Abstract
The low-molecular-weight protein tyrosine phosphatase (LMWPTPase) belongs to a distinctive class of phosphotyrosine phosphatases widely distributed among prokaryotes and eukaryotes. We report here the crystal structure of LMWPTPase of microbial origin, the first of its kind from Mycobacterium tuberculosis. The structure was determined to be two crystal forms at 1.9- and 2.5-Å resolutions. These structural forms are compared with those of the LMWPTPases of eukaryotes. Though the overall structure resembles that of the eukaryotic LMWPTPases, there are significant changes around the active site and the protein tyrosine phosphatase (PTP) loop. The variable loop forming the wall of the crevice leading to the active site is conformationally unchanged from that of mammalian LMWPTPase; however, differences are observed in the residues involved, suggesting that they have a role in influencing different substrate specificities. The single amino acid substitution (Leu12Thr [underlined below]) in the consensus sequence of the PTP loop, CTGNICRS, has a major role in the stabilization of the PTP loop, unlike what occurs in mammalian LMWPTPases. A chloride ion and a glycerol molecule were modeled in the active site where the chloride ion interacts in a manner similar to that of phosphate with the main chain nitrogens of the PTP loop. This structural study, in addition to identifying specific mycobacterial features, may also form the basis for exploring the mechanism of the substrate specificities of bacterial LMWPTPases.
The emergence in recent years of multidrug-resistant strains of Mycobacterium tuberculosis and HIV coinfection has made tuberculosis a serious threat to mankind. Approximately 2 billion people are infected with this dreaded disease, and 8.8 million new cases of tuberculosis occur annually, victimizing 50,000 persons each week (19). The mechanisms of entry and survival of the tubercle bacillus in the host cell are partially known. M. tuberculosis overcomes host defenses by preventing phagosome-lysosome fusion (2), inhibiting phagosome acidification (38), and recruiting as well as retaining tryptophan-aspartate-containing coat proteins on phagosomes to prevent their delivery to lysosomes (14). Kinases and phosphatases are major families of enzymes involved in signal transduction and play key roles in intracellular event coordination (6, 35, 36). The genes encoding a protein tyrosine phosphatase (PTPase) and a protein tyrosine kinase in a bacterial species are often located next to each other, generally as part of large operons. These genes direct the coordinated synthesis of proteins involved in the production or regulation of exopolysaccharides and capsular polysaccharides. There exists a direct relationship between the reversible phosphorylation of proteins at the tyrosine residue and the production of these carbohydrate polymers (10). The Salmonella enterica serovar Typhimurium protein tyrosine phosphatase SptP is an essential component of the system for the interaction of these bacteria with host cells (15). It is anticipated that tyrosine kinases and phosphatases are major virulence determinants (10, 21, 34), and they are considered to be potential drug targets.
Covalent modification of proteins by reversible phosphorylation and dephosphorylation is an essential physiological mechanism for the control of cellular functions. A wide variety of cellular processes, such as cell division, differentiation, and development, are controlled by molecular signals obtained through cascades of phosphorylation and dephosphorylation of proteins (17, 40). Protein phosphorylation and dephosphorylation are controlled by the enzymatic actions of kinases and phosphatases, respectively (20, 28, 31), on serine, threonine, histidine, and tyrosine residues. Histidine kinases are present in prokaryotic signal transduction systems such as the two-component system (20). The dephosphorylation of phosphohistidine takes place with PP1, PP2A, and PP2C (26). PTPases reverse the effect of tyrosine kinases by dephosphorylating the protein tyrosine residue of cellular substrate proteins and are important in cellular signaling processes (17, 29, 36). PTPases in general are divided into the following three families, based on their molecular masses and substrate specificities: the high-molecular-mass PTPase family, whose members have a conserved 30-kDa catalytic domain; the dual-specificity PTPase family (11, 43), whose members are capable of dephosphorylating both serine-threonine and tyrosine residues; and the low-molecular-mass PTPase family, whose members contain a single 18-kDa catalytic domain (44). High-molecular-weight PTPases include the following: the nonreceptor type of phosphatase, such as PTP1B (3, 4, 24); Yersinia PTPase (37, 48); and the receptor-like PTPases, such as CD45, μ, and LAR (16). All these families of PTPases exhibit very low sequence identities, except in a common active-site signature motif, CXXXXXR(S/T), at the phosphate-binding site (44).
The low-molecular-weight PTPases (LMWPTPases) exhibit no apparent sequence homology with the high-molecular-weight phosphatases apart from the minimal PTPase conserved signature motif C(X)5R(S/T) situated near the N terminus (45). The wide distribution of the low-molecular-weight PTPases among organisms from bacteria to humans suggests their pivotal role in cellular functions and substrate specificity (23, 25, 27, 32).
The common catalytic mechanism of dephosphorylation of the substrates by PTPases involves the conserved active-site sequence motif C(X)5R(S/T), which is called the protein tyrosine phosphate-binding loop (PTP loop) (39). Structural studies of eukaryotic LMWPTPases have demonstrated that the phosphate ion is cradled between the catalytic cysteine and arginine side chains, stabilizes the phosphate by hydrogen bonding, and proceeds through a cysteinyl-phosphate intermediate (47). Cysteine acts as the nucleophile, and its thiolate form binds covalently to the phosphate group of the substrate protein along with the side chain of the arginine located 6 residues downstream of the catalytic cysteine residue. Also, the backbone nitrogens of the PTP loop form hydrogen bonds with the phosphate group of the substrate (47). PTPases are active over a broad range of pHs, i.e., from pHs 5.5 to 7.5, but are inhibited by low concentrations of vanadates. The substrate specificity of LMWPTPase might be determined by the charge distribution around the active site, and distinct charge alterations near the active sites of different LMWPTPases are expected to recognize amino acids with different charges as part of potential peptide or protein substrates (8, 12, 42, 46).
Sequence analysis of the M. tuberculosis genome revealed the presence of two mycobacterial PTPases, mycobacterial PtpA (MPtpA) and MPtpB. MPtpA belongs to the LMWPTPase family and is present in slow-growing mycobacterial species as well as fast-growing saprophytes, whereas MPtpB is a 30-kDa protein and is restricted to members of the M. tuberculosis complex. These two PTPases are released by M. tuberculosis into extracellular culture medium and are implicated in the interaction of mycobacteria with the host cell (21). Disruption of the MPtpB gene impairs the ability of M. tuberculosis to survive in guinea pigs and demonstrates the role of PTPases in the pathogenesis of disease caused by M. tuberculosis (34). MPtpA is highly specific for substrates containing phosphotyrosine residues and shows some activity on myelin basic protein and no activity on phosphoserine or phosphothreonine (9). The MPtpA promoter expresses the protein when cloned into the slow-growing Mycobacterium bovis BCG and is also expressed during infection of human macrophages (9).
Here we report the crystal structures of a low-molecular-weight protein tyrosine phosphatase of M. tuberculosis (MPtpA) at 1.9- and 2.5-Å resolutions and present a detailed structural and sequence comparison with other low-molecular-weight protein tyrosine phosphatases of humans (HCPTPA) (46), bovines (BPTP) (39, 45), and Saccharomyces cerevisiae (LTP1) (42) to understand its specific features and role in substrate recognition and binding.
MATERIALS AND METHODS
Preparation and crystallization of MPtpA.
M. tuberculosis H37Rv genomic DNA was used for amplification of the mptpA gene (Rv2234) through PCR. The mptpA gene was amplified using the forward primer ptpA10F (5′-ATCGAGCTCGTGTCTGATCCGCTGCACG-3′), which has a Sac1 restriction site (in boldface), and the reverse primer ptpA10R (5′-ATCCCAAGCTTTCAACTCGGTCCGTTCCG-3′), which has a HindIII restriction site. The amplified product of mptpA was sequentially digested with SacI (New England Biolabs) and HindIII (New England Biolabs), and the resulting fragment was inserted into the pQE30 plasmid (QIAGEN) digested with SacI and HindIII. The pQE30-mptpA plasmid was transformed into the SG13009 strain of Escherichia coli to produce recombinant His-tagged MPtpA protein. Transformants were grown in Luria-Bertani broth containing 100 μg of ampicillin per ml and 50 μg of kanamycin per ml at 37°C until the A595 reached 0.8. Induction was done with isopropyl-1-thio-β-galactopyranoside (IPTG) to a final concentration of 0.1 mM, and cultures were incubated overnight at 15°C. The cells were then harvested by centrifugation at 10,375 × g for 10 min, and the pellet was resuspended in lysis buffer (20 mM Tris HCl [pH 8.2] containing 100 mM NaCl; 10% [vol/vol] glycerol; 0.1 mM [each] leupeptin, pepstatin, aprotinin, and lysozyme; and 0.02 mM phenylmethylsulfonyl fluoride). The cells were lysed by ultrasonication on ice and subsequently centrifuged at 21,036 × g for 40 min. The supernatant was loaded onto a Ni-nitrilotriacetic acid column (Novagen), preequilibrated with buffer A (20 mM Tris HCl [pH 8.0], 20 mM NaCl, and 10% glycerol), and eluted with buffer B (10 mM l-histidine in buffer A [pH 8.0]). The eluted protein was dialyzed overnight against buffer C (10 mM Tris HCl [pH 8.0], 50 mM NaCl, 1 mM dithiothreitol, and 10% glycerol). The protein sample was loaded onto a Q-Sepharose anion-exchange column (Amersham Biosciences Ltd.) preequilibrated with buffer C. The protein was eluted with a linear gradient to 1 M NaCl. The obtained sample was concentrated by using a 50-ml Amicon stirred ultrafiltration cell (model 8050) with a YM 10 membrane, and the concentrated sample was loaded onto a 250-ml Sephadex G-75 column equilibrated with buffer D (10 mM Tris HCl [pH 8.0], 50 mM NaCl, 1 mM dithiothreitol, 5 mM EDTA, 0.02 mM phenylmethylsulfonyl fluoride, and 10% glycerol). All the chemicals were purchased from Sigma Aldrich Chemicals Pvt. Ltd. Fourteen milligrams of MPtpA could be obtained from 4 liters of culture. The eluted protein was concentrated to 12.6 mg/ml for the crystallization trials.
Initial searches for crystallization conditions were made at 277 K using the commercially available sparse-matrix screening kits Crystal Screen and Crystal Screen 2 from Hampton Research. The hanging-drop vapor diffusion method was used, where each drop, containing equal volumes (2 μl) of protein and reservoir solution, was equilibrated against 750 μl of reservoir solution. Crystals were obtained under two different conditions and were classified as MPtpA (A) and MPtpA (B). Rod-shaped MPtpA (A) crystals were obtained from 22% (vol/vol) ethylene glycol with dimensions of 0.1 by 0.1 by 1.0 mm, while diamond-shaped MPtpA (B) crystals were grown in a solution containing 30% (wt/vol) polyethylene glycol 4000, 200 mM ammonium acetate, and 100 mM trisodium citrate dihydrate [pH 5.6] and had dimensions of 0.3 by 0.3 by 0.6 mm.
Data collection.
Data were collected using a MAR Research MAR-345dtb image plate detector with Cu Kα X rays generated by a Rigaku RU-H3R rotating anode X-ray generator equipped with an osmic mirror system and operated at 50 kV and 100 mA. Prior to flash cooling in a liquid nitrogen stream at 100 K for data collection, MPtpA (A) crystals were soaked in 10% glycerol along with the mother liquor for 60 to 120 s. The data were processed using DENZO (33), and the subsequent scaling and merging of intensities were carried out using SCALEPACK (33). Both crystal forms belong to the orthorhombic (P212121) space group and have different cell dimensions. MPtpA (A) and MPtpA (B) crystals diffracted to 1.9- and 2.5-Å resolutions, respectively, and both contained one molecule per asymmetric unit (AU). The solvent contents of MPtpA (A) and MPtpA (B) crystals were 43.7 and 48.2%, respectively. A complete data set was collected from both crystal forms, and the statistical data are given in Table 1.
TABLE 1.
Summary of crystallographic data and refinement statistics
| Data | Crystal structurea
|
|
|---|---|---|
| MPtpA (A) | MPtpA (B) | |
| Crystal parameters | ||
| Space group | P212121 | P212121 |
| Cell parameters | ||
| a (Å) | 40.82 | 49.31 |
| b (Å) | 53.61 | 53.00 |
| c (Å) | 68.49 | 64.15 |
| Solvent content | 43.7 | 48.2 |
| Z (no. of monomers/AU) | 1 | 1 |
| Data processing | ||
| Resolution range (Å) | 25-1.9 | 25-2.5 |
| No. observations | 71,972 (6,691) | 14,498 (719) |
| No. of unique reflections | 12,389 (1,215) | 5,833 (435) |
| Completeness (%) | 99.7 (100.0) | 93.7 (71.9) |
| Multiplicity (%) | 5.8 (5.7) | 2.48 (1.6) |
| Rsymb | 4.7 (6.7) | 6.4 (13.5) |
| I/sigma (I) | 40.4 (23.3) | 13.8 (4.5) |
| Refinement statistics | ||
| Resolution range (Å) | 25-1.9 | 25.0-2.5 |
| No. of reflections (F > 0σ) | ||
| No. of working statistics | 11,693 | 5,517 |
| No. of tests | 616 | 301 |
| R factorc | ||
| Rcryst (%) | 20.1 | 21.2 |
| Rfreed (%) | 22.6 | 27.3 |
| No. of atoms/AU | ||
| Protein | 1,211 | 1,200 |
| Water | 152 | 83 |
| Chloride ion | 1 | 1 |
| Glycerol | 6 | |
| RMS deviation from ideal values | ||
| Bond length (Å) | 0.0048 | 0.0061 |
| Bond angle (°) | 1.18° | 1.23° |
| Ramachandran plot values | ||
| In most-favored regions (%) | 92.6 | 93.3 |
| In additional allowed regions (%) | 7.4 | 6.7 |
| Avg B factors (Å2) | ||
| Protein | 13.8 | 27.7 |
| Water | 24.4 | 31.4 |
Values in parentheses throughout are for the highest resolution shell.
Rsym = Σhkl Σi|Ii(hkl) − <I(hkl)>|/Σhkl Σi Ii(hkl).
R factor = Σhkl||Fo(hkl)| − |Fc(hkl)||/Σhkl|Fo(hkl)|.
Rfree calculated for a random set of 5% of reflections not used in the refinement.
Structure determination.
The structure of MPtpA was determined by the molecular replacement method using the program MOLREP-AUTO MR (41) in the CCP4 suite (1). MPtpA exhibits a sequence homology with HCPTPA of 38%. The structure of MPtpA (A) was determined using HCPTPA polyalanine (Protein Data Bank code, 5PNT) as a search model. The rotation and translation function solutions were obtained by using the search model and the X-ray data in the 25- to 3.5-Å resolution range. The resulting solution had a correlation coefficient and R factor of 0.565 and 41.6%, respectively. After rigid-body refinement followed by a simulated-annealing protocol, the SIGMAA-weighted 2Fo-Fc and Fo-Fc maps were calculated using CNS (7). The two loops, residues 100 to 106 and 113 to 125, and a helix region, residues 150 to 158, were built manually in the electron density by using program O (18). A strong electron density peak of 5.5 σ in the Fo-Fc map was tentatively fitted as a chloride ion based on temperature factor refinement. After a few cycles of refinement with CNS and model building with program O, the structure could be refined to an Rfree and Rcryst of 22.6 and 20.1%, respectively. The PROCHECK program (22) was used to generate the Ramachandran diagram. Assessment of the model quality showed that 92.6% of the residues were in the most favored region and that the rest were in the allowed region. The refinement statistics for the MPtpA (A) crystal are given in Table 1.
The MPtpA (B) crystal structure was also determined by the molecular replacement method using MPtpA (A) as a search model. An electron density in the active-site region mimicking the substrate was modeled as glycerol based on its electron density and the temperature factor of surrounding protein atoms. The final structure was refined to an Rfree and Rcryst of 27.3 and 21.1%, respectively. The refinement statistics of the MPtpA (B) crystal form are shown in Table 1. Residues 4 to 159 and 4 to 158 in the MPtpA (A) and the MPtpA (B) structures were modeled in 2Fo-Fc and Fo-Fc maps, respectively. The temperature factor of the chloride ion was 14.5 Å2 in MPtpA (A), whereas in MPtpA (B), it was 39.8 Å2. The average temperature factor of glycerol in MPtpA (B) was 36.8 Å2.
Protein Data Bank accession numbers.
The coordinates of MPtpA (A) and MPtpA (B) have been deposited in the Protein Data Bank with accession codes 1U2P and 1U2Q, respectively.
RESULTS AND DISCUSSION
Overall structure of MPtpA.
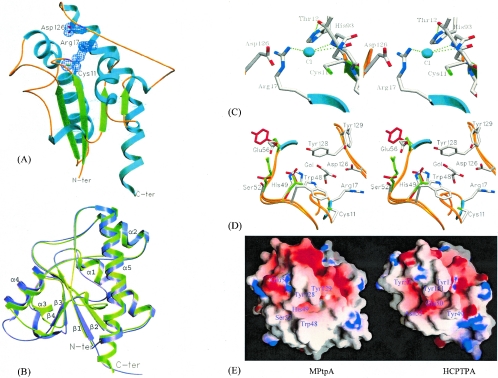
The overall structure of MPtpA (A) was very similar to that of MPtpA (B), and the root-mean-square (RMS) deviation at the Cα positions of MPtpA (A) relative to the RMS of MPtpA (B) was 0.397 Å. Also, there were no differences in core packing or accessible surface area between the two structures. A comparison of the B factors of the protein atoms indicated that MPtpA (A) is more ordered than MPtpA (B), and the overall B factors of protein atoms of MPtpA (A) and MPtpA (B) were 14.96 and 28.20 Å2, respectively. The following discussion is based on MPtpA (A) unless otherwise indicated. The MPtpA structure was composed of a single α/β domain characterized by a central, parallel, four-stranded, twisted β sheet flanked by α helices on both sides. This overall structure has the character of a classic Rossman fold, with two right-handed βαβ motifs contributing to the central four-stranded parallel β-sheet (Fig. 1A). A typical phosphate-binding loop (PTP loop) lies at the C terminus of β1 and the N terminus of α1. The structure-based multiple sequence alignment is shown in Fig. 2. The overall topology of MPtpA closely resembles that of eukaryotic low-molecular-weight PTPases, and the major changes found were in insertions or deletions. Four amino acids were deleted from and 3 amino acids were inserted in the loop regions, i.e., in residues 100 to 107 and 115 to 124, respectively, and the α5 helix contained an additional 6 residues compared to the HCPTPA structure (Fig. 1B). Proline at position 147 (Arg150 in HCPTPA) created a kink in the α5 helix, resulting in the loss of the main chain hydrogen bonds 147N—143O and 148N—144O.
FIG. 1.
Crystal structure of M. tuberculosis low-molecular-weight MPtpA. (A) Ribbon diagram of the structure of MPtpA (A). The disposition of the three invariant catalytic residues is shown with a 2Fo-Fc electron density map contoured at 1.1 σ. N-ter, N terminus; C-ter, C terminus. (B) Structural superposition of MPtpA (A) (green) with HCPTPA (purple). Large structural deviations could be noted among the β4-to-α5 (insertion), α4-to-β4 (deletion), α3-to-β3 regions, and the extended α5 helix. N-ter and C-ter labels apply to the MPtpA structure. (C) Stereo diagram showing the modeled chloride ion in the phosphate-binding site of the active-site region. Thr12, being involved in additional stabilization of the PTP loop through its interaction with His93, is indicated. (D) Superposition of the active-site region of MPtpA (B) with HCPTPA and BPTP shown in stereo view. The residues implicated in substrate specificity are His49 (Glu in HCPTPA and Asn in BPTP), Ser52 (Asn in HCPTPA and Arg in BPTP), and Glu56 (Tyr in HCPTPA and Pro in BPTP) and are indicated with glycerol in the active-site region. The side-chain atoms of MPtpA and glycerol are shown in atomic-color mode (O, N, and S atoms are in red, blue, and green, respectively), while HCPTPA and BPTP side chains are shown in red and green, respectively. The depictions in panels A to D were generated using the SETOR program (13). (E) View of the active site showing the surface charge distributions of MPtpA (A) (left) and HCPTPA (right), which are displayed with crucial residues that determine the indicated substrate specificities. For MPtpA, the crevice leading to the active site is indicated by tryptophan and aromatic residues positioned on either side of the active site. This depiction was generated using the GRASP program (30).
FIG. 2.
Structure-based sequence alignment of the low-molecular-weight PTPases MPtpA, BPTP, HCPTPA, and LTP1, along with those from E. coli, and S. enterica serovar Typhi. The active-site signature motifs of the PTP loop and the variable-loop residues in MPtpA are shown in boxes. The residues presumed to determine substrate specificities and that differ from those of mammalian PTPases are shaded. The residues involved in MPtpA PTP loop stabilization are underlined. This figure was created using the program ALSCRIPT (4).
The RMS deviations in the Cα positions of MPtpA from those of HCPTPA, BPTP, and LTP1 were 0.99, 1.07, and 1.02 Å, respectively, but the RMS deviation between two mammalian LMWPTPases, HCPTPA and BPTPA, was 0.496 Å. MPtpA showed 38, 37, and 33% sequence homologies with HCPTPA, BPTP, and LTP1, respectively, whereas the sequence homology between the mammalian LMWPTPases HCPTPA and BPTP was 82%. These noticeable differences in RMS deviations and sequence homologies indicated that the structure of MPtpA is significantly different from that of mammalian LMWPTPases. Major changes from the HCPTPA structure were observed in the regions between β4 and α5 (insertion), α4 and β4 (deletion), and α3 and β3 (Fig. 1B). The variable loop (residues 44 to 56), attributed to unique catalytic properties of isoenzymes that vary in size (46), was conformationally unchanged (RMS deviation of 0.342 Å in Cα with HCPTPA); however, its residues are not conserved, in contrast to those of mammalian LMWPTPases.
A secondary-structure-based multiple sequence alignment of the MPtpA was made with other prokaryotic LMWPTPases, such as S. enterica serovar Typhi and E. coli, and with eukaryotes, such as Saccharomyces cerevisiae, Homo sapiens, and Bos taurus (Fig. 2). The amino acids Glu22, His65, His71, and Glu136 are highly conserved in the LMWPTPase family. Differences in the variable-loop regions are revealed by structural and multiple sequence alignment. The two proline amino acids in mammalian LMWPTPases (residues 53 and 54 in HCPTPA and residues 54 and 56 in BPTP) are replaced by different amino acids in MPtpA (Fig. 2). The pyrolidine ring of proline restricts this residue to fewer conformations than are available to the other amino acids. As a consequence, the variable loops of mammalian LMWPTPases might be conformationally more stable than that of MPtpA. There is considerable evidence that the variable loop plays a key role in determining the nature of the substrate (42, 46), so the variable loop in MPtpA might help the enzyme to recognize different types of substrates. In addition to these changes in the variable loop specific to MPtpA, the charge distribution around the active site is significantly altered in comparison to that of the mammalian LMWPTPases, due to negatively charged Glu56. The structural analysis shows that Arg65 in HCPTPA (46) and Asn65 in BPTP (45) interact with Glu139 but that these residues, Arg65 and Asn65, are mutated to a hydrophobic residue, Ala64, in MPtpA (Fig. 2), resulting in a loss of interaction with Glu136. Hence, the negative charge on Glu136 is shared only with His65, and the pKa value of His65 in MPtpA might be different from that of HCPTPA and BPTP. Also, the pKa value of His71 of MPtpA differs from that of mammalian LMWPTPases, due to the presence of Thr41 instead of Asp42 in mammalian LMWPTPases (Fig. 2), whereas Glu23 and Asp42 play a crucial role in changing the pKa value of His72 in BPTP (45) and HCPTPA (46). His71 stabilizes the left-handed conformation of Asn14 by interacting with Nɛ2 and, in turn, stabilizes the PTP loop. These differences in the variable loops indicate the different substrate preferences of MPtpA and mammalian LMWPTPases. The specific activity of MPtpA (0.5 μΜ/min/mg) (9) is nearly 200 times lower than that of the LMWPTPases (95 to 100 μΜ/min/mg) (46) when the substrate p-nitrophenyl phosphate is used; this difference can be attributed to different amino acids around the active site.
Structure of the active site.
Despite the overall similarities in the active-site architectures and, hence, in the enzymatic mechanisms, significant differences between prokaryotic and eukaryotic LMWPTPases, mammalian LMWPTPases in particular, can be noted. These differences, along with mycobacterium-specific changes, are described in detail below. The active site of MPtpA reveals the presence of the PTP loop (residues 11 to 18) with two active-site residues at positions 11 and 17, an aspartic acid residue at position 126, and a group of hydrophobic residues that form the wall of the active-site cavity (Fig. 1A and D). The favorable binding of the tyrosine ring of the substrate and the substrate selectivity of the enzyme are duly attributed to the facts that the active-site cavity is deep and the residues lining the walls of the cavity are mainly aromatic and hydrophobic in nature (Fig. 1D and E). The P loops are formed by the active-site consensus sequence motifs, CTGNICRS in MPtpA and CLGNICRS in mammalian low-molecular-weight PTPases (46). The Leu12Thr substitution (underlined in the MPtpA sequence) may play a major role in PTP loop stabilization. Leu12 is a hydrophobic residue present along with the charged residues at the surface of the molecule facing the solvent region in mammalian LMWPTPases, and a change to a polar residue may be stabilizing. Also, the side chain Oγ1 of Thr12 makes a hydrogen-bonding interaction with the side chain Nɛ2 of His93 (Fig. 1C), present only in bacterial LMWPTPases, whereas it is replaced by Asn in all mammalian LMWPTPases (Fig. 2). The side chain Nδ1 of His93, in turn, interacts with the main chain nitrogen and side chain Oδ1 of Asp90. The PTP loop is a well-ordered region in both structures and has low temperature factors for both its main chain and side chain atoms. The temperature factors of the main chain and side chain atoms in the PTP loop were 10.4 and 10.65 Å2, respectively, in the MPtpA (A) structure. These temperature factors are lower than the overall temperature factors of the main chain and side chain atoms of the MPtpA (A) structure, which were 12.31 and 15.32 Å2, respectively. An extensive network of H bonds in the three-dimensional structure, in accordance with a lower temperature factor, stabilizes the PTP loop conformation. However, the backbone structure of the PTP loop in this enzyme is very similar to that of other protein tyrosine phosphatases whose structures are known (42, 45, 46).
The PTP loop contains the residue Asn14, which is in a left-handed helical conformation (φ = 59°; ψ = 34°), as is evident from the Ramachandran plot. This residue forms a network of H bonds with conserved residues Ser18, Ser42, and His71, which help to stabilize the left-handed conformation and thus the overall structure of the PTP loop, allowing it to adopt the most favorable geometry for substrate binding and transition state stabilization. Asn14 is highly conserved in all LMWPTPases. Arg17 is a key residue for substrate binding in MPtpA and is equivalent to Arg18 in HCPTPA. The PTP loop residues are arranged in such a way that all the amide protons between Cys11 and Ser18 face the center of the loop, and they are well positioned for phosphate binding; Arg17 also contributes two hydrogen bonds for interaction with phosphate oxygen. Both of the structures exhibited a chloride ion at the phosphate-binding region of the active site that interacts with the main chain nitrogens of Thr12 and Gly13 and with the side chain NH2 of Arg17 in MPtpA (A) (Fig. 1C). In the case of the MPtpA (B) structure, the position of the chloride ion is shifted by 3.45 Å with respect to that of MPtpA (A), and the chloride ion interacts with the main chain nitrogens of Ile15 and Cys16 due to the additional binding of the glycerol moiety in the active site.
Comparison of the active site with those of the other eukaryotic LMWPTPases indicated that MPtpA also has a crevice leading from one side of the molecule into the active site. The variable loop forms one wall and floor of the crevice, and the loop between β4 and α5 containing the catalytically active residue Asp 126 forms the other wall. The crevice formed by Trp48, Tyr128, and Tyr129 creates a hydrophobic surface for the incoming phosphotyrosine substrate (Fig. 1D and E). The side chains of these residues line the wall near the active site. The charge pattern observed in the variable-loop region of MPtpA is different than that of others. The presence in the variable loop of His49 and Ser52 in MPtpA compared to the presence of Glu50 and Asn53 in HCPTPA and that of Asn50 and Arg53 in BPTP indicates the change in pattern of charge distribution around the wall of the crevice leading to the aromatic region at the active site (46) (Fig. 1D and E). The fact that these two residues are in close proximity to the active site, where phosphotyrosine binds, indicates that these two residues at positions 49 and 52 may be crucial in determining the substrate specificity. In bacterial LMWPTPases, His49 is either conserved (except in Streptococcus spp. and Bifidobacterium spp., where a Glu is present) or deleted, in contrast to what occurs with mammalian LMWPTPases, where this position is occupied by different amino acids (Fig. 2). The glutamate at position 91 in mammalian LMWPTPases is replaced by a positively charged residue, arginine, in MPtpA. In addition to this substitution, the presence of charged or polar residues (for example, Asp in MPtpA, where the side chain carboxylate group of Asp faces the active site) at position 123 in most bacterial LMWPTPases (except Photorhabdus spp.), compared to the presence of a hydrophobic residue, Ile, around the active site in mammalian LMWPTPases, alters the charge distribution significantly (Fig. 2). Thus, this unique charge distribution contributed by Asp123, Arg91, and His49 around the active site along with aromatic residues Trp48, Tyr128, and Tyr129 might have a role in the recognition of amino acids with different charges as part of potential peptide or protein substrates, in contrast to what is observed in mammalian LMWPTPases. This distinction can also explain the difference in the specific activities of bacterial and mammalian LMWPTPases towards the substrate p-nitrophenyl phosphate (9, 46).
We have presented here the structure of a low-molecular-weight protein tyrosine phosphatase in two crystal forms. This is the first structural report of a bacterial representative of LMWPTPases. Even though the structural comparison of the MPtpA with the eukaryotic LMWPTPases revealed overall structural similarities, features specific to bacterial enzymes and to mycobacteria could be discerned. These features include additional stabilization of the PTP loop and significant differences in the residues surrounding the active-site region besides common hydrophobic residues lining the active site. Though the structure of MPtpA is similar to those of other LMWPTPases, it may have a different substrate specificity that may lead to different physiological functions. Thus, this structural work provides a solid platform for exploring the mechanisms of the substrate specificities of MPtpA and other bacterial LMWPTPases that may be identified by biochemical and/or genetic screens.
Acknowledgments
This work was supported by the Indo-Norwegian Programme for Institutional Cooperation (INPIC) grant IND-040. E.R. and P.A.M are Senior Research Fellows of the Council of Scientific and Industrial Research (India). R.S. is a Wellcome Trust International Senior Research Fellow in Biomedical Science in India.
Dominique Caugant at the Norwegian Institute of Public Health is acknowledged for supplying DNA from M. tuberculosis. We thank Debabrata Das for support and encouragement.
REFERENCES
- 1.Acta Crystallographica D. 1994. Collaborative computational project number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., and P. D. Hart. 1975. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J. Exp. Med. 142:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barford, D., A. J. Flint, and N. K. Tonks. 1994. The crystal structure of human protein tyrosine phosphatase 1B. Science 263:1397-1404. [PubMed] [Google Scholar]
- 4.Barton, G. J. 1993. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 6:37-40. [DOI] [PubMed] [Google Scholar]
- 5.Bilwes, A. M., J. Den Hertog, T. Hunter, and J. P. Noel. 1996. Structural basis for inhibition of receptor protein-tyrosine phosphatase-alpha by dimerization. Nature 382:555-559. [DOI] [PubMed] [Google Scholar]
- 6.Blenis, J. 1993. Signal transduction via the MAP kinases—proceed at your own RSK. Proc. Natl. Acad. Sci. USA 90:5889-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. Delano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Cirri, P., T. Fiaschi, P. Chiarugi, G. Camici, G. Manao, G. Raugei, and G. Ramponi. 1996. The molecular basis of the differing kinetic behavior of the two low molecular mass phosphotyrosine protein phosphatase isoforms. J. Biol. Chem. 271:2604-2607. [DOI] [PubMed] [Google Scholar]
- 9.Cowley, S. C., R. Babakaiff, and Y. Av-Gay. 2002. Expression and localization of the Mycobacterium tuberculosis protein tyrosine phosphatase PtpA. Res. Microbiol. 153:233-241. [DOI] [PubMed] [Google Scholar]
- 10.Cozzone, A. J., C. Grangeasse, P. Doublet, and B. Duclos. 2004. Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 181:171-181. [DOI] [PubMed] [Google Scholar]
- 11.Denu, J. M., G. Zhou, Y. Guo, and J. E. Dixon. 1995. The catalytic role of aspartic acid-92 in a human dual-specific protein-tyrosine-phosphatase. Biochemistry 34:3396-3403. [DOI] [PubMed] [Google Scholar]
- 12.Evans, B., P. A. Tishmack, C. Pokalsky, M. Zhang, and R. L. Van Etten. 1996. Site-directed mutagenesis, kinetic, and spectroscopic studies of the P-loop residues in a low molecular weight protein tyrosine phosphatase. Biochemistry 35:13609-13617. [DOI] [PubMed] [Google Scholar]
- 13.Evans, S. V. 1993. SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. Model. 11:134-138. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari, G., H. Langen, M. Naito, and J. Pieters. 1999. A coat protein on phagosomes involved in the intracellular survival of mycobacteria. Cell 97:435-447. [DOI] [PubMed] [Google Scholar]
- 15.Fu, Y., and J. E. Galan. 1998. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol. Microbiol. 27:359-368. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann, K. M., N. K. Tonks, and D. Barford. 1997. The crystal structure of domain 1 of receptor protein-tyrosine phosphatase μ. J. Biol. Chem. 272:27505-27508. [DOI] [PubMed] [Google Scholar]
- 17.Hunter, T. 1995. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signalling. Cell 80:225-236. [DOI] [PubMed] [Google Scholar]
- 18.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 19.Kamholz, S. L. 2002. Drug resistant tuberculosis. J. Assoc. Acad. Minor. Phys. 13:53-56. [PubMed] [Google Scholar]
- 20.Klumpp, S., and J. Krieglstein. 2002. Phosphorylation and dephosphorylation of histidine residues in proteins. Eur. J. Biochem. 269:1067-1071. [DOI] [PubMed] [Google Scholar]
- 21.Koul, A., A. Choidas, M. Treder, A. K. Tyagi, K. Drlica, Y. Singh, and A. Ullrich. 2000. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 182:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 23.Li, Y., and W. R. Strohl. 1996. Cloning, purification, and properties of a phosphotyrosine protein phosphatase from Streptomyces coelicolor A3(2). J. Bacteriol. 178:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohse, D. L., J. M. Denu, N. Santoro, and J. E. Dixon. 1997. Roles of aspartic acid-181 and serine-222 in intermediate formation and hydrolysis of the mammalian protein-tyrosine-phosphatase PTP1. Biochemistry 36:4568-4575. [DOI] [PubMed] [Google Scholar]
- 25.Manao, G., L. Pazzagli, P. Cirri, A. Casell, G. Camici, G. Cappugi, A. Saeed, and G. Ramponi. 1992. Rat liver low M(r) phosphotyrosine protein phosphatase isoenzymes: purification and amino acid sequences. J. Protein Chem. 11:333-345. [DOI] [PubMed] [Google Scholar]
- 26.Matthews, H. R. 1995. Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins: a possible regulator of the mitogen activated protein kinase cascade. Pharmacol. Ther. 67:323-350. [DOI] [PubMed] [Google Scholar]
- 27.Modesti, A., P. Cirri, F. Raugei, L. Carraresi, F. Magherini, G. Manao, G. Camici, and G. Ramponi. 1995. Expression, purification and kinetic behaviour of fission yeast low Mr protein-tyrosine phosphatase. FEBS Lett. 375:235-238. [DOI] [PubMed] [Google Scholar]
- 28.Murray, A. W. 1992. Creative blocks: cell-cycle checkpoints and feedback controls. Nature 359:599-604. [DOI] [PubMed] [Google Scholar]
- 29.Neel, B. G., and N. K. Tonks. 1997. Protein tyrosine phosphatases in signal transduction. Curr. Opin. Cell Biol. 9:193-204. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls, A. N., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11:281-296. [DOI] [PubMed] [Google Scholar]
- 31.Nurse, P. 1990. Universal control mechanism regulating onset of M-phase. Nature 334:503-508. [DOI] [PubMed] [Google Scholar]
- 32.Ostanin, K., C. Pokalsky, S. Wang, and R. L. Van Etten. 1995. Cloning and characterization of a Saccharomyces cerevisiae gene encoding the low molecular weight protein tyrosine phosphatase. J. Biol. Chem. 270:18491-18499. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 34.Singh, R., V. Rao, H. Shakila, R. Gupta, A. Khera, N. Dhar, A. Singh, A. Koul, Y. Singh, M. Naseema, P. R. Narayanan, C. N. Paramasivan, V. D. Ramanathan, and A. K. Tyagi. 2003. Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol. Microbiol. 50:751-762. [DOI] [PubMed] [Google Scholar]
- 35.Stein, E., A. A. Lane, D. P. Ceretti, H. O. Shoecklnann, A. D. Schroff, R. L. Van Etten, and O. T. Daniel. 1998. Eph receptors discriminate specific ligand oligomers to determine alternative signaling complexes, attachment, and assembly responses. Genes Dev. 12:667-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streuli, M. 1996. Protein tyrosine phosphatases in signaling. Curr. Opin. Cell Biol. 8:182-188. [DOI] [PubMed] [Google Scholar]
- 37.Stuckey, J. A., H. L. Schubert, E. B. Fauman, Z.-Y. Zhang, J. E. Dixon, and M. A. Saper. 1994. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 Å and the complex with tungstate. Nature 370:571-575. [DOI] [PubMed] [Google Scholar]
- 38.Sturgill-Koszycki, S., P. H. Schlesinger, P. Chakraborty, P. L. Haddix, H. L. Collins, A. K. Fok, R. D. Allen, S. L. Gluck, J. Heuser, and D. G. Russell. 1994. Lack of acidification in mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263:678-681. [DOI] [PubMed] [Google Scholar]
- 39.Su, X.-D., N. Taddei, M. Stefani, G. Ramponi, and P. Nordlund. 1994. The crystal structure of a low-molecular-weight phosphotyrosine phosphatase. Nature 370:575-578. [DOI] [PubMed] [Google Scholar]
- 40.Tonks, N. K., and B. G. Neel. 1996. From form to function: signaling by protein tyrosine phosphatases. Cell 87:365-368. [DOI] [PubMed] [Google Scholar]
- 41.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]
- 42.Wang, S., L. Tabernero, M. Zhang, E. Harms, R. L. Van Etten, and C. V. Stauffacher. 2000. Crystal structure of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate. Biochemistry 39:1903-1914. [DOI] [PubMed] [Google Scholar]
- 43.Yuvaniyama, J., J. M. Denu, J. E. Dixon, and M. A. Saper. 1996. Crystal structure of the dual specificity protein phosphatase VHR. Science 272:1328-1331. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, M., C. V. Stauffacher, and R. L. Van Etten. 1995. In W. Merlevede (ed.), Advances in protein phosphatases, vol. 9, p. 1-23. Leuven University Press, Leuven, Belgium. [Google Scholar]
- 45.Zhang, M., R. L. Van Etten, and C. V. Stauffacher. 1994. Crystal structure of bovine heart phosphotyrosyl phosphatase at 2.2-Å resolution. Biochemistry 33:11097-11105. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, M., C. V. Stauffacher, D. Lin, and R. L. Van Etten. 1998. Crystal structure of a human low molecular weight phosphotyrosyl phosphatase. J. Biol. Chem. 273:21714-21720. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Z., E. Harms, and R. L. Van Etten. 1994. Asp129 of low molecular weight protein tyrosine phosphatase is involved in leaving group protonation. J. Biol. Chem. 269:25947-25950. [PubMed] [Google Scholar]
- 48.Zhang, Z.-Y., and J. E. Dixon. 1993. Active site labeling of the Yersinia protein tyrosine phosphatase: the determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry 32:9340-9345. [DOI] [PubMed] [Google Scholar]