional attenuation and that attenuator 2, located in the pyrR1-pyrB intergenic region (Fig. 2A and Fig. 4), is functional in vivo. The role of attenuator 1, located between the transcriptional start site and pyrR1 (Fig. 2A), is not clear, since no mutations affecting this structure were obtained. This model will be discussed in view of the different mutants altered in pyrR1 and in the pyr operon mRNA 5′-end sequence, in particular in attenuator 2.
Comparison of wild-type PyrR sequences with the altered L. plantarum PyrR1 proteins.
Different PyrR1 mutations modulated the levels of deregulation of pyrimidine biosynthesis, as various levels of pyrimidine excretion were observed (Table 1), suggesting that the mutated protein activities were affected in vivo. These defects could be due to the change of essential residues implied in mRNA binding, coregulator binding, or dimerization as well as from protein misfolding or reduced mutated pyrR expression. The effect of point mutations on PyrR1 function was examined in view of other gram-positive PyrR proteins which have been characterized as functional regulators, such as in Enterococcus faecalis (7), Lactococcus lactis (13), and B. subtilis (23).
The functional domains deduced from the extensive studies in B. subtilis PyrR (8, 20, 23) are reported on the PyrR protein sequence alignments in Fig. 6. Since L. plantarum PyrR1 has 58% identity with B. subtilis PyrR, the effects of PyrR1 mutations were discussed by comparison with the B. subtilis PyrR mutational analysis (8, 20). Mutants such as U3, U13, and U16 share relatively high excretion levels (5, 2, and 2 μg/ml, respectively) and harbor mutations in highly conserved residues localized in the dimer loop (Fig. 6). Such residues were mutated in B. subtilis mutants: H140A compared to H138R in U13, and R141Q compared to R139C in U16. The B. subtilis R141Q mutants was correctly folded, which led the authors to conclude that this region would be required for dimerization or mRNA binding.
Three mutations were found in the α3 helix. Only its N-terminal part is conserved and was identified in B. subtilis PyrR as part of the phosphoribosyltransferase active site involved in coregulator PRPP/UMP binding. As found for the B. subtilis PyrR, the D106Y mutation also reduced L. plantarum PyrR1 repressor function, since AE1026 was a Urar prototroph that excreted pyrimidines. In mutant U41, the R113W mutation is also located in the conserved phosphoribosyltransferase domain of helix α3, but the role of this residue has not previously been studied. In the less conserved region of helix α3, the M120I mutation in mutant U6 conferred a Urar prototroph phenotype with a low excretion level compared to AE1026 and U41. Thus, mutation M120I would only moderately reduce the repressor function of PyrR1.
Near the phosphoribosyltransferase domain, a disordered flexible loop was observed in the B. subtilis PyrR dimeric structure that would be required for coregulator binding. Mutations in the conserved residues of this flexible loop (U9 and U22 or U29) (Fig. 6) gave mutants that showed higher levels of pyrimidine excretion (2 and 1 μg/ml; Table 1). The G45R mutation in U5 (Fig. 6) is expected to affect the PPi loop. This mutation could therefore lead to impaired coregulator binding in PyrR1. Finally, a point mutation, D168N (mutant U15), and a short deletion of three residues (170 to 172 in mutant U12) were localized in the C-terminal part. No similar B. subtilis mutations were obtained in this region, and no particular function has been proposed for this region. Mutations in conserved regions of L. plantarum PyrR1 and B. subtilis PyrR (flexible loop; phosphoribosyltransferase domain; dimer loop/mRNA binding, Fig. 6) led to deregulation, suggesting that these proteins are functionally similar.
pyr operon mRNA 5′-end sequence is recruited for pyr transcription attenuation.
Pyrimidine-controlled transcription attenuation was lost in mutants harboring mutations in attenuator 2 localized between pyrR1 and pyrB (summarized in Fig. 4). These mutations either increased or decreased pyr operon transcription. The first set of mutations led to pyrimidine-independent increased transcription by destabilization of the anti-antiterminator aat2 RNA loop so that termination was inefficient. The second set of mutations led to pyrimidine-independent decreased transcription by destabilizing the antiterminator at2 RNA loop so that terminator t2 formation was favored. This will be discussed below.
Increased transcription readthrough after the pyrR1 gene was correlated to mutations in the RNA aat2 loop. Strains with mutations in attenuator 2 had increased pyrimidine nucleotide excretion (AE1021, U1, and U4 in Table 1). In mutant U1, a 13-nucleotide-long deletion (circled nucleotides in Fig. 4A) resulted in the loss of the proposed aat2 RNA loop. Within this aat2 RNA loop, the terminal hexaloop (underlined in Fig. 4A) needed to be conserved to observe regulation with point mutations in the hexaloop itself (mutant AE1021, CAaAGA, and mutant U4, CAGAuA; mutation indicated in lowercase) or at its stem (mutants U23 and U26; Fig. 4A). Thus, the aat2 loop and in particular the hexaloop seem to be required to terminate transcription after pyrR1 and inhibit de novo pyrimidine nucleotide synthesis in vivo.
Previous in vitro experiments suggested that B. subtilis PyrR tightly binds to a 28-nucleotide-long minimal RNA that contains the hexaloop and its stem (2). Based on alignments of known RNA PyrR-binding sites, a consensus PyrR-binding loop was proposed with the terminal sequence CNGNGA (2). Within this hexaloop consensus, the two G's were replaced individually in mutants AE1021 and U4 and led to loss of repression. These data, combined with secondary-structure similarities with other PyrR-binding loops (data not shown), strongly suggest that in L. plantarum as in B. subtilis, the anti-antiterminator loop, and in particular the terminal hexaloop, is involved in PyrR binding and transcription attenuation of the pyr operon.
Increased transcription termination after pyrR1 was found in the high-CO2-requiring mutant AE1023 that harbors an unstable at2 RNA loop. As expected from such a mutant that would have reduced transcription of the pyr operon, no pyrimidine nucleotide excretion was observed (Table 1). Finally, the measured intracellular UTP and CTP pools were reduced in mutant AE1023 compared to the wild type (Table 4). We conclude that impairment of the at2 loop leads to the loss of pyrimidine-controlled transcription termination of the pyr operon. Despite the fact that AE1023 and mutant U33 are high-CO2-requiring auxotrophs (Table 3), mutant U33 has slightly increased intracellular UTP and CTP pools (Table 4, compare strains FB335 and U33). Strain U33 has a U→G mutation in the second attenuation site. This mutation is predicted to partially destabilize terminator t2 as well as antiterminator at2 (Fig. 4A and B). The position of this mutation 27-nucleotides downstream of the aat2 loop may also be a transcriptional pausing site (25). Thus, how pyrimidine regulation is lost in Urar mutant U33 remains unclear.
Carbamoyl phosphate pools determine the ability of L. plantarum to grow in normal air without CO2 enrichment.
Carbamoyl phosphate is required for arginine and pyrimidine biosynthesis. The pyrimidine-regulated CPS-P can produce carbamoyl phosphate for either arginine or pyrimidine biosynthesis, but the resulting pyrimidine nucleotide pools are lower in a strain that harbors only CPS-P (Table 4, compare the ΔcarAB mutant strain to the wild-type strain). Therefore, in the genetic context of FB335, the reduced carbamoyl phosphate pool limits UMP synthesis. From the differences in the concentrations of UTP and CTP in the wild-type and FB335, we deduced that CPS-A activity provides about one third of the carbamoyl phosphate required for arginine and pyrimidine synthesis in the conditions tested (liquid DLP medium without agitation). The lower UMP and CTP pools had no effect on FB335 growth in DLA medium since FB335 was prototrophic for the pyrimidines and arginine and strain FB335 and the wild-type strain had similar generation times (Table 4). On the other hand, when the pyrimidine nucleotide pools dropped by more than a factor of 2, such as in strain AE1023 (Table 4), the growth of L. plantarum on DLA became dependent on higher levels of supplied CO2, as AE1023 has the high-CO2-requiring phenotype (Table 4). Thus, CO2 may be the limiting reactant in carbamoyl phosphate synthesis in normal air.
This hypothesis is in agreement with the following genetic data. In the FB335 genetic context, the need for CO2 was increased in cells that needed to produce more carbamoyl phosphate due to inefficient recycling of preformed uracil derived from RNA degradation. This was found in 11 of the 40 Urar mutants selected in this work, which harbored mutations in the upp gene, encoding uracil phosphoribosyltransferase (EC 2.4.2.9). These upp mutants had the high-CO2-requiring phenotype and grew only in CO2-enriched conditions (unpublished data). Increased CPS-P concentrations resulting from constitutive high pyr operon transcription conferred the ability to grow in air (mutants with impaired pyrR1 or altered RNA aat2 loops). We therefore conclude that L. plantarum growth is prevented when the intracellular carbamoyl phosphate pool is low. Reduced carbamoyl phosphate pools may result from inadequate CPS activity because of depletion in bicarbonate or from low CPS expression, as observed in strain AE1023.
Is PyrR1 only partially controlling pyr operon expression?
Higher amounts of pyrimidine excretion up to 15 μg/ml were observed when, in addition to pyrR1 deletion, the anti-antiterminator loop aat2 was deleted in mutant U25. The mutants U17 and U40, missing only the pyrR1 gene, excreted 7 to 8 μg of pyrimidine nucleotides per ml. This suggested that pyrR1 deletion alone does not lead to a total derepression phenotype and that the aat2 loop would be formed even in the absence of PyrR1 binding. On the contrary, deletion of pyrR from the B. subtilis chromosome resulted in constitutive, elevated expression of aspartate transcarbamylase, which is encoded by the third gene of the pyr operon (24).
Unlike that of B. subtilis, the L. plantarum genome harbors a second putative repressor gene named pyrR2 (10). In this study, no spontaneous mutations were found in pyrR2. This may suggest that the screening procedure prevented pyrR2 mutant selection or that the role of pyrR2 in pyrimidine regulation may be minor compared to that of pyrR1. Site-directed mutagenesis of the pyrR loci is required to test their involvement in pyr operon regulation. This issue is currently being investigated.
Acknowledgments
We gratefully acknowledge Céline Sutter for technical assistance and Jean-Claude Hubert for valuable discussions on pyrimidine metabolism in Lactobacillus plantarum.
F.B. received a grant from the EU (Major Research Infrastructure grant) for her stay at the Danish Center for Advanced Food Studies.
REFERENCES
- 1.Beck, C. F., J. Neuhard, E. Thomassen, J. L. Ingraham, and E. Kleker. 1974. Salmonella typhimurium mutants defective in cytidine monophosphate kinase (cmk). J. Bacteriol. 120:1370-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonner, E. R., J. N. D'Elia, B. K. Billips, and R. L. Switzer. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res. 29:4851-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouia, A., F. Bringel, L. Frey, A. Belarbi, A. Guyonvarch, B. Kammerer, and J. C. Hubert. 1990. Cloning and structure of the pyrE gene of Lactobacillus plantarum CCM 1904. FEMS Microbiol. Lett. 57:233-238. [DOI] [PubMed] [Google Scholar]
- 4.Bringel, F., L. Frey, S. Boivin, and J. C. Hubert. 1997. Arginine biosynthesis and regulation in Lactobacillus plantarum: the carA gene and the argCJBDF cluster are divergently transcribed. J. Bacteriol. 179:2697-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bringel, F., and J. C. Hubert. 2003. Extent of genetic lesions of the arginine and pyrimidine biosynthetic pathways in Lactobacillus plantarum, L. paraplantarum, L. pentosus, and L. casei: prevalence of CO2 dependent auxotrophs and characterization of deficient arg genes in L. plantarum. Appl. Environ. Microbiol. 69:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elagöz, A., A. Abdi, J. C. Hubert, and B. Kammerer. 1996. Structure and organisation of the pyrimidine biosynthesis pathway genes in Lactobacillus plantarum: a PCR strategy for sequencing without cloning. Gene 182:37-43. [DOI] [PubMed] [Google Scholar]
- 7.Ghim, S. Y., C. C. Kim, E. R. Bonner, J. N. D'Elia, G. K. Grabner, and R. L. Switzer. 1999. The Enterococcus faecalis pyr operon is regulated by autogenous transcriptional attenuation at a single site in the 5′ leader. J. Bacteriol. 181:1324-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghim, S. Y., and R. L. Switzer. 1996. Mutations in Bacillus subtilis PyrR, the pyr regulatory protein, with defects in regulation by pyrimidines. FEMS Microbiol. Lett. 137:13-18. [DOI] [PubMed] [Google Scholar]
- 9.Horvath, P. 2000. Dynamique, évolution et expression de génomes de bactéries lactiques: cas du métabolisme des pyrimidines chez Lactobacillus plantarum CCM 1904. Thèse de doctorat. Université Louis-Pasteur, Strasbourg, France.
- 10.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu, Y., R. J. Turner, and R. L. Switzer. 1995. Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J. Bacteriol. 177:1315-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinussen, J., P. Glaser, P. S. Andersen, and H. H. Saxild. 1995. Two genes encoding uracil phosphoribosyltransferase are present in Bacillus subtilis. J. Bacteriol. 177:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinussen, J., J. Schallert, B. Andersen, and K. Hammer. 2001. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 183:2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinussen, J., S. L. Wadskov-Hansen, and K. Hammer. 2003. Two nucleoside uptake systems in Lactococcus lactis: competition between purine nucleosides and cytidine allows for modulation of intracellular nucleotide pools. J. Bacteriol. 185:1503-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masson, A., B. Kammerer, and J. C. Hubert. 1994. Selection and biochemical studies of pyrimidine-requiring mutants of Lactobacillus plantarum. J. Appl. Bacteriol. 77:88-95. [Google Scholar]
- 16.Mechold, U., and H. Malke. 1997. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J. Bacteriol. 179:2658-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuhard, J., and R. A. Kelln. 1996. Biosynthesis and conversion of pyrimidines, p. 580-599. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Linn, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 18.Nicoloff, H., and F. Bringel. 2003. ISLpl1 is a functional IS30-related insertion element in Lactobacillus plantarum that is also found in other lactic acid bacteria. Appl. Environ. Microbiol. 69:5832-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicoloff, H., J. C. Hubert, and F. Bringel. 2000. In Lactobacillus plantarum, carbamoyl phosphate is synthesized by two carbamoyl-phosphate synthetases (CPS): carbon dioxide differentiates the arginine-repressed from the pyrimidine-regulated CPS. J. Bacteriol. 182:3416-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savacool, H. K., and R. L. Switzer. 2002. Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J. Bacteriol. 184:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxild, H. H., and P. Nygaard. 1991. Regulation of levels of purine biosynthetic enzymes in Bacillus subtilis: effects of changing purine nucleotide pools. J. Gen. Microbiol. 137:2387-2394. [DOI] [PubMed] [Google Scholar]
- 22.Switzer, R. L., R. J. Turner, and Y. Lu. 1999. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acids Res. Mol. Biol. 62:329-367. [DOI] [PubMed] [Google Scholar]
- 23.Tomchick, D. R., R. J. Turner, R. L. Switzer, and J. L. Smith. 1998. Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure 6:337-350. [DOI] [PubMed] [Google Scholar]
- 24.Turner, R. J., Y. Lu, and R. L. Switzer. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 176:3708-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, H., and R. L. Switzer. 2003. Transcriptional pausing in the Bacillus subtilis pyr operon in vitro: a role in transcriptional attenuation? J. Bacteriol. 185:4764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]