Abstract
Using gene expression reporter vectors, we examined the activity of the spoIIE promoter in wild-type and spo0A-deleted strains of Clostridium acetobutylicum ATCC 824. In wild-type cells, the spoIIE promoter is active in a transient manner during late solventogenesis, but in strain SKO1, where the sporulation initiator spo0A is disrupted, no spoIIE promoter activity is detectable at any stage of growth. Strains 824(pMSpo) and 824(pASspo) were created to overexpress spoIIE and to decrease spoIIE expression via antisense RNA targeted against spoIIE, respectively. Some cultures of strains 824(pMSpo) degenerated during fermentations by losing the pSOL1 megaplasmid and hence did not produce the solvents ethanol, acetone, and butanol. The frequent degeneration event was shown to require an intact copy of spoIIE. Nondegenerate cultures of 824(pMSpo) exhibited normal growth and solvent production. Strain 824(pASspo) exhibited prolonged solventogenesis characterized by increased production of ethanol (225%), acetone (43%), and butanol (110%). Sporulation in strains harboring pASspo was significantly delayed, with sporulating cells exhibiting altered morphology. These results suggest that SpoIIE has no direct effect on the control of solventogenesis and that the changes in solvent production in spoIIE-downregulated cells are mediated by effects on the cell during sporulation.
The gram-positive, obligate anaerobe Clostridium acetobutylicum was used for the industrial production of the solvents acetone and butanol for over 60 years in the 20th century. With chemical synthesis of acetone and butanol proving significantly more economic, there are no reported industrial fermentation plants using C. acetobutylicum operational in the world today (11). However, over the last 20 years, the genetics and biochemistry of C. acetobutylicum have been investigated in detail, as we try to understand and improve upon the processes that control the production of solvents.
The genes aad, ctfA, ctfB, and adc are required for solvent production and are located on a 192-kb megaplasmid designated pSOL1 (8, 27). ctfA and ctfB code for a multifunctional coenzyme A (CoA) transferase, which transfers the CoA moiety from acetoacetyl-CoA to acetate or butyrate (40). Subsequently, acetoacetate is decarboxylated to form acetone, and acetyl-CoA and butyryl-CoA are converted to ethanol and butanol, respectively, via an aldehyde intermediary (26). For a detailed description of clostridial biochemistry, see the paper by Mitchell (26). It has been shown that C. acetobutylicum can lose pSOL1, rendering the cells unable to produce solvents. Cells and strains that have lost pSOL1 are termed “degenerate” (8, 28).
Whereas much is known about the biochemistry of C. acetobutylicum metabolism and the genes and proteins that catalyze these processes, relatively little is known about the genetic control of the expression of these genes. Recently, it was shown that a homologue to Bacillus subtilis stage 0 sporulation protein A (Spo0A) controls both the onset of solventogenesis and the process of sporulation in Clostridium beijerinckii and C. acetobutylicum (18, 31) In strain SKO1 of C. acetobutylicum, where spo0A is inactivated, acetone and butanol production are reduced to 2 and 8% of wild-type levels, respectively. Furthermore, SKO1 cells fail to sporulate and form extended filaments of conjoined rods, compared to the typical morphology of sporulating cells, which are swollen with developing endospores visible within (18). Studies have also shown that there are a considerable number of likely homologues in C. acetobutylicum to sigma factors and other proteins required for sporulation in B. subtilis (29, 33).
It appears that a cascade of sigma factors and stages similar to those involved in B. subtilis sporulation are present in C. acetobutylicum. The control of solventogenesis, which does not occur in B. subtilis, is genetically linked to the control of sporulation in C. acetobutylicum, as shown by the spo0A studies.
It has been suggested that solventogenesis and sporulation may be genetically uncoupled at some point during early sporulation (17), although as yet there are no reports of any attempts to do so. If solventogenesis could be genetically separated from sporulation, this would serve as an interesting and important illustration of the complexity of bacterial genetic control. Additionally, it may prove useful in the bioengineering of strains of C. acetobutylicum for use in industry; e.g., a strain that could produce solvents but that would never sporulate would be ideal for large-scale continuous fermentations.
An attempt to inactivate the genes coding for the sigma factors σE and σ F and the processing enzyme now designated SpoIIGA in C. acetobutylicum generated strains exhibiting a two- to threefold elevation of acetone and butanol production. The effects on sporulation were not reported (41).
In B. subtilis, stage II sporulation protein E (SpoIIE) is a membrane-bound serine phosphatase that exhibits multiple functions. It is localized to the septum that forms between the mother cell and developing forespore during stage II of sporulation and is essential for this asymmetric division, through interactions with the cytokinetic protein FtsZ (4, 22). The phosphatase activity of SpoIIE is required for the dephosphorylation of the anti-anti-sigma factor, SpoIIAA. In a dephosphorylated form, SpoIIAA binds the anti-sigma factor SpoIIAB, which is no longer able to bind to and inhibit the sigma factor σF. The liberated σF in the forespore is now active, and the development of the spore continues (2, 12, 20).
Sequence analysis of SpoIIE from B. subtilis reveals considerable homology to the eukaryotic protein serine/threonine phosphatase 2C (PP2C) family (5). The N terminus of SpoIIE has 10 membrane-spanning domains, whereas the C terminus is cytoplasmic and forms the catalytic domain of the enzyme (3, 9). A central, oligomerization domain is involved in the association of SpoIIE with FtsZ (13). PP2C-type phosphatases have been implicated in signaling cascades in animals, plants, yeasts, fungi, and bacteria (5).
The gene designated CAC3205 in C. acetobutylicum has been identified as spoIIE (26; GenBank accession number NC_003030). Comparative hydropathy analysis revealed that the N terminus of SpoIIE in C. acetobutylicum likely forms a membrane-spanning domain similar to that in B. subtilis (2). The C-terminal catalytic domain of SpoIIE in C. acetobutylicum also exhibits conservation of critical amino acids. The Asp-610 and Asp-628 residues have been shown to be conserved throughout a range of bacterial and eukaryotic PP2C-like phosphatases and form a metal ion binding pocket within the active site of human PP2C (9). The two conserved regions surrounding the invariant Asp-746, Gly-747, and Asp-795 have also been identified in SpoIIE homologues and PP2C phosphatases in Schizosaccharomyces pombe, cows, mice, humans, and Arabidopsis thaliana. Mutation of these invariant residues to alanine also causes a severe decrease in sporulation efficiency in B. subtilis (1, 34). All the invariant amino acids are conserved in the C. acetobutylicum homologue of SpoIIE.
We chose to investigate the role of SpoIIE in the control of solventogenesis and sporulation in C. acetobutylicum. Our initial studies focused on the activity of the spoIIE promoter, using a chloramphenicol acetyltransferase or β-galactosidase reporter system (7, 35, 36, 39).
Having determined the expression profile of spoIIE, we transformed wild-type C. acetobutylicum with the spoIIE overexpression vector pMSpo or with the pASspo vector harboring an antisense RNA construct targeted against spoIIE. We examined the effects of spoIIE misexpression on solvent production, sporulation, and cell morphology.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used are listed in Table 1.
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| C. acetobutylicum ATCC 824 | Wild type | ATCC |
| C. acetobutylicum SKO1 | Δspo0A MLSr | 18 |
| E. coli DH10β | mcrA ΔmcrBC recA1 Strr | NEB |
| Plasmids | ||
| pCATP | MLSr OriII ColElori catP | 35 |
| pCATspo | MLSr OriII ColElori catP, spoIIE promoter | This study |
| pSC12lacZ | Cmr OriII ColElori lacZ′ | 43 |
| pHT3 | Apr MLSr ColElori repL lacZ | 7, 39 |
| pThilac | Thir OriII ColElori lacZ | This study |
| pTLspo | Thir OriII ColElori lacZ, spoIIE promoter | This study |
| pIMP1 | Apr MLSr ColElori repL | 23 |
| pMSpo | Apr MLSr ColElori repL spoIIE | This study |
| pMSpoD | Apr MLSr ColElori repL spoIIE′ | This study |
| pSOS94 | ptb promoter, Apr MLSr ColElori repL | 38 |
| pASsos | ptb promoter, Apr MLSr ColElori repL | This study |
| pASspo | ptb promoter, Apr MLSr ColElori repL | This study |
spo0A, deletion of spo0A; MLSr, macrolide-lincosamide-streptogramin B resistant; mcrA ΔmcrBC, methylcytosine-specific restriction system abolished; recAl, homologous recombination abolished; Strr, streptomycin resistant; OriII repL, gram-positive origin of replication; ColEl, gram-negative origin of replication; catP, chloramphenicol acetyltransferase open reading frame; Cmr, chloramphenicol resistant; lacZ′, truncated, nonfunctional copy of lacZ; Apr, ampicillin resistant; lacZ, functional, promoterless β-galactosidase gene; Thir, thiamphenicol/chloramphenicol resistant due to functional copy of catP with a complete promoter; spoIIE, intact copy of spoIIE including promoter; spoIIE′, spoIIE with partial deletion of open reading frame. ATCC, American Type Culture Collection, Manassas, VA.; NEB, New England Biolabs, Beverly, Mass.
Growth conditions.
Escherichia coli was grown in Luria-Bertani medium aerobically at 37°C (25). For recombinant strains, liquid or agar-solidified medium was appropriately supplemented with ampicillin (100 μg/ml), erythromycin (200 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (35 μg/ml). Strains were stored at −80°C in medium supplemented with 50% glycerol.
C. acetobutylicum was grown in clostridial growth medium (CGM) with an initial glucose content of 50g/liter (∼280 mM) anaerobically at 37°C (19). For recombinant strains, liquid or agar-solidified medium was appropriately supplemented with erythromycin (40 μg/ml) or the chloramophenicol alternative thiamphenicol (25 μg/ml). Strains were stored as horse serum-supplemented lyophilized stocks at room temperature or at −80°C in medium supplemented with 10% glycerol. For the sporulation and morphology assays, strains were grown on agar-solidified CBM supplemented with erythromycin (40 μg/ml) anaerobically at 37°C (30).
DNA isolation and manipulation.
Plasmids were purified from E. coli by using the QIAprep Miniprep protocols. DNA was purified from agarose gels by using the QIAquick Gel Extraction kit, and PCR products or enzymatically manipulated DNA was purified by using the QIAquick PCR Purification kit (all from QIAGEN Inc., Valencia, Calif.). Plasmids were purified from C. acetobutylicum according to the protocol developed by Harris (16). Genomic DNA was purified from C. acetobutylicum by using the Genomic DNA Purification kit from Puregene (Gentra Systems, Minneapolis, Minn.).
All commercial enzymes used in this study (Taq polymerase, restriction endonucleases, calf intestinal phosphatases, T4 DNA ligase, and the Klenow fragment of DNA polymerase I) were used according to the manufacturers' recommendations.
Automated DNA sequencing was performed by LoneStar automated DNA sequencing (LoneStar Laboratories Inc., Houston, Tex.).
Construction of reporter vectors pCATspo and pTLspo.
The entire intergenic region upstream of the spoIIE open reading frame was amplified by PCR using primers incorporating terminal BamHI sites (shown underlined), spoprom (CCGGGATCCATCAACATCCCCAATCTATAAACC) and sporev (AGCGGATCCACACTACCAAGTCAAGAAGCTTTCAC). The resulting ∼0.25-kb fragment was ligated into BamHI-digested, dephosphorylated pCATP (33) to form vector pCATspo. The correct orientation of the spoIIE promoter upstream of catP in pCATspo was confirmed by automated DNA sequencing. pCATspo is shown in Fig. 1.
FIG. 1.
Antisense construct to spoIIE. The spoIIE antisense construct consists of oligonucleotide spoasastop (upper DNA strand) and spoasbtm (lower DNA strand). The single underlined GATC forms the 5′ BamHI “sticky” end. The black-shaded region includes 38 bases of the spoIIE open reading frame followed by 16 bases of the upstream leader sequence, shown shaded in dark gray. The bold region forms the 17-base rho-independent terminator region from an antisense RNA targeted against the glnA gene, found naturally in Clostridium saccharobutylicum NCP262 (10).
Plasmid pSC12lacZ (43) was designed as a thiamphenicol-resistant reporter vector, but a prematurely truncated copy of lacZ was accidentally incorporated into the vector during its construction. pSC12lacZ was digested with PstI, treated with the Klenow fragment of DNA polymerase I, and then digested with BamHI. The 2.2-kb fragment containing the gram-positive and gram-negative origins of replication and the thiamphenicol resistance cassette CATP was purified. pHT3 (7, 36) was digested with NotI, treated with the Klenow fragment of DNA polymerase I, and then digested with BamHI. The 3.8-kb fragment containing the complete lacZ open reading frame was purified and ligated into the 2.2-kb fragment of pSC12lacZ to form pThilac. The spoIIE promoter fragment was excised from pCATspo, using BamHI and XhoI, and ligated into BamHI/XhoI-digested pThilac to form pTLspo.
Construction of vectors pMSpo, pMSpoD, and pASspo.
Primers sporev and spofor (AGCGGATCCACATATTGATAACATCATTT ATCAACAAAAACA) were used to amplify by PCR the entire spoIIE open reading frame including all upstream and downstream intergenic bases from C. acetobutylicum genomic DNA. The 2.7-kb PCR fragment was digested with BamHI and ligated into BamHI-digested pIMP1 (23) to form vector pMSpo. Vector pMSpo was digested with SphI and BbsI to excise a 1.4-kb fragment of the spoIIE open reading frame located 180 bp downstream from the spoIIE start codon. The remaining 6.1-kb fragment of pMSpo was treated with the Klenow fragment of DNA polymerase I and circularized to form pMSpoD.
The antisense vector targeted against spoIIE was designed according to the method of Desai and Papoutsakis (10, 37, 38). Oligonucleotides spoastop and spoasbtm were diluted to a concentration of 0.5 μg/μl. Nine microliters of the spoastop and 9 μl of the spoasbtm oligonucleotides were mixed with 2 μl of 10× STE buffer (100 mM Tris-HCl, 500 mM NaCl, 10 mM EDTA [pH 8.0]) and placed in a water bath set to 94°C. The water bath was allowed to cool to room temperature overnight, during which time the oligonucleotides annealed to form the antisense construct shown in Fig. 1. Vector pSOS94 (GenBank accession number AY187685) was digested with BamHI and SfoI, and the 5.0-kb fragment was purified. This fragment was treated with the Klenow fragment of DNA polymerase I and circularized to form the control vector pASsos. The spoIIE antisense construct and the 5.0-kb fragment of pSOS94 were ligated through the BamHI cohesive ends, treated with the Klenow fragment of DNA polymerase I, and circularized to form vector pASspo. Correct construction of pASsos and pASspo was confirmed by automated sequencing using primer ASseq (TTACGAAGTAAATAAGTCTAGTGTGTTAGA), which hybridizes to pSOS94 between 148 and 118 bases upstream of the ptb promoter.
DNA transformation of C. acetobutylicum.
Prior to transformation into C. acetobutylicum, plasmid DNA must be methylated by the Φ3TI methyltransferase to prevent restriction by the clostridial endonuclease Cac824I (24). Methylation was achieved by transformation of the required plasmid into E. coli DH10β harboring vector pDHKM (43), which carries an active copy of the Φ3TI methyltransferase gene. Electrotransformation of wild-type C. acetobutylicum or strain SKO1 was carried out according to the protocol developed by Mermelstein et al. (23). Positive transformants were isolated on agar-solidified CGM supplemented with the appropriate antibiotic, and transformations were confirmed by plasmid DNA purification. In this article, wild-type transformants are designated 824 and SKO1 transformants are designated SK, with the transformed plasmid written in parentheses following 824 or SK. All vectors used harbor the OriII gram-positive origin of replication derived from plasmid pIM13, giving an expected copy number of eight plasmids per cell (21, 23).
Batch fermentations of C. acetobutylicum.
Single colonies of transformed C. acetobutylicum were grown in closed-cap batch fermentations of 100 ml of CGM supplemented with the appropriate antibiotic at 37°C in a Forma Scientific anaerobic chamber (Thermo Forma, Marietta, Ohio). To allow for differences in lag time following inoculation, zero hour (T0) was determined when the culture had reached an optical density at 600 nm (OD600) of 0.1. Fermentations were allowed to proceed for 120 h.
For cell growth and product formation assays, 1.5-ml samples were taken at the time points specified. Cell growth was quantified by measuring the OD600 using a Beckman DU64 spectrophotometer external to the anaerobic chamber (Beckman Coulter, Inc., Fullerton, Calif.). Samples were then centrifuged at 16,000 × g for 15 min at room temperature in a Sorvall Biofuge PICO (Kendro Laboratory Products, Newtown, Conn.), and the supernatant was collected. A 1-ml volume of supernatant was acidified with 20 μl of 50% H2SO4, and a 5-μl sample was injected into a Hewlett-Packard 5890 Series II gas chromatograph for solvent content analysis (Hewlett-Packard Company, Palo Alto, Calif.).
Degeneracy tests.
To test whether strains of C. acetobutylicum had degraded and lost the pSOL1 plasmid, the activity of amylase was monitored. amyP, encoding α-amylase, is present on pSOL1 and hence would be lost if the cell degrades. This renders the cell unable to metabolize starch (18, 32). After 24 h of growth in batch culture, 100-μl samples of cell suspension were spread on three agar-solidified 2XYTGMA plates to test for α-amylase activity as previously described (18). Approximately 200 individual colonies for each batch culture were examined on 2XYTGMA plates.
Samples of 10 ml were taken from the same cultures after 120 h of growth, and genomic DNA was extracted. The pSOL1 megaplasmid copurifies with the genomic DNA. PCRs were performed using the purified DNA as a template. Primers adhEleft (AATATAATAGGTTGGATAGATGAAC) and adhEright (TTTGTTAATTAAGAGATCTACCTTT), specific for adhE located on the pSOL1 megaplasmid, were used for PCR to confirm the presence or absence of pSOL1. Additionally, primers sinRfor (AGCGGATCCACATGTTATCAATCCATTCCAT TAACATC) and sinRrev (AGCGGATCCACACAATTTCTTCGCCTCCCTATAC), specific for sinR located on the chromosome, were used for the control PCR.
Enzyme activity assays.
To assay for chloramphenicol acetyltransferase (CAT) activity, samples of 5 ml were taken at the time points specified, the OD600 was recorded, and the sample was prepared and assayed for CAT activity as previously described (35, 36).
To assay for β-galactosidase activity, samples of 5 ml were taken at the time points specified and centrifuged at maximum speed at 4°C for 20 min in a Sorvall NT6000B centrifuge. Subsequent sample preparation and β-galactosidase activity assays were performed as previously described (37).
To assay for the activity of the multifunctional CoA transferase (CoAT), 100-ml cultures of the wild type and strains 824(pASsos) and 824(pASspo) were grown until approximately 3 h after stationary phase was reached, as determined by OD600 readings. A crude extract of CoAT was prepared, and CoAT activity was assayed according to the methods of Wiesenborn et al. (40).
For all assays, enzyme specific activity was quantified as units of enzyme per milligram of protein extracted (6).
Morphology and sporulation assay.
Strains harboring pASsos and pASspo were grown simultaneously in liquid medium and transferred after 16 to 24 h of growth to CBM plates supplemented with erythromycin. At 24-, 48-, 72-, and 140-h intervals, cells were picked from the medium plates using a sterile toothpick and resuspended in 20 μl of liquid CGM supplemented with 10% glycerol. Samples were frozen at −80°C until all samples had been collected. Previous freeze-thaw tests had confirmed that the freezing process had no effect on cell morphology.
A thawed cell sample of 2 μl was mixed with 8 μl of CGM on a glass microscope slide, allowed to dry, and heat fixed by passing briefly through a flame. A glass coverslip was sealed over the sample, and the cells were examined by conventional light microscopy at a magnification of ×1,000 using a Zeiss Axioplan II microscope fitted with a NeoFluar 100× oil immersion objective lens (Carl Zeiss MicroImaging Inc., Thornwood, N.Y.). Cells were photographed with a CoolSnap H2 camera (Photometrics, Tucson, Ariz.) controlled by Metamorph software (Universal Imaging Corporation, Downington, Pa.).
RESULTS
Reporter vector assays of spoIIE expression.
Acetone and butanol formation and CAT activity in strain 824(pCATspo) and CAT activity in the control strain 824(pCATP), are shown in Fig. 2. The solvent profile for 824(pCATP) (data not shown) was very similar to that of 824(pCATP), indicating that the cultures of each strain were at similar stages of their life cycles when samples were taken.
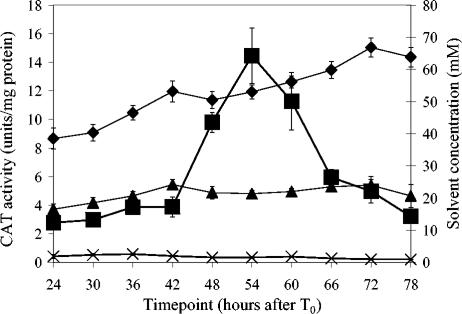
FIG. 2.
CAT activities in strains 824(pCATspo) and 824(pCATP). CAT activities are shown in strain 824(pCATspo)(▪) and the control strain 824(pCATP)(×). The solvent concentrations for the 824(pCATspo) fermentations are represented by the following symbols: acetone (▴) and butanol (♦). Data are means ± 1 standard error. For acetone and butanol, n = 6; for 824(pCATP) CAT activity, n = 3; for 824(pCATspo) CAT activity, n = 6.
CAT levels in 824(pCATspo) were detectable at a basal level of approximately 3 U of CAT/mg of protein. Between 42 and 54 h of growth, CAT expression increased at a uniform rate to a maximum of approximately 14 U of CAT/mg of protein at 54 h. Over the course of the next 24 h, CAT levels returned to their basal levels. These data show that the spoIIE promoter is active during mid- to late solventogenesis, which is the stage of growth where the cells are transitioning from vegetative growth to sporulation.
We were obligated to use the β-galactosidase reporter system, which utilizes a thiamphenicol resistance marker, to compare spoIIE expression in wild-type ATCC 824 and the SKO1 strain, because SKO1 is erythromycin resistant. In both control strains 824(pThilac) and SK(pThilac), β-galactosidase activity was less than 6 U/mg of protein in all samples (data not shown). In strain 824(pTLspo), β-galactosidase activity was detectable during late solventogenesis, reaching a maximum of ∼250 U/mg of protein from 66 to 78 h of growth after T0. This was approximately 12 h later than the CAT activity maximum in 824(pCATspo). Additionally, β-galactosidase activity continued for over 48 h, whereas CAT activity lasted 30 h. These differences may be a reflection of the variability in growth of and the stability of proteins in C. acetobutylicum. β-Galactosidase activity in SK(pTLspo) was not different from that observed in the control strains and remained less than 1.3 U/mg of protein in all samples, indicating that spoIIE is not expressed in the spo0A mutant strain SKO1.
Product formation in fermentations of strains 824(pIMP1), 824(pMSpoD), and 824(pMSpo).
In the control strain 824(pIMP1) and strain 824(pMSpoD), there were no significant differences in product formation throughout the course of the 120-h fermentations. In fermentations of 824(pMSpo), individual cultures were found to be either solventogenic or nonsolventogenic, and data for 824(pMSpo) are therefore divided into those cultures which produced solvents, designated 824(pMSpo)+, and those which did not, designated 824(pMSpo)0. The data for product formation after 120 h of growth of strains 824(pIMP1) and 824(pMSpoD) and both solventogenic and nonsolventogenic populations of strain 824(pMSpo) are shown in Table 2.
TABLE 2.
Product formation after 120-h fermentation of strains 824(pIMP1), 824(pMSpoD), and 824(pMSpo)a
| Strain | Product formation (mM) (mean ± SD)
|
||||
|---|---|---|---|---|---|
| Ethanol (mM) | Acetone (mM) | Acetate (mM) | Butanol (mM) | Butyrate (mM) | |
| 824(pIMP1) (n = 4) | 7.25 ± 0.48 | 24.50 ± 0.87 | 12.50 ± 1.32 | 69.75 ± 3.50 | 5.00 ± 0.41 |
| 824(pMSpoD) (n = 4) | 7.00 ± 0.71 | 25.50 ± 1.94 | 11.75 ± 0.48 | 72.75 ± 2.93 | 3.50 ± 0.29 |
| 824(pMSpo)+ (n = 4) | 7.25 ± 0.95 | 27.00 ± 2.12 | 10.50 ± 0.65 | 69.50 ± 4.01 | 3.25 ± 0.48 |
| 824(pMSpo)0 (n = 3) | 0.00 ± 0.00 | 0.00 ± 0.00 | 17.67 ± 0.88 | 0.00 ± 0.00 | 37.33 ± 2.03 |
Data for strain 824(pMSpo) is divided into those cultures which produced solvents, designated 824(pMSpo)+, and those which did not, designated 824(pMSpo)0.
In cultures of 824(pIMP1), 824(pMSpoD), and 824(pMSpo)+,product formation did not differ significantly between the strains. Acetone and butanol concentrations reached maximums of 25 to 27 mM and ∼70 mM, respectively, which are typical for fermentations of this scale. After 120 h, most of the acetate and butyrate had been reassimilated into acetone and butanol, leaving final concentrations of 10 to 12 mM acetate and less than 5 mM butyrate. No ethanol, acetone, or butanol was produced in cultures of 824(pMSpo)0. This resulted in the accumulation of acids, and hence acetate and butyrate levels were elevated by ∼33 and ∼400%, respectively, after 120 h of growth.
All cultures were tested for degeneracy, using the α-amylase tests and PCR procedure described earlier. The proportion of colonies of 824(pIMP1), 824(pMSpoD), and 824(pMSpo)+ found to exhibit α-amylase activity was greater than 99.5%, compared to less than 0.5% of colonies from the cultures of 824(pMSpo)0. PCR tests confirmed that the pSOL1 megaplasmid was absent from all three cultures of 824(pMSpo)0. This high-frequency degeneration event correlated with the presence of an intact copy of the spoIIE open reading frame.
Product formation in fermentations of strains 824(pASsos) and 824(pASspo).
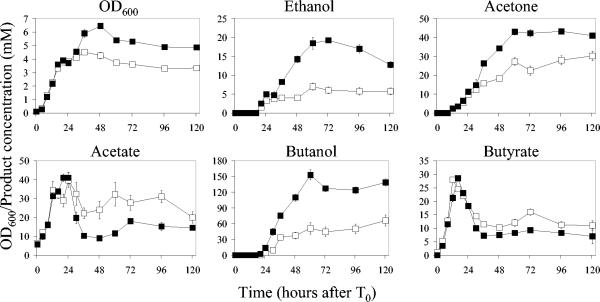
Growth and product formation in 120-h fermentations of strains 824(pASsos) and 824(pASspo) are shown in Fig. 3. Cultures of 824(pASspo) grew significantly better than 824(pASsos), with a maximum OD600 of ∼6.5 compared to ∼4.5 for the plasmid control strain observed after 36 to 48 h of growth. Maximum acetate concentrations in both strains were similar at ∼40 mM after 24 h of growth. However, acetate levels decreased rapidly in 824(pASspo) as acetate was reassimilated, to a minimum of ∼9 mM after 48 h of growth. Acetate production increased again after 48 h, which coincided with the concentration of acetone reaching a maximum of ∼45 mM, a level at which it remained for the rest of the fermentation. This is 50% greater than the maximum acetone concentration of ∼ 30 mM observed in 824(pASsos) after 120 h of growth.
FIG. 3.
Growth and product formation in fermentations of strains 824(pASsos) and 824(pASspo). The measured quantity for each profile of strain 824(pASsos) (□) and strain 824(pASspo) (▪) is shown above each graph. Data are means ± 1 standard error. For each datum point, n = 4.
Butyrate production in both strains did not differ significantly, but this was not reflected in butanol production. In the control strain, butanol production followed a typical pattern, reaching a maximum of ∼66 mM after 120 h of growth. 824(pASspo) exhibited a rapid increase in butanol production between 16 and 64 h of growth, after which time the concentration of butanol remained constant for the rest of the fermentation. A maximum butanol concentration of ∼153 mM was recorded in 824(pASspo), which is a 132% increase compared to the control strain.
The activity of CoAT in the wild type was 0.092 ± 0.003 U/mg of protein, whereas CoAT activity in both strains 824(pASsos) and 824(pASspo) was slightly lower, at 0.079 ± 0.018 and 0.074 ± 0.013 U/mg of protein, respectively.
Morphology and sporulation of strains 824(pASsos) and 824(pASspo).
At 24 h of growth, cells of strains 824(pASsos) and 824(pASspo) were morphologically similar. Cells in all stages of division were visible, and in neither strain were sporulating cells observed. After 48 h of growth, many 824(pASspo) cells were observed in the process of dividing, whereas the majority of 824(pASsos) cells were single rods with the occasional sporulating cell identified, exhibiting a typical swollen, cigar-shaped morphology, with the developing spore clearly visible in the center of the cell. After 72 h, many 824(pASsos) cells were seen to be sporulating, but in cultures of 824(pASspo), single cells of the clostridial form were observed, with no sporulating cells identified. Additionally, some individual cells exhibited an abnormal morphology, such that they were elongated two- to threefold compared to the control (data not shown). Examination of several different cultures of 824(pASspo) indicated that these elongated cells are common and that they were not vegetative cells undergoing division that were misidentified.
After 140 h of growth, sporulating cells or free endospores dominated the 824(pASsos) culture, with very few vegetative cells observed. In contrast, very few sporulating cells were observed in cultures of 824(pASspo), with the majority of cells appearing to be in the clostridial growth state. Of those cells which were sporulating, some cells exhibited abnormal morphology. In strain 824(pASspo), many cells exhibited the typical swollen phenotype associated with spore production, but no developing spore could be observed within the cell. In other cells of strain 824(pASspo), the developing spore was present but was located at one pole of the cell, with the remainder of the cell being exceptionally elongated.
DISCUSSION
The data from the CAT and β-galactosidase assays of the spoIIE promoter constructs are in strong agreement with the timing and initiation of spoIIE transcription in B. subtilis (15, 42). Expression from pCATspo transformed into wild-type cells is temporally restricted to mid- to late solventogenesis, at which time the cells are expected to be transitioning between solventogenic growth and the onset of sporulation. Expression from pTLspo transformed into wild-type cells is also observed during late solventogenesis, but no expression is detectable at any stage from pTLspo transformed into strain SKO1. As SKO1 is inactivated for spo0A, we can conclude that as in B. subtilis, spo0A is required for the correct expression of spoIIE in C. acetobutylicum (42). These data do not show that expression spoIIE in C. acetobutylicum is directly controlled by spo0A, but we intend to use the pCATspo reporter system to further investigate regulatory elements within the spoIIE promoter.
The fermentation of strain 824(pMSpoD) did not differ significantly from that of the control strain 824(pIMP1). It can be concluded therefore that any differences in growth or product formation observed in strain 824(pMSpo) are due to the presence of an intact copy of spoIIE. Fermentations of strain 824(pMSpo) were either identical to the control strains, or they degenerated. The data presented from the fermentations of 824(pMSpoD) and 824(pMSpo) are typical of many time course assays performed. Degeneration was never observed in strain 824(pMSpoD), but degeneration was entirely unpredictable in strain 824(pMSpo). Growths of 824(pMSpo) where all, none, or a variable proportion of the cultures have degenerated have been observed. Interestingly, in cultures which do not degenerate, growth and product formation is not altered, suggesting that the overexpression of spoIIE does not directly affect solvent production and that the degeneration event is mediated through another means. In B. subtilis, it has been shown that the chromosome is translocated across the septum from the mother cell to the forespore during the early stages of sporulation, and this event may be responsible for establishing SpoIIE phosphatase activity in the forespore (14). It is possible that the elevated expression of spoIIE may disrupt the correct translocation of genomic material in C. acetobutylicum and that this may lead to the loss of the pSOL1 megaplasmid. However, spoIIE expression is elevated only during sporulation and remained at basal levels during the exponential growth phase when normal cell division is occurring, and further work would be necessary to determine the exact mechanism by which the pSOL1 plasmid is lost in strains harboring plasmid pMSpo.
In strain 824(pASspo), cell optical density remains high throughout growth and solvent levels are significantly elevated. Microscopy showed that sporulation is delayed in this strain and also that cell morphology is affected by decreased expression of spoIIE. There is no significant difference in CoAT activity between the wild type, 824(pASsos), and 824(pASspo), suggesting that the molecular mechanism for elevated solvent production in strain 824(pASspo) is not due to increased activity of solventogenic enzymes. Based upon all these data, we propose the following model for the mode of action of SpoIIE in C. acetobutylicum.
SpoIIE does not directly affect solventogenesis but controls sporulation in a manner similar to that in B. subtilis. The spoIIE promoter assays show that spoIIE is transcribed long after solventogenesis has commenced, and nondegenerate cultures of 824(pMSpo) do not exhibit any changes in solventogenesis. The degeneration event itself does not directly control solventogenesis but indirectly through the loss of adhE, ctfA, ctfB, and adc on the pSOL1 plasmid (8).
In strain 824(pASspo), the absence of SpoIIE causes sporulation to be blocked at stage II. As sporulation cannot proceed, the cell remains in a clostridial, solventogenic state, thus allowing solvent production to proceed for a longer time and for solvents to accumulate more rapidly and to a higher concentration. In this manner, decreased expression of spoIIE indirectly causes altered solvent production by extending the solventogenic growth phase. The misshaped sporulating cells of strain 824(pASspo) observed after 140 h of growth on CBM may reflect the integral membrane function of SpoIIE.
We have shown that the control of solventogenesis and sporulation can be genetically uncoupled in C. acetobutylicum. The genetic deletion of spo0A causes defects in both solvent production and sporulation, whereas antisense RNA to spoIIE causes defects solely in sporulation (18). It seems likely that factors within the sporulation cascade between spo0A and spoIIE activation are responsible for the genetic separation of solventogenesis and sporulation. This presents an excellent opportunity for further investigation of how and where solventogenesis and sporulation may be separated in C. acetobutylicum.
From a bioengineering perspective, strain 824(pASspo) exhibits extremely high solvent production and may be suitable for development for industrial usage. We intend to examine strain 824(pASspo) under a variety of controlled conditions in a bioreactor to optimize performance.
Acknowledgments
This work was supported by U.S. Department of Agriculture grant 00-35504-9269, National Science Foundation grant BES-0001288, and Robert A. Welch Foundation grant C-1268 (G.N.B.).
REFERENCES
- 1.Adler, E., A. Donella-Deana, F. Arigoni, L. A. Pinna, and P. Stragler. 1997. Structural relationship between a bacterial developmental protein and eukaryotic PP2C protein phosphatases. Mol Microbiol. 23:57-62. [DOI] [PubMed] [Google Scholar]
- 2.Arigoni, F., A. M. Guerout-Fleury, I. Barak, and P. Stragier. 1999. The SpoIIE phosphatase, the sporulation septum and the establishment of forespore-specific transcription in Bacillus subtilis: a reassessment. Mol. Microbiol. 31:1407-1415. [DOI] [PubMed] [Google Scholar]
- 3.Arigoni, F., K. Pogliano, C. D. Webb, P. Stragier, and R. Losick. 1995. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science 270:637-640. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 5.Bork, P., N. P. Brown, H. Hegyi, and J. Schultz. 1996. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 5:1421-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Burchhardt, G., and H. Bahl. 1991. Cloning and analysis of the beta-galactosidase-encoding gene from Clostridium thermosulfurogenes EM1. Gene 106:13-19. [DOI] [PubMed] [Google Scholar]
- 8.Cornillot, E., R. V. Nair, E. T. Papoutsakis, and P. Soucaille. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, A. K., N. R. Helps, P. T. Cohen, and D. Barford. 1996. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 15:6798-6809. [PMC free article] [PubMed] [Google Scholar]
- 10.Desai, R. P., and E. T. Papoutsakis. 1999. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl. Environ. Microbiol. 65:936-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durre, P. 2001. From Pandora's box to cornucopia: clostridia—a historical perspective, p. 1-17. In H. Bahl and P. Durre (ed.), Clostridia—biotechnology and medical applications. Wiley-VCH, New York, N.Y.
- 12.Feucht, A., L. Abbotts, and J. Errington. 2002. The cell differentiation protein SpoIIE contains a regulatory site that controls its phosphatase activity in response to asymmetric septation. Mol. Microbiol. 45:1119-1130. [DOI] [PubMed] [Google Scholar]
- 13.Feucht, A., R. A. Daniel, and J. Errington. 1999. Characterization of a morphological checkpoint coupling cell-specific transcription to septation in Bacillus subtilis. Mol. Microbiol. 33:1015-1026. [DOI] [PubMed] [Google Scholar]
- 14.Frandsen, N., I. Barak, C. Karmazyn-Campelli, and P. Stragier. 1999. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 13:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, P., J. Westpheling, and P. Youngman. 1988. Characterization of the promoter region of the Bacillus subtilis spoIIE operon. J. Bacteriol. 170:1598-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, L. M. 2001. Cloning and characterization of the Clostridium acetobutylicum ATCC824 gene encoding the Spo0A transcription regulator and its role in controlling solvent formation and sporulation-specific gene expression. Ph.D. dissertation. Northwestern University, Evanston, Ill.
- 17.Harris, L. M. 1997. Fermentation characterization of Clostridium acetobutylicum ATCC824 recombinant strains. MS thesis. Northwestern University, Evanston, Ill.
- 18.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmanis, M. G. N., and S. Gatenbeck. 1984. Intermediary metabolism in C. acetobutylicum: levels of enzymes involved in the formation of acetate and butyrate. Appl. Environ. Microbiol. 47:1277-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroos, L., B. Zhang, H. Ichikawa, and Y. T. Yu. 1999. Control of sigma factor activity during Bacillus subtilis sporulation. Mol. Microbiol. 31:1285-1294. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. Y., L. D. Mermelstein, and E. T. Papoutsakis. 1993. Determination of plasmid copy number and stability in Clostridium acetobutylicum ATCC 824. FEMS Microbiol. Lett. 108:319-323. [DOI] [PubMed] [Google Scholar]
- 22.Lucet, I., A. Feucht, M. D. Yudkin, and J. Errington. 2000. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J. 19:1467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mermelstein, L., N. Welker, G. Bennett, and E. Papoutsakis. 1992. Expression of cloned homologous fermentation genes in Clostridium acetobutylicum ATCC 824. Bio/Technology 10:190-195. [DOI] [PubMed] [Google Scholar]
- 24.Mermelstein, L. D., and E. Papoutsakis. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage Φ3TI methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59:1077-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Mitchell, W. J. 1998. Physiology of carbohydrate to solvent conversion by clostridia. Adv. Microb. Physiol. 39:31-130. [DOI] [PubMed] [Google Scholar]
- 27.Nair, R. V., E. M. Green, D. E. Watson, G. N. Bennett, and E. T. Papoutsakis. 1999. Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J. Bacteriol. 181:319-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair, R. V., and E. T. Papoutsakis. 1994. Expression of plasmid-encoded aad in Clostridium acetobutylicum M5 restores vigorous butanol production. J. Bacteriol. 176:5843-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Brien, R. W., and J. G. Morris. 1971. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J. Gen. Microbiol. 68:307-318. [DOI] [PubMed] [Google Scholar]
- 31.Ravagnani, A., K. C. Jennert, E. Steiner, R. Grunberg, J. R. Jefferies, S. R. Wilkinson, D. I. Young, E. C. Tidswell, D. P. Brown, P. Youngman, J. G. Morris, and M. Young. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172-1185. [DOI] [PubMed] [Google Scholar]
- 32.Sabathe, F., C. Croux, E. Cornillot, and P. Soucaille. 2002. amyP, a reporter gene to study strain degeneration in Clostridium acetobutylicum ATCC 824. FEMS Microbiol Lett. 210:93-98. [DOI] [PubMed] [Google Scholar]
- 33.Santangelo, J. D., A. Kuhn, A. Treuner-Lange, and P. Durre. 1998. Sporulation and time course expression of sigma-factor homologous genes in Clostridium acetobutylicum. FEMS Microbiol. Lett. 161:157-164. [DOI] [PubMed] [Google Scholar]
- 34.Schroeter, R., S. Schlisio, I. Lucet, M. Yudkin, and R. Borriss. 1999. The Bacillus subtilis regulator protein SpoIIE shares functional and structural similarities with eukaryotic protein phosphatases 2C. FEMS Microbiol. Lett. 174:117-123. [DOI] [PubMed] [Google Scholar]
- 35.Scotcher, M. C., K. X. Huang, M. L. Harrison, F. B. Rudolph, and G. N. Bennett. 2003. Sequences affecting the regulation of solvent production in Clostridium acetobutylicum. J. Ind. Microbiol. Biotechnol. 30:414-420. [DOI] [PubMed] [Google Scholar]
- 36.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 37.Tummala, S. B., S. G. Junne, and E. T. Papoutsakis. 2003. Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J. Bacteriol. 185:3644-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 2003. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J. Bacteriol. 185:1923-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiesenborn, D. P., F. B. Rudolph, and E. T. Papoutsakis. 1989. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl. Environ. Microbiol. 55:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong, J., and G. N. Bennett. 1996. Recombination-induced variants of Clostridium acetobutylicum ATCC 824 with increased solvent production. Curr. Microbiol. 32:349-356. [DOI] [PubMed] [Google Scholar]
- 42.York, K., T. J. Kenney, S. Satola, C. P. Moran, Jr., H. Poth, and P. Youngman. 1992. Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J. Bacteriol. 174:2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, Y., L. A. Nurman, A. Chuang, M. Monroe-Augustus, M. Lyristis, M. L. Harrison, F. B. Rudolph, and G. N. Bennett. 2003. Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 69:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]