Abstract
PpsR from the anoxygenic phototrophic bacterium Rhodobacter sphaeroides has been known as an oxygen- and light-dependent repressor of bacteriochlorophyll and carotenoid biosynthesis genes and puc operons involved in photosystem development. However, the putative PpsR-binding sites, TGTN12ACA, are also located upstream of numerous nonphotosystem genes, thus raising the possibility that the role of PpsR is broader. To characterize the PpsR regulon, transcriptome profiling was performed on the wild-type strain grown at high and low oxygen tensions, on the strain overproducing PpsR, and on the ppsR mutant. Transcriptome analysis showed that PpsR primarily regulates photosystem genes; the consensus PpsR binding sequence is TGTcN10gACA (lowercase letters indicate lesser conservation); the presence of two binding sites is required for repression in vivo. These findings explain why numerous single TGTN12ACA sequences are nonfunctional. In addition to photosystem genes, the hemC and hemE genes involved in the early steps of tetrapyrrole biosynthesis were identified as new direct targets of PpsR repression. Unexpectedly, PpsR was found to indirectly repress the puf and puhA operons encoding photosystem core proteins. The upstream regions of these operons contain no PpsR binding sites. Involvement in regulation of these operons suggests that PpsR functions as a master regulator of photosystem development. Upregulation of the puf and puhA operons that resulted from ppsR inactivation was sufficient to restore the ability to grow phototrophically to the prrA mutant. PrrA, the global redox-dependent activator, was previously considered indispensable for phototrophic growth. It is revealed that the PrrBA and AppA-PpsR systems, believed to work independently, in fact interact and coordinately regulate photosystem development.
Rhodobacter sphaeroides is a facultatively phototrophic anoxygenic alphaproteobacterium. Under high oxygen tension, whether in the light or in the dark, this bacterium uses aerobic respiration for energy generation. When oxygen tension decreases, R. sphaeroides induces synthesis of photosynthetic apparatus, an alternate energy generation system functional under anoxic-light conditions (28, 30, 32, 36). The photosynthetic apparatus is comprised of one type II photosystem (PS). Development of PS involves synthesis of photosynthetic pigments, bacteriochlorophyll (Bchl) and carotenoids (Crt), membrane proteins of the reaction center (RC), and two light-harvesting (LH) complexes as well as assembly factors. The RC and LH complexes are housed in the specialized intracytoplasmic membrane system. A decrease in oxygen tension triggers a significant increase in transcription of PS genes, which is required for PS development. Major regulatory components involved in oxygen control of PS development have been identified previously (1, 44). However, precise roles of individual regulators and interactions between the regulatory pathways are not yet fully understood. This work is aimed at characterizing the role of one major transcriptional regulator, PpsR.
The PpsR protein from R. sphaeroides and its homologs from other anoxygenic phototrophic proteobacteria (12, 34, 39) have been known to repress the transcription of a subset of PS genes, i.e., those involved in biosynthesis of Bchl and Crt, bch and crt genes, respectively (15, 33). These genes are located in the so-called R. sphaeroides PS gene cluster (6). In addition, PpsR represses transcription of the puc1 and puc2 operons encoding components of the LHII complex and located apart from the PS gene cluster (15, 45). Upstream regions of these genes contain a TGTN12ACA palindrome, believed to represent a PpsR consensus binding site (15, 26).
R. sphaeroides PpsR binds DNA most strongly under oxic conditions. Two mechanisms are apparently involved in controlling the strength of DNA binding. One mechanism involves oxidation-reduction of two cysteine residues, Cys251 and Cys424 (19, 29). In the aerobically grown cells, these residues are believed to be oxidized and form an intramolecular disulfide bond, whereas in the anaerobically grown cells, they exist as free thiols. PpsR in the oxidized form was shown to bind DNA in vitro approximately twofold stronger than PpsR in the reduced form (29). Recently, the validity of this mechanism was challenged (5), and its importance for repression in vivo remains to be tested.
The second mechanism involves interactions between PpsR and the AppA protein. AppA was identified by Gomelsky and Kaplan (16) and shown to act as an antirepressor of PpsR in vivo (17). The ability of AppA to directly interact with PpsR and its antirepressor function were subsequently confirmed in vitro (29). AppA is unique in that it represents the first known protein that senses and integrates such different environmental stimuli as oxygen and light (3, 20, 29). The mechanism of light sensing by AppA through the novel type flavin adenine dinucleotide (FAD)-binding domain designated BLUF (14, 18) is emerging (24, 25). The mechanism of oxygen sensing remains elusive.
The survey of the sequenced R. sphaeroides genome (28) (http://genome.ornl.gov/microbial/rsph) revealed that the putative PpsR binding site, TGTN12ACA, is present upstream of numerous non-PS genes. This observation posed the question of whether the role of PpsR extends beyond the PS genes mentioned above. This prompted us to determine the scope of the R. sphaeroides PpsR regulon and to refine the role of PpsR in PS development.
(Preliminary findings reported here were presented previously [L. Gomelsky, O. V. Moskin, and M. Gomelsky, XI Int. Symp. Phototroph. Prokaryotes, Tokyo, Japan, abstr. P-165, 2003].)
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. R. sphaeroides strains were grown in Sistrom's minimal medium A with succinate as the carbon source at 30°C (7). For DNA microarray experiments, 60-ml cultures were grown in 100-ml glass culture tubes under continuous vigorous sparging with defined gas mixtures (39 to 48 ml min−1 per tube). The following gases were used: 20% O2, 79% N2, and 1% CO2 (high oxygen) and 0.5% O2, 98.5% N2, and 1% CO2 (low oxygen). Dissolved oxygen tension was measured by using an oxygen microelectrode (Microelectrodes, Inc., Bedford, N.H.) in the cell-free medium as well as in the sparged cultures at an A600 of 0.20. It was found to be identical, suggesting that oxygen dissolution rates in the existing setup are sufficient to maintain saturating and constant oxygen levels throughout bacterial growth.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| R. sphaeroides | ||
| 2.4.1 | Wild type | Laboratory collection |
| APP11 | 2.4.1 appA::Tpr | 16 |
| PPS2-4 | 2.4.1 ppsR (Cys251-Ser, Cys424-Ser) | This study |
| PRRA1 | 2.4.1 prrA::Tpr | 11 |
| RPS1 | 2.4.1 prrA::TprppsR::Kmr | This study |
| P. denitrificans ATCC 17741 | Wild type | American Type Culture Collection |
| E. coli | ||
| DH5α phe | Strain used for cloning | 11 |
| S17-1 | Tra+ strain used for plasmid mobilization | 37 |
| Plasmids | ||
| pRK415 | Tcr IncP, vector | 23 |
| pPNs | pRK415::ppsR | 15 |
| pLX1 | IncQ, promoterless lacZYA:: ΩSmr-Spr | 15 |
| pLXhemC | pLX1 containing hemC::lacZYA | This study |
| pLXhemE | pLX1 containing hemE::lacZYA | This study |
| p714SmH::Kmr::mob | Suicide mobilizable plasmid, ppsR::ΩKmr | 16 |
The anaerobic phototrophic cultures were grown in front of light bulbs (10 W m−2) in tightly capped 15-ml tubes top-filled with Sistrom's minimal medium. These were seeded with inocula grown under anaerobic-dark-dimethyl sulfoxide (DMSO) conditions. The anaerobic-dark-DMSO cultures were grown in tightly capped tubes wrapped with aluminum foil in medium supplemented with 10% (vol/vol) Luria broth (LB) and 0.3% (vol/vol) DMSO. Prior to transfer to anaerobic phototrophic conditions, the anaerobic-dark-DMSO cultures were centrifuged, and traces of LB and DMSO were removed by washes with Sistrom’s medium.
Paracoccus denitrificans ATCC 17741 was grown in Sistrom's minimal medium at 30°C. Ten-milliliter cultures were grown in 125-ml flasks on a rotating shaker. Antibiotics were used for R. sphaeroides and P. denitrificans, where necessary, at the following concentrations: tetracycline, 1 μg ml−1; streptomycin and spectinomycin, 50 μg ml−1 (each).
Escherichia coli strains were grown at 30°C in LB medium supplemented with the following concentrations of antibiotics: ampicillin, 100 μg ml−1; tetracycline, 10 μg ml−1; streptomycin and spectinomycin, 50 μg ml−1 (each) for strain S17-1, 25 μg ml−1 (each) for strain DH5α.
Spectroscopy and enzymatic assays.
Spectra of the soluble extracts from various R. sphaeroides strains were recorded as previously described (16). Briefly, cells were pelleted, resuspended in 10 mM phosphate buffer (pH 7.2), and passed through a French pressure cell, followed by removal of cell debris by a 15-min centrifugation at 20,000 × g. Soluble cell extracts from various strains were adjusted with the buffer to identical A600 values, and UV-visible spectra were recorded with a Shimatzu UV-1601PC spectrophotometer.
Assays for β-galactosidase activity in P. denitrificans were performed by using o-nitrophenyl-β-d-galactopyranoside with cells permeabilized with chloroform and sodium dodecyl sulfate as described previously (15).
Genetic manipulations and cloning.
Plasmid mobilization into R. sphaeroides and P. denitrificans was performed by using E. coli S17-1 as the donor as described previously (15).
The R. sphaeroides ppsR prrA double mutant, designated RPS1, was constructed by replacement of the wild-type ppsR gene in the prrA null mutant strain PRRA1 (11) with the ppsR::ΩKmr allele essentially as described previously (17). For this purpose, the suicide plasmid p714SmH::Kmr::mob was used (17) (Table 1). The structure of the double mutant was verified by Southern blot hybridization. The mutant PPS2-4, carrying two point mutations in the chromosomal ppsR gene that result in amino acid substitutions Cys251→Ser and Cys424→Ser, was used as a strain with decreased PpsR repressor activity. Details on the construction and characterization of this mutant strain will be presented elsewhere.
Plasmids pLXhemC (hemC::lacZ) and pLXhemE (hemE::lacZ), used to monitor transcription from the hemC and hemE promoters, were constructed as follows. The 0.73-kb fragment containing the intergenic region between two divergently transcribed genes, hemC (RSP0679) and hemE (RSP0680), as well as portions of the coding sequences of these genes was PCR-amplified from R. sphaeroides genomic DNA. The following primers were used for amplification, HemCE-Xb, 5′-gctctagaTGTCCTTCATCGAATGGACCG, and HemCE-Spe, 5′-aggactagtTGGATGTCGTCGCGGCCCTTC, where capital letters correspond to the R. sphaeroides sequence. The amplified fragment was digested with XbaI and SpeI, for which sites were created at the termini of the PCR fragment, and cloned into the digested-with-XbaI vector pLX1 that contains a strong transcription-translation terminator upstream of the reporter gene (15). When cloned in one orientation, the fragment resulted in the hemC::lacZ transcriptional fusion (plasmid pLXhemC), while in the opposite orientation, it resulted in the hemE::lacZ transcriptional fusion (pLXhemE).
RNA extraction and R. sphaeroides genechip hybridization.
R. sphaeroides cells in the early exponential phase (A600 of 0.16 to 0.20) were collected and used for RNA extraction. Three independently grown replicates from each R. sphaeroides strain grown under a given condition were used. The details on RNA extraction; cDNA synthesis, fragmentation, and labeling; and DNA microarray, genechip, structure, and hybridization were described by us previously (13, 32). The genechips were hybridized by using a fluidics station and scanned with a genechip scanner (Affymetrix, Santa Clara, Calif.) at the University of Colorado Cancer Center Microarray Core Facility according to specifications provided by the manufacturer.
Genechip data analysis.
For genechip data analysis, robust multiarray analysis with quantile normalization was used (22) (http://www.stat.berkeley.edu/users/bolstad/RMAExpress/RMAExpress.html). The GeneSpring, version 4.2, software package (Silicon Genetics, Redwood City, Calif.) was used for data presentation. The experimental reproducibility (r values) between three independent replicates from each strain grown at the same conditions were in the range of 0.96 to 0.99. The expression data are deposited in the Gene Expression Omnibus database (www.ncbi.nih.gov/projects/geo), platform GPL162.
Unreliably measured and unchanged genes were filtered out by using two criteria. Criterion one was used to filter out unreliable changes. We retained only genes whose expression values from each replicate of a strain of interest (ai) differed by at least 15% from values for the same gene from each replicate of strain 2.4.1 grown at 20% oxygen (aj), i.e., ai ≥ 1.15 aj or ai ≤ 0.85 aj. Expression in 2.4.1 grown at 20% oxygen was used as the baseline throughout this study. For genes that passed the first criterion, average values from all replicates were derived. Relative changes were calculated based on these average values. These are presented in the figures and text. Criterion two was used to filter out potentially physiologically meaningless changes among genes whose expression passed the reliability criterion. We defined meaningful changes as those where an average expression value for a given gene in a strain of interest (āi) compared to the average value for the same gene in strain 2.4.1 grown at 20% oxygen (āj) followed either of the following guidelines: āi ≥ 1.50 āj or āi ≤ 0.67 āj.
To increase the number of potentially meaningful changes that did not pass the two criteria, we retained for analysis those genes that belonged to clearly identifiable operons if several genes from the operon passed only the first criterion. This is justified because (i) according to our observations and those of others, the magnitude of expression changes measured by genechips are often lower than that measured by alternative techniques, e.g., reverse transcription-PCR (21, 27, 32, 42), and (ii) the primary statistical analysis tool applied here, robust multiarray analysis, has been shown to produce a low percentage of false positives but a relatively high percentage of false-negative changes compared to other methods (22).
qPCR.
Aliquots of cDNA samples used for genechip hybridization, prior to fragmentation, were used for quantitative reverse transcription-mediated real-time PCR (qPCR). The iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, Calif.) with SYBR Green chemistry was used to monitor amplification and to quantify the amount of PCR products. The primers and conditions used for probe amplification were described earlier; expression levels of the rpoZ gene were used for normalization (13). Additional primers for qPCR were selected as follows: bchC forward, ACCTTGGCCTTCAGACCATC, and reverse, ATCTGCCCGGTGTAGAACAG; hemC forward, CCCGTCTGAAGAAGCTGAAC, and reverse, GCATTTCCTCGGACTCGAT; hemE forward, GACAACCTTCATCGGCTTTG, and reverse, GTCGGTGTCCTTCAGCTTGT; prrA forward, GGCTGAGGATCTGGTATTCG, and reverse, AAACCCCGCTTCTCCATC; puhA forward, TGGTGTGACTGCTTTTGGAA, and reverse, CATGTTCTCGGTCTGGAGGT. Each qPCR reaction was performed in triplicate; average data from two to four experimental replicates are reported.
RESULTS
Characterization of strains used for transcriptome profiling.
The R. sphaeroides genome sequence was searched for the TGTN12ACA palindromes believed to represent the PpsR repressor binding site (15, 26). Unexpectedly, 240 palindromes (containing no mismatches) were identified; many of them are present in the noncoding regions upstream of open reading frames (data not shown). Only a small number of these sites are located in the PS gene cluster. Four sites are located upstream of the two PS operons, puc1 and puc2, outside the PS cluster. If all identified palindromes were subject to PpsR binding, the role of PpsR as a regulator specific to a subset of PS genes would have to be reconsidered. Alternatively, TGTN12ACA may be insufficient for PpsR repression in vivo.
To determine the scope of the PpsR regulon, the requirements for PpsR repression, and the role of the PpsR repressor in PS development, we carried out the genome-wide transcription analysis with the following strains: 2.4.1, wild type; 2.4.1(pPNs), 2.4.1 overexpressing the ppsR gene; the ppsR point mutant, PPS2-4 (Table 1), which expresses a less-active repressor (see below).
The ppsR null mutant could not be used in the experiment because of its genetic instability in the presence of high oxygen (17). All strains were grown at high oxygen tension (20% sparged oxygen). Under these conditions, the PpsR repressor activity is high; therefore, PS gene expression is low. The wild-type strain was also grown at low oxygen (0.5% sparged oxygen); when repressor activity is low, PS gene expression is high (17, 29).
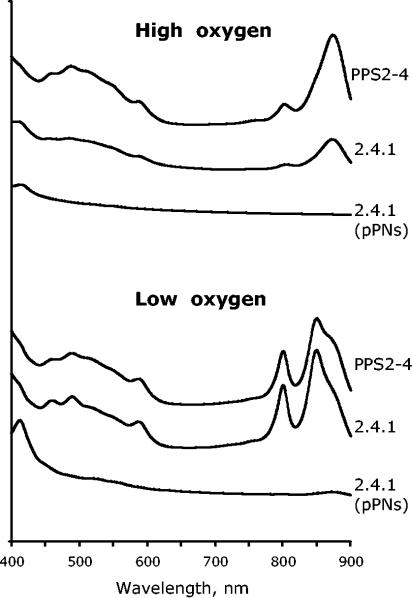
To characterize the effect of various PpsR activities on the formation of PS, we recorded the abundance of photosynthetic complexes in these strains grown at both high and low oxygen tensions (Fig. 1). The wild-type strain produced low levels of PS at high oxygen and high levels at low oxygen. Strain 2.4.1(pPNs) produced no or barely detectable photocomplexes under high or low oxygen, i.e., it was essentially irresponsive to decreased oxygen. In contrast, strain PPS2-4, partially impaired in PpsR activity, produced significant levels of photocomplexes under high oxygen and levels similar to those of the wild type under low oxygen. Thus, strains chosen for transcriptome analysis represent a broad spectrum of PpsR activities. An additional noteworthy point is that increased PpsR abundance in strain 2.4.1(pPNs) grown under high oxygen results in the significantly lower expression of many PS genes.
FIG. 1.
Photosynthetic complexes of the R. sphaeroides strains grown at high (20%) and low (0.5%) oxygen.
PpsR regulon revealed by transcriptome profiling.
Genes whose expression met the following three criteria were considered PpsR-dependent: (i) higher in strain 2.4.1 with 0.5% oxygen than in strain 2.4.1 with 20% oxygen; (ii) higher in PPS2-4 than in strain 2.4.1 with 20% oxygen; (iii) lower in strain 2.4.1(pPNs) than in strain 2.4.1 with 20% oxygen. Three major classes of genes met these criteria.
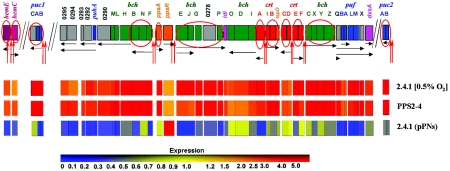
The first class involved PS genes belonging to transcriptional units whose upstream regions contained putative PpsR-binding sites, TGTN12ACA. These genes were shown or anticipated to be directly repressed by PpsR (13, 15, 16, 31, 40, 45). Most bch and crt genes and operons located in the PS cluster as well as puc1 and puc2 operons located outside the PS cluster belong to this class (Fig. 2; see Table S1 in the supplemental material).
FIG. 2.
(Upper panel) R. sphaeroides PpsR regulon. Each gene is represented by a box colored according to its function. Green, bch genes; red, crt genes; blue, genes encoding structural polypeptides of photocomplexes; grey, genes encoding assembly factors or proteins of unknown function; orange, genes encoding regulatory factors; pink, genes encoding enzymes common to Bchl and ubiquinone biosynthesis; magenta, protoporphyrin IX biosynthesis genes. PpsR-binding sites are shown as red vertical arrows. Putative transcripts are shown as black horizontal arrows. Genes that are either known or predicted to be directly repressed by PpsR are circles. (Lower panels) Relative expression of PpsR-dependent genes measured by genechips (compared to that in the wild-type strain grown at high [20%] oxygen). Expression levels are according to the presented color scheme. The expression of every gene in 2.4.1 grown at high (20%) oxygen is assigned a value of 1 (data not shown).
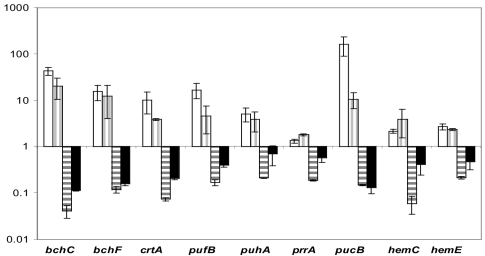
Expression of some PS genes met criteria i and ii but did not meet criterion iii, e.g., bchC (RSP0263), bchY (RSP0261), crtF (RSP0264), crtC (RSP0267), crtD (RSP0266), bchN (RSP0285), and ppaA (RSP0283). Analysis of the absolute expression levels revealed that their expression in 2.4.1 at 20% oxygen was very low, often near detection level by DNA microarrays. Therefore, higher PpsR abundance in strain 2.4.1(pPNs) did not result in the significant expression decrease. As shown here and in the referenced studies, most genes in this category are directly repressed by PpsR, e.g., ppaA (13); bchY belongs to the bchCXYZ operon (repressed by PpsR as shown below); and bchN belongs to the bchFNB operon (15) (Fig. 2). A more-sensitive qPCR performed on one of the genes, bchC, confirmed a significant decrease in its expression in strain 2.4.1(pPNs) compared to 2.4.1 (Fig. 3). Therefore, all genes in this group are, most likely, directly repressed by PpsR. Thus, DNA microarray data confirmed the known fact that PpsR is a regulator of bch, crt, and puc genes.
FIG. 3.
Expression changes (plotted as relative changes with a logarithmic scale) of selected PpsR-dependent genes as measured by qPCR. The expression of every gene in 2.4.1 grown at high (20%) oxygen (data not shown) is assigned a value of 1. White, 2.4.1 grown at low (0.5%) oxygen; vertical stripes, PPS2-4; horizontal stripes, 2.4.1(pPNs); black, APP11. Expression values are derived from RNA from two to four independent experiments, with each done in three replicates. Error bars represent standard deviations.
The second class involved two divergently transcribed genes, hemC and hemE, that do not belong to the PS gene cluster and that were unexpected to be under the control of PpsR (Fig. 2). hemC and hemE encode porphobilinogen deaminase and uroporphyrinogen decarboxylase, respectively, which are involved in protoporphyrin IX biosynthesis. Protoporphyrin IX is a common precursor of Bchl and heme. Increased expression of hemC and hemE would seem reasonable given the higher demand for Bchl under low oxygen. The intergenic region between hemC and hemE contains a TGTN12ACA palindrome, i.e., a putative PpsR-binding site (see Table S1 in the supplemental material). Below we show that PpsR directly represses hemC and hemE transcription.
The third class involved genes whose expression followed all criteria described above; however, their upstream regions did not contain PpsR binding sites (Fig. 2). For example, mRNA levels of the puf operon (RSP0258-0255) located at one end of the PS gene cluster were severalfold increased in strain PPS2-4 and significantly decreased in strain 2.4.1(pPNs) compared to 2.4.1 with 20% oxygen. A similar expression pattern was observed for puhA (RSP0291) and genes downstream of puhA, RSP0292-0295, which are located at the other end of the PS gene cluster (Fig. 2). We showed previously that PpsR does not directly repress puf::lacZ expression (15). The mechanism(s) by which PpsR affects puf and puhA operon expression remains elusive (see Discussion). The fact that PpsR activity affects expression of the RC and LHI proteins, in addition to expression of Bchl, Crt, and LHII, suggests that PpsR functions as a master regulator of PS development. Physiological significance of this hypothesis is tested below.
A separate group involved non-PS genes whose expression changed significantly in some but not all strains with perturbed PpsR activities. The upstream regions of these genes contain no putative PpsR-binding sites. Therefore, they must be affected by PpsR indirectly, e.g., through the effects of PpsR on expression of other regulatory proteins (see Discussion).
Verification of genechip data by qPCR.
To verify the results of DNA microarrays, we assessed gene expression by qPCR of selected genes (Fig. 3). The qPCR data were in good agreement with the DNA microarray results confirming all observed changes. However, the relative changes measured by the two techniques were not identical, with the microarray data usually underestimating changes detected by qPCR. Importantly, qPCR data verified dependence on PpsR of the hemC and hemE genes, two newly identified targets of PpsR.
To confirm the significance of expression changes observed in strain 2.4.1(pPNs), we used an additional strain, i.e., APP11 (Table 1). This strain harbors a null mutation in the appA antirepressor gene, which in effect results in a higher level of the active PpsR repressor in APP11 than in strain 2.4.1. As seen in Fig. 3, the changes in expression in 2.4.1(pPNs) and APP11 are qualitatively identical, with the latter strain having generally smaller relative changes. This is consistent with the comparison of transcriptome profiles of these two strains measured by genechips and reported previously (4).
Refined requirements for PpsR-mediated repression.
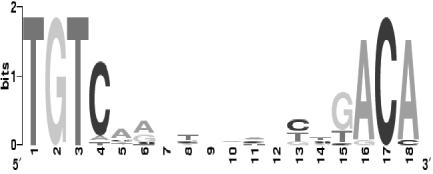
The DNA microarray data showed that only a small fraction of the 240 putative PpsR binding sites, TGTN12ACA, identified in the R. sphaeroides genome are functional. Based on the sequences of predicted repressor binding sites that are identified upstream of the genes whose expression is PpsR dependent (see Table S1 in the supplemental material), we refined the consensus sequence for PpsR binding as TGTcN10gACA (lowercase letters indicate lesser conservation) (Fig. 4).
FIG. 4.
Sequence logo of the consensus sequence for PpsR binding created by using WebLogo (http://weblogo.berkeley.edu) (8).
We found that all PpsR-dependent genes contain two repressor binding sites, one of which has no more than a single mismatch with the refined consensus, whereas the other one may contain up to three mismatches (Fig. 4; see Table S1 in the supplemental material). Given the high sequence similarity in the DNA binding domains of the PpsR homologs from anoxygenic phototrophic proteobacteria, we believe that the identified consensus will be applicable to the homologs of R. sphaeroides PpsR.
Regulation of hemC and hemE gene expression by PpsR.
Until this study, no R. sphaeroides genes other than those in the PS gene cluster and two puc operons were known to be regulated by the PpsR repressor. The indication from genechip (Fig. 2) and qPCR (Fig. 3) data that hemC and hemE expression could be directly regulated by PpsR was therefore unexpected and investigated further.
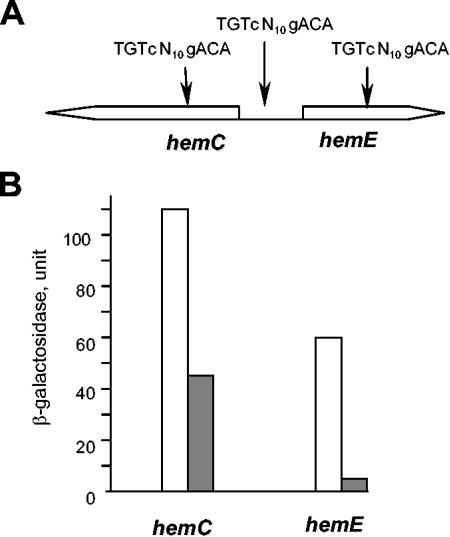
The intergenic region between the divergently transcribed hemC and hemE genes contains one TGTgN10cACA palindrome, whose 5′ end is located 35 bp upstream of the start codon of hemC and 84 bp upstream of the start codon of hemE (Fig. 5A; see Table S1 in the supplemental material). The location of the palindrome suggests that PpsR may regulate transcription of both of these genes directly by interfering with either RNA polymerase binding or transcription elongation. However, all PS genes directly regulated by PpsR appear to contain two repressor binding sites. No second palindrome, with up to three mismatches, is present between hemC and hemE. Interestingly, two TGTgN10cACA palindromes are located within coding regions of these genes, i.e., 236 bp downstream of the start codon of hemC and 284 bp downstream of the start codon of hemE (Fig. 5A). The probability of three palindromes occurring by chance in an approximately 0.5-kb DNA fragment of R. sphaeroides genome is extremely low (approximately 2 × 10−5). This suggests that one or both palindromes located in the coding regions is/are involved in repression; the molecular details remain to be investigated.
FIG. 5.
(A) Scheme of genetic organization of the hemC-hemE region. Putative PpsR binding sites are shown. (B) Expression of the hemC::lacZ and hemE::lacZ transcriptional fusions in P. denitrificans containing in trans either vector pRK415 (white bars) or plasmid pPNs (pRK415::ppsR) (grey bars). One unit of β-galactosidase is equal to 1 nmol of o-nitrophenyl-β-d-galactopyranoside ΔA600−1 ml of culture−1 min−1. Average data from three independent experiments are shown, with standard deviations not exceeding 13% of the averages.
To test whether or not PpsR directly represses hemC and hemE expression, we constructed hemC::lacZ and hemE::lacZ transcriptional fusions (plasmids pLXhemC and pLXhemE, respectively), both containing the fragment that incorporates all three TGTgN10cACA palindromes. The hemC::lacZ and hemE::lacZ fusions were transferred into the heterologous host, P. denitrificans ATCC 17741, which belongs to the same alpha subdivision of proteobacteria as R. sphaeroides. P. denitrificans is a close relative of R. sphaeroides; however, it is nonphototrophic and lacks PS genes and their regulators. P. denitrificans lends itself as a useful host for investigating the effects of individual regulators of PS gene expression (13, 15, 17).
lacZ expression was compared in strains containing either plasmid pPNs, which overexpresses PpsR, or empty vector pRK415, i.e., ATCC 17741(pLXhemC, pPNs) versus ATCC 17741(pLXhemC, pRK415) and ATCC 17741(pLXhemE, pPNs) versus ATCC 17741(pLXhemE, pRK415) (Fig. 5B). The presence of PpsR in P. denitrificans resulted in the significant repression of expression of both lacZ fusions, which is similar to what was observed for other PpsR-dependent genes tested (15, 17). This suggests that PpsR directly represses expression of hemC and hemE in R. sphaeroides.
Role of PpsR in PS development.
One of the unexpected findings of this study was that PpsR affected expression of the puf and puhA operons encoding RC and LHI complex proteins, which comprise the core of the PS unit. This suggests that the role of PpsR is not limited to regulation of Crt and Bchl biosynthesis and expression of the LHII complex, as is commonly perceived. The transcriptome data summarized in Fig. 2 suggest that PpsR is a master regulator of PS development. However, do increased levels of puf and puhA that result from inactivation of PpsR have physiological significance, e.g., are they sufficient for phototrophic growth?
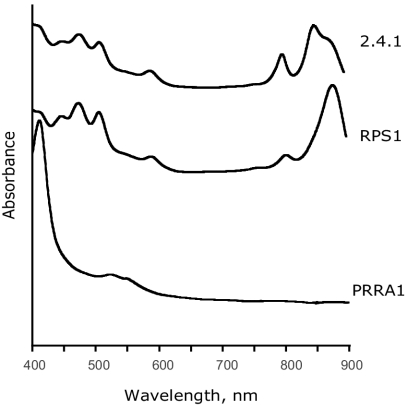
Under the anoxic conditions, puf and puhA expression is activated by PrrA, the global redox-dependent activator of gene expression (10, 11, 44). A prrA null mutant is phototrophically incompetent (11). To separate the effects of PrrA and PpsR on puf and puhA expression, we constructed a prrA ppsR double mutant, RPS1, and assayed (i) its ability to grow phototrophically and (ii) the abundance of the LHI complex (using absorption at 875 nm) encoded by the puf operon. The abundance of the RC complex is difficult to assess because its absorption maximum (805 nm) overlaps with the maximum of the LHII complex (800 nm) (Fig. 6).
FIG. 6.
Photosynthetic complexes of the R. sphaeroides strains grown under anaerobic phototrophic conditions at 10 W of white light m−2 (2.4.1 and RPS1) or under anaerobic-dark-DMSO conditions (PRRA1). Cultures were started from the inoculum grown under anaerobic-dark-DMSO conditions. Strain PRRA1 is unable to grow phototrophically.
We found that, in contrast to the prrA mutant PRRA1, the prrA ppsR double mutant RPS1 grows phototrophically, albeit much slower than 2.4.1. Spectral analysis of the phototrophically grown strain RPS1 revealed that the LHI complex is abundant (Fig. 6). The direct comparison of LHI abundance in RPS1 and PRRA1 is impossible because the latter strain does not grow phototrophically. However, comparison of the photosynthetic spectral complexes from cultures grown under the anaerobic-dark-DMSO conditions showed high levels of LHI in strain RPS1 and absence of LHI in PRRA1 (Fig. 6 and data not shown). The fact that elevated puf and puhA expression in RPS1 (along with elevated expression of other PS genes) was sufficient for phototrophic growth in the absence of PrrA provides support for physiological significance of regulation of these operons by PpsR. This supports the notion that PpsR is a master regulator of PS development.
Does the AppA-PpsR regulatory pathway affect other regulators of PS gene expression?
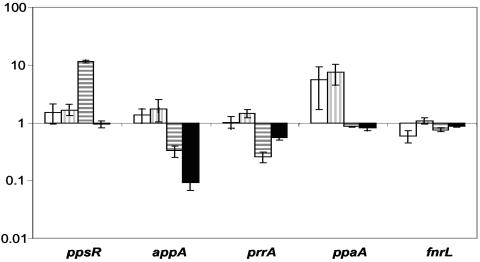
Data from this study link the AppA-PpsR pathway to other regulators of PS gene expression. One such link is dependence of expression of regulatory genes on the activity of PpsR. The expression patterns of most influential regulators of PS gene expression (44) are summarized in Fig. 7.
FIG. 7.
Expression changes (plotted as relative changes with a logarithmic scale) of selected regulatory genes controlling PS gene expression as measured by genechips. The expression of every gene in 2.4.1 grown at high (20%) oxygen (data not shown) was assigned a value of 1. White, 2.4.1 grown at low (0.5%) oxygen; vertical stripes, PPS2-4; horizontal stripes, 2.4.1(pPNs); black, APP11.
Expression of three regulatory factors significantly responded to the level of PpsR repressor activity. Expression of the ppaA gene encoding an apparent aerobic activator of PS genes was higher in strain PPS2-4 than in 2.4.1. PpsR-dependent expression of ppaA is consistent with our earlier reports (4, 13). Expression of the appA gene was significantly downregulated in strain 2.4.1(pPNs), suggesting that not only does this strain contain more PpsR protein expressed from plasmid pPNs, it also contains significantly less antirepressor AppA.
Surprisingly, expression of the prrA gene was found to respond to the level of PpsR activity in the most consistent manner. The prrA mRNA levels were low (approximately 25% of the wild-type levels) in 2.4.1(pPNs) and high (approximately 200% of the wild-type levels) in PPS2-4 (Fig. 3 and 7). The decrease in prrA expression in 2.4.1(pPNs) was corroborated by lower prrA expression (approximately 50% of the wild-type levels) in the appA null mutant, APP11 (Fig. 3 and 7). The mechanism of dependence of prrA expression on PpsR remains elusive. It is obviously indirect, since there are no PpsR binding sites upstream of prrA. The PpsR-dependent pattern of prrA expression and strikingly different phenotypes of the prrA single mutant and prrA ppsR double mutant pose interesting questions regarding interactions between the PrrBA and AppA-PpsR systems. These are discussed below.
DISCUSSION
Requirements for PpsR-mediated repression.
Analysis of the complete R. sphaeroides genome identified 240 putative PpsR binding sites, TGTN12ACA. The transcriptome profiling reported here revealed that only a small fraction of these sites is functional in vivo (see Table S1 in the supplemental material). Based on the repressor binding sites located upstream of the genes whose expression is PpsR dependent, we refined the consensus sequence for PpsR binding as TGTcN10gACA (Fig. 4).
We found that all PpsR-dependent genes contain two repressor binding sites, one of which has no more than a single mismatch with the refined consensus, whereas the other one may contain up to three mismatches. It has been shown that the PpsR repressor homolog from Rhodobacter capsulatus, CrtJ, which shares 52% identity (72% similarity) with PpsR, binds to a single palindrome in vitro with a relatively low affinity. However, the presence of two palindromes results in much stronger repression, apparently due to physical interactions between two CrtJ-DNA complexes. The binding sites may be located in the immediate vicinity of each other or separated by an intervening DNA loop (9, 35). Our expression data in vivo provide support to these in vitro observations and suggest that the presence of a single PpsR binding site is likely to be insufficient for functional repression. This is consistent with the fact that PpsR exists as a tetramer in solution (19). Therefore, a dimer or a tetramer of PpsR bound to one site must interact with a dimer or tetramer bound to another site to ensure repression in vivo.
Newly discovered members of the PpsR regulon.
The vast majority of PpsR-dependent genes are located in the PS gene cluster or upstream of the puc operons, thus confirming the preexisting notion that PpsR is a regulator of PS gene expression. In addition to these genes, we identified two new groups of genes that belong to the PpsR regulon. One group includes hemC and hemE genes. Their products are involved in protoporphyrin IX biosynthesis and therefore contribute to Bchl and heme production. Only one binding site is located in the intergenic region between these genes, while two putative sites are located in the coding sequences. Whether one or both sites located in the coding regions are necessary for repression has yet to be determined. We showed that, despite the unusual arrangement of the PpsR binding sites, PpsR directly represses expression of hemC and hemE. A recent report published after submission of the manuscript found that hemC and hemE expression in the related organism, R. capsulatus, is dependent on the PpsR homolog, CrtJ (38).
In R. sphaeroides, the expression of most hem genes is upregulated in response to deprivation of oxygen (32, 36). It is peculiar that various steps of the protoporphyrin IX pathway are regulated by different transcriptional factors, e.g., PpsR is responsible for upregulation of hemC and hemE, PrrBA is responsible for upregulation of hemA, and FnrL is responsible for upregulation of hemN and hemZ (31, 43). To achieve coordinate upregulation of the hem pathway, one may anticipate that activities of these regulators must be coordinated. The microarray data for the first time revealed that such coordination exists between the PrrBA and AppA-PpsR regulatory systems. However, the scope and molecular mechanisms of coordination of activities of oxygen-dependent transcriptional factors in R. sphaeroides have yet to be explored.
PpsR as a master regulator of PS development.
One of the unexpected outcomes of the DNA microarray studies was identification of the third group of PpsR-dependent genes, i.e., those whose upstream regions don't contain PpsR binding sites. Importantly, these genes included the puf and puhA operons encoding structural proteins and assembly factors of the RC and LHI complexes, which comprise the core of the R. sphaeroides PS. Involvement of PpsR in regulation of expression of these complexes, in addition to regulation of the Bchl and Crt biosynthetic pathways and LHII, suggests that PpsR is a master regulator of PS development. Consistent with this interpretation is our observation that inactivation of PpsR was sufficient for restoration of phototrophic growth in the prrA null mutant (Fig. 6). Up to this study, PrrA was believed to be essential for PS growth. Further supporting this conclusion is the fact that overexpression of either PpsR or an appA null mutation is sufficient to render cells unable to grow phototrophically, even in the presence of intact PrrA (Fig. 1) (17).
How does PpsR affect expression of the puf and puhA operons? One possibility is that transcript levels of genes that contain no PpsR binding sites are increased due to the readthrough from the PpsR-dependent promoters of the upstream genes. Long multioperon transcripts termed superoperons have been detected in a related bacterium, R. capsulatus, e.g., bchCXYZ-Q-pufBALMX and bchFNBHLM-orf1477-puhA (2, 41). This possibility remains viable. However, the existence of superoperons in R. sphaeroides has not been shown, and their physiological significance, if any, remains unknown.
Another hypothesis is that PpsR affects the expression of a regulatory factor(s) that controls puf and puhA gene expression. Several independent lines of evidence support this hypothesis. One is based on the observed differential dynamics of changes in PS gene expression in response to changing environment (4). Blue-light irradiation of the semiaerobically grown culture results in a decreased expression of the vast majority of PS genes. Blue light is perceived by the antirepressor AppA, whose affinity to PpsR decreases upon illumination, thus increasing the level of free repressor (3, 4, 17, 29). We found that levels of those PS transcripts that are directly regulated by PpsR decrease fast after exposure to blue light. The levels of puf and puhA transcripts also decrease; however, they do so after a significant delay. The delayed response of puf and puhA operons compared to the response of genes directly repressed by PpsR implies that a change in the level of an additional regulatory factor takes place during the delay (4).
Another line of evidence is based on the fact that expression of the puf::lacZ transcriptional fusion containing no PpsR-binding sites is significantly lower in the appA null mutant, APP11, than in 2.4.1 (16). Note that we showed that PpsR does not directly affect puf expression (15) and that AppA affects transcription exclusively through the PpsR pathway (4). The identity of the regulatory factor of puf and puhA expression remains unknown; however, PpaA and PrrA are prime candidates based on the dependence of their expression on the PpsR activity (Fig. 3 and 7).
Interactions between the AppA-PpsR and PrrA regulatory pathways of PS gene expression.
The dependence of prrA gene expression on PpsR activity provides one link between the AppA-PpsR and PrrBA regulatory pathways, which up to this study were believed to work independently. The phototrophic growth of the prrA ppsR double mutant provides another, perhaps even more intriguing, link. The fact that the prrA mutant does not grow phototrophically under anaerobic conditions, whereas the prrA ppsR double mutant does, suggests the following. (i) The reason for phototrophic incompetence of the prrA mutant lies primarily in its inability to upregulate PS gene expression and not in defects in numerous other processes that are controlled by PrrA (10, 44). (ii) Phototrophic incompetence of the prrA mutant stems from its inability to inactivate PpsR. This means that the PrrBA system somehow controls the activity or expression of the PpsR repressor or the activity or expression of the AppA antirepressor. Our unpublished data suggest that the latter possibility is true. While the details of the interdependence of the AppA-PpsR and PrrBA pathways remain to be unraveled, it is clear that these two major regulatory pathways communicate with one another to achieve coordinate regulation of PS development.
Supplementary Material
Acknowledgments
This work was supported by grants NIH NCRR P20 RR15640 (COBRE) and DOE DE-FG02-01ER63232.
We are grateful to the staff of the DNA Microarray Core Facility at the UCHSC Cancer Center in Denver for genechip hybridizations. The real-time reverse transcription-PCR was performed at the Macromolecular Core Facility of the University of Wyoming.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bauer, C., S. Elsen, L. R. Swem, D. L. Swem, and S. Masuda. 2003. Redox and light regulation of gene expression in photosynthetic prokaryotes. Philos. Trans. R. Soc. Lond. B 358:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, C. E., J. J. Buggy, Z. M. Yang, and B. L. Marrs. 1991. The superoperonal organization of genes for pigment biosynthesis and reaction center proteins is a conserved feature in Rhodobacter capsulatus: analysis of overlapping bchB and puhA transcripts. Mol. Gen. Genet. 228:443-454. [DOI] [PubMed] [Google Scholar]
- 3.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. The single flavoprotein, AppA, from Rhodobacter sphaeroides integrates both redox and light signals. Mol. Microbiol. 45:827-836. [DOI] [PubMed] [Google Scholar]
- 4.Braatsch, S., O. V. Moskvin, G. Klug, and M. Gomelsky. 2004. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J. Bacteriol. 186:7726-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, S. H., S. H. Youn, S. R. Lee, H. S. Yim, and S. O. Kang. 2004. Redox property and regulation of PpsR, a transcriptional repressor of photosystem gene expression in Rhodobacter sphaeroides. Microbiology 150:697-706. [DOI] [PubMed] [Google Scholar]
- 6.Choudhary, M., and S. Kaplan. 2000. DNA sequence analysis of the photosynthesis region of Rhodobacter sphaeroides 2.4.1. Nucleic Acids Res. 28:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Comp. Physiol. 49:25-68. [DOI] [PubMed] [Google Scholar]
- 8.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsen, S., S. N. Ponnampalam, and C. E. Bauer. 1998. CrtJ bound to distant binding sites interacts cooperatively to aerobically repress photopigment biosynthesis and light harvesting II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 273:30762-30769. [DOI] [PubMed] [Google Scholar]
- 10.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68:263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eraso, J. M., and S. Kaplan. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 176:32-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giraud, E., J. Fardoux, N. Fourrier, L. Hannibal, B. Genty, P. Bouye, B. Dreyfus, and A. Vermeglio. 2002. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417:202-205. [DOI] [PubMed] [Google Scholar]
- 13.Gomelsky, L., J. Sram, O. V. Moskvin, I. M. Horne, H. N. Dodd, J. M. Pemberton, A. G. McEwan, S. Kaplan, and M. Gomelsky. 2003. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology 149:377-388. [DOI] [PubMed] [Google Scholar]
- 14.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497-500. [DOI] [PubMed] [Google Scholar]
- 15.Gomelsky, M., and S. Kaplan. 1995. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J. Bacteriol. 177:1634-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomelsky, M., and S. Kaplan. 1995. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 177:4609-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomelsky, M., and S. Kaplan. 1997. Molecular genetic evidence suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 179:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomelsky, M., and S. Kaplan. 1998. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel FAD binding domain. J. Biol. Chem. 273:35319-35325. [DOI] [PubMed] [Google Scholar]
- 19.Gomelsky, M., I. M. Horne, H. J. Lee, J. M. Pemberton, A. G. McEwan, and S. Kaplan. 2000. Domain structure, oligomeric state, and mutational analysis of PpsR, the Rhodobacter sphaeroides repressor of photosystem gene expression. J. Bacteriol. 182:2253-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, Y., S. Braatsch, L. Osterloh, and G. Klug. 2004. A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc. Natl. Acad. Sci. USA 101:12306-12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holland, M. J. 2002. Transcript abundance in yeast varies over six orders of magnitude. J. Biol. Chem. 277:14363-14366. [DOI] [PubMed] [Google Scholar]
- 22.Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 24.Kraft, B. J., S. Masuda, J. Kikuchi, V. Dragnea, G. Tollin, J. M. Zaleski, and C. E. Bauer. 2003. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: a novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry 42:6726-6734. [DOI] [PubMed] [Google Scholar]
- 25.Laan, W., M. A. van der Horst, I. H. van Stokkum, and K. J. Hellingwerf. 2003. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine dinucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: a key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochem. Photobiol. 78:290-297. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. K., and S. Kaplan. 1995. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides. Analysis of the cis-acting downstream regulatory sequence. J. Biol. Chem. 270:20453-20458. [PubMed] [Google Scholar]
- 27.Lee, J. M., S. Zhang, S. Saha, S. Santa Anna, C. Jiang, and J. Perkins. 2001. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 183:7371-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackenzie, C., M. Choudhary, F. W. Larimer, P. F. Predki, S. Stilwagen, J. P. Armitage, R. D. Barber, T. J. Donohue, J. P. Hosler, J. Newman, J. P. Shapleigh, R. E. Sockett, J. Zeilstra-Ryalls, and S. Kaplan. 2001. The home stretch, a preliminary analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 70:19-41. [DOI] [PubMed] [Google Scholar]
- 29.Masuda, S., and C. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110:613-623. [DOI] [PubMed] [Google Scholar]
- 30.McEwan, A. G. 1994. Photosynthetic electron transport and anaerobic metabolism in purple non-sulfur phototrophic bacteria. Antonie Leeuwenhoek 66:151-164. [DOI] [PubMed] [Google Scholar]
- 31.Oh, J. I., J. M. Eraso, and S. Kaplan. 2000. Interacting regulatory circuits involved in orderly control of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 182:3081-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pappas, C. T., J. Sram, O. V. Moskvin, P. S. Ivanov, R. C. Mackenzie, M. Choudhary, M. L. Land, F. W. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 186:4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 176:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponnampalam, S. N., and C. E. Bauer. 1997. DNA binding characteristics of CrtJ. A redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 272:18391-18396. [DOI] [PubMed] [Google Scholar]
- 35.Ponnampalam, S. N., S. Elsen, and C. E. Bauer. 1998. Aerobic repression of the Rhodobacter capsulatus bchC promoter involves cooperative interactions between CrtJ bound to neighboring palindromes. J. Biol. Chem. 273:30757-30761. [DOI] [PubMed] [Google Scholar]
- 36.Roh, J. H., W. E. Smith, and S. Kaplan. 2004. Effects of oxygen and light intensity on transcriptome expression in Rhodobacter sphaeroides 2.4.1. Redox active gene expression profile. J. Biol. Chem. 279:9146-9155. [DOI] [PubMed] [Google Scholar]
- 37.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 38.Smart, J. L., J. W. Willett, and C. E. Bauer. 2004. Regulation of hem gene expression in Rhodobacter capsulatus by redox and photosystem regulators RegA, CrtJ, FnrL, and AerR. J. Mol. Biol. 342:1171-1186. [DOI] [PubMed] [Google Scholar]
- 39.Steunou, A. S., C. Astier, and S. Ouchane. 2004. Regulation of photosynthesis genes in Rubrivivax gelatinosus: transcription factor PpsR is involved in both negative and positive control. J. Bacteriol. 186:3133-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeliseev, A. A., and S. Kaplan. 1995. A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 270:21167-21175. [DOI] [PubMed] [Google Scholar]
- 41.Young, D. A., C. E. Bauer, J. C. Williams, and B. L. Marrs. 1989. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol. Gen. Genet. 218:1-12. [DOI] [PubMed] [Google Scholar]
- 42.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeilstra-Ryalls, J., and S. Kaplan. 1996. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: regulation through alterations in the cellular redox state. J. Bacteriol. 178:985-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeilstra-Ryalls, J., and S. Kaplan. 2004. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell. Mol. Life Sci. 61:417-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng, X., M. Choudhary, and S. Kaplan. 2003. A second and unusual pucBA operon of Rhodobacter sphaeroides 2.4.1: genetics and function of the encoded polypeptides. J. Bacteriol. 185:6171-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.