Abstract
An isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter was constructed in Myxococcus xanthus. The single-copy pilA gene encodes pilin, the monomer unit of M. xanthus type IV pili. To vary the level of pilA expression, we cloned its promoter in front of the lac operator, and a plasmid containing the construct was inserted into the chromosome of a ΔpilA strain. Induction of pilin expression increased smoothly as the dose of IPTG added to the culture was increased. IPTG-induced pilin rescued S motility of the ΔpilA strain to wild-type levels. The rate of S-motile swarming was found to be proportional to the number of pili (shear-sensitive pilin) produced rather than to the level of total pilin. In fact, S motility was not rescued until the total level of pilin was more than 50% of the wild-type level. This observation implies that a threshold concentration of pilin must be exceeded before the shear-sensitive material (pili) is polymerized in M. xanthus.
The polar type IV pili of the gram-negative δ-proteobacterium Myxococcus xanthus help it to swarm over the surface of a solid growth medium, like agar (4, 5, 8). The pili also help M. xanthus build fruiting bodies when it begins to starve (7). In addition to the pili, which are called the social (S) engine and which are located at the leading pole of a gliding cell, M. xanthus has engines for adventurous motility located at the trailing pole of the cell (21). Swarming and fruiting body construction depend on coordinating these two polar gliding engines. Pili are long filamentous appendages that are helical polymers that contain tens of thousands of copies of the protein subunit pilin, the pilA gene product (15). To glide, the polar type IV pili of a M. xanthus cell attach to fibrils on other cells and then retract, which pulls the piliated cell forward (14). On both sides of the pilA gene, 16 other pil genes cluster together in the M. xanthus genome. The cluster includes two two-component systems, the pilS-pilR and pilS2-pilR2 systems (18). Little is known about the role of these regulators except that pilR is necessary for pilA expression (24), and a pilR2 knockout mutant is unable to swarm (2).
M. xanthus is particularly useful for investigating the molecular mechanisms that underlie gliding motility and its coordination during fruiting body development (7). Transposon insertions of transcriptional reporters and nonpolar in-frame gene deletions have permitted the assignment of functions to several motility genes. Conditional expression of proteins with a nontoxic inducer has been lacking, however, which has limited expression studies and has hindered investigation of the pilus assembly process. Previous studies of the Escherichia coli tac promoter/lac repressor/operator in M. xanthus showed that isopropyl-β-d-thiogalactopyranoside (IPTG) could be used as an inducer of gene expression in M. xanthus (12). Unfortunately, expression of the tac promoter is expected to depend on σA, the major sigma factor of M. xanthus, and σA is lost during the development of M. xanthus (3). The native CarQRS system has also been tested as a means for controlling gene expression in M. xanthus (12). However, this system, which is induced by high-intensity light, interferes with development (13). Here we report a new adaptation of the E. coli lac repressor and operator that was used to construct a chromosome-based system for inducible regulation of gene expression that can function during both growth and development in M. xanthus. With this system, a single-copy gene can be removed from its endogenous regulation by placing it under the transcriptional control of an IPTG-inducible promoter. To test the regulation, we employed the well-characterized pilA (pilin) promoter that depends on the alternative sigma factor σ54 for transcription. This promoter was chosen because it is expressed at a high level during both growth and development (24). To quantify promoter activity, we tested the proportionality among the level of pilA expression, the amount of pili assembled, and the rate of S-motile swarming in M. xanthus.
MATERIALS AND METHODS
Bacterial strains and growth.
Table 1 lists the strains employed. M. xanthus strains were cultured in liquid CTT medium (1% Casitone, 10 mM Tris-HCl, 8 mM MgSO4, 1 mM KPO4 [pH 7.6]) or on CTT agar plates. E. coli strains were cultured in Luria-Bertani medium. Antibiotics were added when appropriate at the following concentrations: kanamycin, 40 μg/ml; carbenicillin, 100 μg/ml; and tetracycline, 10 μg/ml.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant genotype, phenotype, and/or sequence | Reference or source |
|---|---|---|
| M. xanthus strains | ||
| DK1622 | Wild type | 8 |
| DK10410 | ΔpilA | 24 |
| DK10404 | DK 1622::pilR-Ω3163 | 24 |
| DK10415 | ΔpilS | 23 |
| DK12600 | DK10410::pLOJ7 | This study |
| DK12601 | DK10410::pLOJ7/pLOJ6 | This study |
| DK12602 | DK1622::pLOJ5 | This study |
| DK12603 | DK1622::pLOJ5/pLOJ6 | This study |
| DK12604 | DK1622::pLOJ5/pLOJ28 | This study |
| DK12612 | DK10404::pLOJ7 | This study |
| E. coli strains | ||
| TOP10 | Invitrogen | |
| C600 | Wild-type lacZ gene | |
| Plasmids | ||
| pBluescriptII KS(+) | Cloning vector, Ampr | Stratagene |
| pBGS18 | Cloning vector, Kanr | 17 |
| pSWU30 | Cloning vector, Mx8 attP Tcr | 22 |
| pET28 | Cloning vector, lacIq gene, Kanr | Novagen |
| pLOJ1 | pilA promoter, Ampr | This study |
| pLOJ2 | Leader with one lac operator, Kanr | This study |
| pLOJ4 | Inducible pilA promoter, Kanr | This study |
| pLOJ5 | Inducible lacZ gene, one operator, Kanr | This study |
| pLOJ6 | lacIq, Mx8 attP Tcr | This study |
| pLOJ7 | Inducible pilA gene, Kanr | This study |
| pLOJ15 | pilA promoter, Ampr | This study |
| pLOJ16 | pilA promoter-lacI, Ampr | This study |
| pLOJ28 | pilA promoter-lacI, Tcr | This study |
| Primersa | ||
| 1 | 5′-GGCCCAAGCTTGATGGCACCGTCATGTTGGACG-3′ | |
| 2 | 5′-CGGGATCCCCCGCGGATGGGATTAGCACTA-3′ | |
| 3 | 5′-CGGGATCCGGGAATTGTGAGCGGATAACAATTCCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGAGGTACCCC-3′ | |
| 4 | 5′-GGGGTACCTCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGAGGGGAATTGTTATCCGCTCACAATTCCCGGATCCCG-3′ | |
| 5 | 5′-GTACCATGCGCGTCTCGCGATTCAAC-3′ | |
| 6 | 5′-GGAATTCGGCCCGAAGAACAGTGGACC-3′ | |
| 7 | 5′-CGGGGTACCATGACCATGATTACGGATTCACTGGC-3′ | |
| 8 | 5′-CCTGCCCGGTTATTATTATTTTTGACACC-3′ | |
| 10 | 5′-GGGATATCCTCAGAGAAGGTTGCAACGGGG-3′ | |
| 11 | 5′-TCCCCCGGGCATGTGAAACCAGTAACGTTATACGATGTCGC-3′ | |
| 12 | 5′-CGGAATTCCGCACCTGTCCTACGAGTTGC-3′ | |
| 13 | 5′-CAGTTGGTTAGAACGCCGGC-3′ | |
| 14 | 5′-CTGATTCTCCCGAGCCCCAC-3′ | |
| 15 | 5′-CAGCACCTTGCGCTCACGTC-3′ | |
| 16 | 5′-GGGCAAAACCCGAGCCTCTC-3′ |
Use of the primers is described in Materials and Methods.
Construction of an IPTG-inducible pilA promoter.
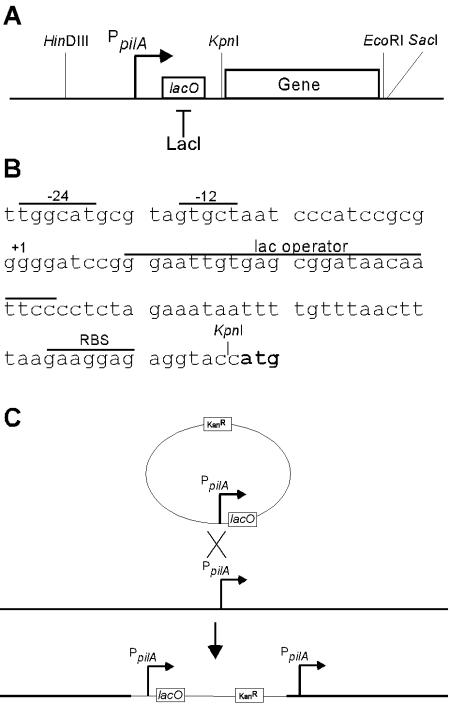
A pilA promoter fragment (PpilA) extending from −855 to 1 bp relative to the transcriptional start site was PCR amplified from genomic DNA of DK1622 by using primers 1 and 2 (Table 1). The resulting PCR product was digested with HindIII-BamHI and cloned into HindIII-BamHI-digested pBluescript II KS(+) (Stratagene), resulting in plasmid pLOJ1. Next, a leader fragment containing the lac operator (lacO) and a ribosome binding site (RBS) was constructed by annealing 79-bp primers 3 and 4 (Table 1). Briefly, 100-pmol portions of the two primers were mixed in water, boiled for 5 min, and allowed to cool to room temperature. The DNA fragment was extracted from a 2% agarose gel, digested with BamHI-KpnI, and cloned into BamHI-KpnI-digested pBGS18 (17), resulting in plasmid pLOJ2.
Plasmid pLOJ4 was constructed by cloning the HindIII-BamHI pilA promoter fragment from pLOJ1 into pLOJ2 digested with HindIII-BamHI. Thus, plasmid pLOJ4 contained the full-length pilA promoter followed by a leader sequence containing lacO and the RBS (PpilA-lacO-RBS). A gene that was to be controlled from this inducible promoter could be inserted 6 bp downstream of the RBS by using a unique KpnI restriction site as the upstream site and either a SacI or EcoRI site as the downstream site. In order to clone the pilA gene, plasmid pLOJ4 was first opened with KpnI, and the 3′ overhang was removed by Klenow treatment. The blunted vector was then digested with EcoRI. The pilA gene was PCR amplified by using primers 5 and 6, followed by digestion with EcoRI, and was cloned into digested pLOJ4, resulting in plasmid pLOJ7. In this plasmid, the pilA start codon was located 7 bp downstream of the RBS. Expression of the pilA gene was controlled by the PpilA-lacO-RBS cassette.
To clone the lacZ gene downstream of the PpilA-lacO-RBS cassette, plasmid pLOJ4 was first opened with EcoRI, and this was followed by filling in the 5′ overhang by Klenow treatment. The blunted vector was then digested with KpnI. The lacZ gene was PCR amplified from genomic DNA of E. coli strain C600 by using primers 7 and 8, digested with KpnI, and cloned into blunt-KpnI pLOJ4, resulting in plasmid pLOJ5.
Construction of plasmids encoding the lac repressor lacI.
In order to repress expression from PpilA-lacO-RBS, the lac repressor gene, lacI, was isolated from plasmid pET28 (Novagen) containing the lacI gene and its promoter as an XbaI-FspI fragment. This fragment was inserted into the Mx8 attP vector pSWU30 digested with XbaI-SmaI, resulting in plasmid pLOJ6. In plasmid pLOJ28, the pilA promoter is used to drive expression of the lacI gene. As with the construction of pLOJ1 described above, primers 1 and 10 (Table 1) were used to PCR amplify the pilA promoter, including its native leader sequence. The fragment was digested with HindIII and EcoRV (Fig. 1A) and cloned into pBluescript KSII(+), resulting in pLOJ15. The lacI gene was PCR amplified from plasmid pET28 by using primers 11 and 12 (Table 1). The fragment was digested with SmaI and EcoRI and inserted into pLOJ15 opened with EcoRV and EcoRI, generating pLOJ16. The HindIII-EcoRI cassette containing the pilA promoter-lacI gene was subsequently transferred to pSWU30, resulting in pLOJ28.
FIG. 1.
Assembly of an IPTG-inducible promoter. (A) Schematic drawing showing the pilA promoter from −855 to 1 bp inserted in front of a leader sequence containing the lac operator for binding of and repression by the lac repressor LacI. The KpnI and EcoRI restriction sites were used to insert a gene of interest (often pilA). (B) Base sequence of the leader and the lac operator. The sequence is preceded by the −24 and −12 boxes of the σ54-dependent pilA promoter and the transcriptional start site at position 1. The locations of the lac operator and the RBS are indicated. The translational start site ATG is indicated by boldface type. (C) Insertion of the inducible promoter into the M. xanthus chromosome by homologous recombination within the pilA locus of DK10410. After electroporation of the plasmid into M. xanthus cells, a single homologous crossover produced a tandem duplication of the pilA promoter in which the upstream chromosomal region (thick line) drove the expression of the inducible promoter, whereas the 855-bp upstream region cloned in the vector (thin line) drove the expression of the native pilA gene, most often a ΔpilA allele.
Construction of M. xanthus strains.
Plasmids pLOJ5 and pLOJ7 were introduced into M. xanthus strains by electroporation (10). Because none of the plasmids used in this work can replicate in M. xanthus, drug-resistant electroporants resulted from a single-crossover homologous recombination event that incorporated the plasmid into the chromosome (Fig. 1C). The structure of each strain was analyzed by Southern blotting to verify that integration of the plasmid had occurred at the pilA locus and that only one copy of the plasmid was present.
Plasmids pLOJ6 and pLOJ28 were introduced into M. xanthus strains by electroporation. The correct integration at the Mx8 attB site resulting from a single-crossover site-specific recombination event was confirmed by PCR by using primers 13, 14, 15, and 16 specific for the 5′ and 3′ ends of the attP and attB regions (Table 1).
Induction during growth and development.
Cells were grown in liquid CTT medium to a density of 5 × 108 cells/ml (100 Klett units), sedimented, and resuspended in CTT medium to a density of 5 × 109 cells/ml. For induction during growth, 2.5 × 108 cells were spotted on 0.5× CTT agar plates with different amounts of IPTG. The cells were incubated at 32°C. At different times, aliquots were harvested and stored at −70°C until all samples were collected. For induction during development, the cells were washed and resuspended in MC7 buffer (10 mM morpholinepropanesulfonic acid [MOPS], 1 mM CaCl2 [pH 7.0]), and 1.25 × 108 cells were incubated at 32°C in 400 μl of MC7 buffer with different amounts of IPTG in microtiter wells with a diameter of 15 mm. At different times, aliquots were harvested and stored at −70°C until all samples were collected.
Pilus preparation.
Cells were prepared and collected as described above for induction during growth. Pili were purified as described by Wall et al. (19). Briefly, the cell samples were resuspended in TPM buffer (10 mM Tris-HCl, 8 mM MgSO4, 1 mM KPO4 [pH 7.6]) in 1.5-ml microcentrifuge tubes. Each suspension was then agitated at the maximum speed with a tabletop vortex mixer to shear the pili off, and this was followed by sedimentation at 16,000 × g for 5 min. The supernatant was transferred to a fresh tube. Pili were precipitated by addition of 100 mM MgCl2 and incubation on ice for 2 h, followed by sedimentation at 16,000 × g for 15 min at 4°C.
Immunoblotting and measurement of β-galactosidase activity.
For immunoblotting, the total protein in the harvested cell aliquots was measured, and the cells were resuspended in sodium dodecyl sulfate gel-loading buffer with β-mercaptoethanol and boiled for 5 min. For whole-cell analysis, 1 or 0.5 μg of protein from each sample was separated on a 12% polyacrylamide gel. For purified pilus analysis, the total protein in cell samples was measured prior to purification of pili. Pili purified from 100 μg of total protein from input cells were separated on a 12% polyacrylamide gel. The proteins were transferred to Immobilon P transfer membranes (Millipore) with a semidry blotting apparatus. The blots were then probed with rabbit anti-PilA serum diluted 1:2,000, followed by peroxidase-conjugated goat anti-rabbit immunoglobulin G (Boehringer Mannheim) diluted 1:10,000. The blots were developed with a chemiluminescence reagent (Perkin-Elmer) by using autoradiography film exposure times ranging from 5 s to 2 min. The band intensities from immunoblots were measured by using NIH imageJ 1.30 (http://rsb.info.nih.gov/ij/). Samples to be tested for the specific activity of β-galactosidase were resuspended in 800 μl of TPM buffer and sonicated for 20 s with a 50-W microtip sonicator at 40% output capacity, and this was followed by sonication for 3 min at 50% output capacity in a 150-W cup horn cooled with ice water. The β-galactosidase specific activities of the samples (in nanomoles of o-nitrophenol produced from o-nitrophenol-β-d-galactoside per minute per milligram of protein) were then determined as described by Kroos et al. (11).
Examination of the swarming zone.
Cells were grown in liquid CTT medium to a density of 5 × 108 cells/ml (100 Klett units), sedimented, and resuspended in CTT medium to a density of 5 × 109 cells/ml. Then 2.5 × 107 cells were spotted on 0.5× CTT-0.4% agar plates with different amounts of IPTG. The plates were incubated at 32°C and photographed after 48 h.
Cell-cell agglutination assay.
Cells were grown overnight in CTT medium with or without 1 mM IPTG to a density of 5 × 108 cells/ml. In order to quantify agglutination, the optical density at 550 nm (OD550) of 800 μl of cells that were initially suspended at an OD550 of ∼0.6 was monitored every 15 min for 120 min. Briefly, the cells were diluted in 800 μl (final volume) of CTT medium to obtain a calculated OD550 of ∼0.6. The cell suspensions were transferred to cuvettes and placed in a Pharmacia LKB Ultrospec Plus spectrophotometer, and the OD550 of each sample was measured every 15 min for 120 min as described by Wu et al. (25).
RESULTS
Construction of an IPTG-inducible promoter for M. xanthus.
We selected the native M. xanthus pilA promoter to test inducibility because it is known to have a high level of expression during both growth and development (24). Since previous studies showed that 851 bp of DNA upstream of the transcriptional start site is sufficient for normal pilA gene expression (24), a fragment from −855 to +1 bp relative to the transcriptional start site of pilA was used. This promoter was rendered IPTG inducible by replacing the native pilA leader sequence with a leader sequence containing the lac operator (lacO) and an RBS from E. coli (Fig. 1A and B). Expression from this promoter, designated PpilA-lacO-RBS, was tested by using both pilA and lacZ as reporter genes. Since wild-type M. xanthus contains neither β-galactosidase nor its repressor, the E. coli lac repressor gene, lacI, was cloned in a separate plasmid (pLOJ6) for introduction into M. xanthus.
An IPTG-inducible pilA gene is able to restore S motility to a ΔpilA strain while normal dependence on PilR is retained.
To analyze the expression from PpilA-lacO-RBS-pilA in M. xanthus, strain DK12600 was constructed. In this strain plasmid pLOJ7 (Table 1) was integrated into the native pilA promoter on the chromosome of DK10410 (ΔpilA) by homologous recombination, creating a tandem duplication, as shown in Fig. 1C. The ΔpilA strain had a deletion of 187 internal codons of the pilA gene, but it retained the upstream regulatory region and normal reading frame (24). Southern blotting verified that the plasmid had integrated within the pilA locus and, importantly, that only one copy of the plasmid had integrated (data not shown). Expression of the pilA gene in DK12600 was investigated by semiquantitative immunoblotting (see Materials and Methods). The same total amount of protein isolated from DK1622 (wild type), DK10410 (ΔpilA), and DK12600 (PpilA-lacO-RBS-pilA) cells was applied to the gel. The ΔpilA control illustrated the specificity of the antibody used to develop the blot for pilin. As shown in Fig. 2A, the production of pilin in DK12600 was comparable to that in DK1622. This shows that PpilA-lacO-RBS-pilA supports a level of pilin protein production that is similar to the level observed for a single copy of the native pilA gene in a wild-type strain.
FIG. 2.
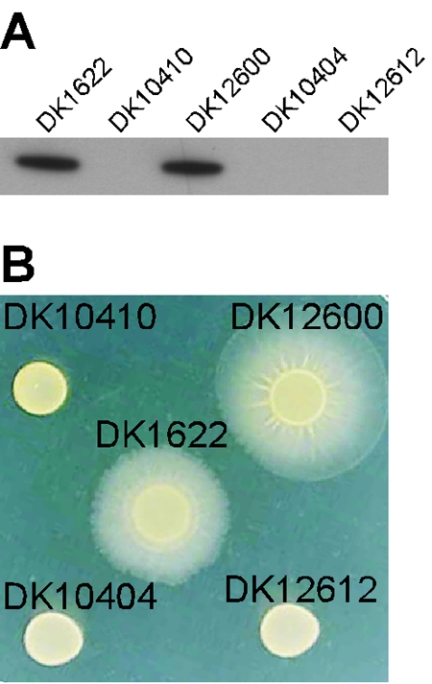
Expression of PpilA-lacO-RBS-pilA and rescue of S motility. (A) Immunoblot showing expression of pilin from PpilA-lacO-RBS-pilA. Total cell lysates were prepared from 2.5 × 108 cells spotted on 0.5× CTT agar plates, and aliquots were harvested after 8 h of incubation at 32°C. One microgram of total protein isolated from DK1622 (wild type), DK10410 (ΔpilA), DK12600 (DK10410::PpilA-lacO-RBS-pilA), DK10404 (pilR-Ω3163), or DK12612 (DK10404::PpilA-lacO-RBS-pilA) was loaded in each lane and reacted with anti-PilA antibody. The results are representative of the results of two independent experiments. (B) S motility assay. The abilities of strains DK1622 (wild type), DK10410 (ΔpilA), DK12600 (DK10410::PpilA-lacO-RBS-pilA), DK10404 (pilR-Ω3163), and DK12612 (DK10404::PpilA-lacO-RBS-pilA) to swarm by using S motility were investigated. A total of 2.5 × 107 cells were spotted on 0.5× CTT-0.4% agar plates and incubated at 32°C. The plates were inspected and photographed after 48 h. The results are representative of the results of two independent experiments.
Because pili are essential for S motility in M. xanthus (8), we measured the capacity of DK12600 cells to swarm by S motility. To do this, 2.5 × 107 cells of DK1622, DK10410, and DK12600 were spotted on 0.5× CTT-0.4% agar plates (Fig. 2B). S motility predominates on soft agar (16). The diameter of each swarm indicated the rate of swarming, and the diameters were measured relative to the diameter observed for wild-type DK1622 on the same plate. DK12600 swarmed as well as or better than DK1622 by this measure, verifying that pilin produced from the PpilA-lacO-RBS-pilA promoter can efficiently assemble into pili and can rescue the S motility that is lacking in the pilA deletion strain DK10410 (Fig. 2B).
Expression of the pilA gene in M. xanthus depends on an NtrC-like response regulator, PilR (24). PilR sets the pilA expression level as the second member of a two-component system with the adjacent PilS protein, the presumptive sensor for this system (24). To determine whether the PpilA-lacO-RBS construct retained the PilR dependence of the wild type, strain DK12612 was constructed by integrating plasmid pLOJ7 into the chromosome of a pilR disruption mutant, DK10404 (pilR-Ω3163). The genomic structure of DK12612 was verified by Southern blotting. The production of pilin was measured by immunoblotting, and neither DK12612 nor DK10404 produced detectable amounts of pilin (Fig. 2A). Moreover, both strains lacked S motility as measured by the swarm diameters (Fig. 2B). These observations show that expression from PpilA-lacO-RBS-pilA depends on PilR, just as expression from the native pilA promoter does. Thus, insertion of the lac operator into the leader sequence of the pilA promoter does not interfere with normal pilin expression or its dependence on PilR; both are qualitatively like the wild-type pilA characteristics.
Dose-dependent induction of pilA during growth.
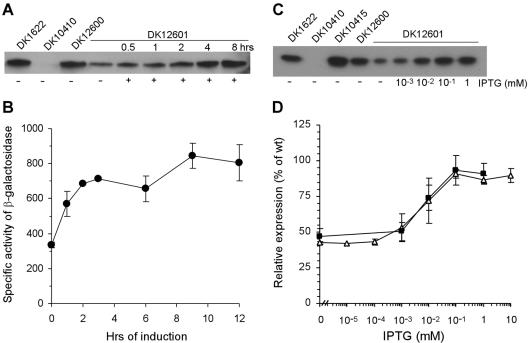
To measure the extent to which pilin production from PpilA-lacO-RBS-pilA was repressed by LacI and derepressed or induced by IPTG, strain DK12601 was constructed. Plasmid pLOJ6, containing the lacI gene from plasmid pET28 and the Mx8 attP region, was inserted into the chromosomal Mx8 attB region of DK12600 by site-specific recombination to obtain DK12601. Since the Mx8 attB region is distant from the pil region, it is not expected to influence the pilA promoter, and the structure of strain DK12601 was verified by PCR as described in Materials and Methods. Induction of pilA in DK12601 was monitored after incubation with 1 mM IPTG by pilin immunoblotting of 1 μg of total protein isolated from cells of DK1622, DK12600, and DK12601 (Fig. 3A). Without IPTG, some pilin was produced, but the amount was less than one-half the amount produced by the pilA+ strain (DK1622). In contrast to the dependence on PilR shown in Fig. 2, the level of production shown in Fig. 3A suggests that PpilA-lacO-RBS-pilA was not fully repressed by the Lac repressor provided. Nevertheless, when the cells were incubated with 1 mM IPTG, production of pilin increased within 0.5 to 1 h and reached the pilA+ levels by 8 h (Fig. 3A), showing that LacI repression can be fully relieved by 1 mM IPTG. IPTG-induced production of β-galactosidase from the constructed promoter had a time course very similar to that of the production of pilin (Fig. 3B), which supports the reliability of the immunoblot assays.
FIG. 3.
Induction of pilin in strain DK12601 grown vegetatively. (A) Immunoblot showing the time course of IPTG-induced expression of pilin from PpilA-lacO-RBS-pilA. Total cell lysates were prepared from 2.5 × 108 cells spotted on 0.5× CTT agar plates with or without 1 mM IPTG, as indicated at the bottom. Aliquots were harvested after incubation at 32°C for 0.5 to 8 h. One microgram of total protein isolated from DK1622 (wild type), DK10410 (ΔpilA), DK12600 (DK10410::PpilA-lacO-RBS-pilA), or DK12601 (DK10410::PpilA-lacO-RBS-pilA, lacI) was loaded in each lane and reacted with anti-PilA antibody. The results are representative of the results of three independent experiments. (B) IPTG induction of PpilA-lacO-RBS-lacZ during growth. Specific activities of β-galactosidase were determined with 2.5 × 108 cells of DK12603 (DK1622::PpilA-lacO-RBS-lacZ, lacI) spotted on 0.5× CTT agar plates with 1 mM IPTG and incubated at 32°C for different times. (C) Immunoblot showing the induction of pilin expression from PpilA-lacO-RBS-pilA as a function of IPTG concentration. Total cell lysates were prepared from 2.5 × 108 cells spotted on 0.5× CTT agar plates with different amounts of IPTG. Aliquots were harvested after incubation at 32°C for 12 h. Then 0.5 μg of total protein isolated from DK1622 (wild type), DK10410 (ΔpilA), DK10415 (ΔpilS), DK12600 (DK10410::PpilA-lacO-RBS-pilA), or DK12601 (DK10410::PpilA-lacO-RBS-pilA lacI) was loaded in each lane and reacted with anti-PilA antibody. The results are representative of the results of three independent experiments. (D) Relative expression of PpilA-lacO-RBS-pilA and PpilA-lacO-RBS-lacZ. Specific activities of β-galactosidase were determined with cells of DK12603 (DK1622::PpilA-lacO-RBS-lacZ lacI) spotted on 0.5× CTT agar plates with different amounts of IPTG and incubated for 12 h at 32°C. The values are expressed relative to the expression of DK12602 (DK1622::PpilA-lacO-RBS-lacZ) (defined as 100%) and are plotted as a function of the IPTG concentration. Each value is the average of the values from two independent experiments, and the error bars indicate standard deviations. The expression of pilin from PpilA-lacO-RBS-pilA in DK12601 in panel C was quantitated by measuring band intensities, and the values are expressed relative to pilin expression in DK1622 and plotted as a function of the IPTG concentration. Each value is the average of the values from three independent experiments, and the error bars indicate standard deviations. Symbols: ▵, β-galactosidase (PpilA-lacO-RBS-lacZ); ▪, pilin (PpilA-lacO-RBS-pilA). wt, wild type.
Dose-dependent induction from PpilA-lacO-RBS-pilA was examined by immunoblotting 0.5-μg portions of total protein from cells incubated for 12 h with concentrations of IPTG ranging from 10−3 to 1 mM (Fig. 3C). The production of pilin was quantified by measuring the band intensities on immunoblots from three independent experiments; the results of one of these experiments are shown in Fig. 3C. The average results of three experiments are shown as a function of IPTG concentration in Fig. 3D. The lowest concentration of IPTG (10−3 mM) was not sufficient to induce pilin production significantly above the uninduced level (47% ± 6% of the wild-type level) (Fig. 3D). However, 10−2 mM IPTG induced pilin production at approximately 74% ± 9% of the wild-type level, while 10−1 and 1 mM induced 93% ± 10% and 91% ± 7%, respectively, of the wild-type levels of pilin (Fig. 3D). These data show that the level of expression from PpilA-lacO-RBS-pilA increases progressively with the amount of IPTG added.
A similar IPTG dose-dependent induction was observed when we measured β-galactosidase activity in strains containing PpilA-lacO-RBS-lacZ (DK12602 and DK12603) (Fig. 3D). The results show that there was identical dose-dependent induction for pilin and lacZ (Fig. 3C and D) and that the expression could be increased approximately twofold by increasing the amount of IPTG added. This demonstrated the inducibility of the PpilA-lacO-RBS construct in M. xanthus.
Induction of lacZ during development.
M. xanthus development poses a problem for repression by LacI in pLOJ6. The lacI gene in pLOJ6 is transcribed from its native E. coli σ70 promoter, but its σ70 homolog in M. xanthus, σA, is lost after 12 to 15 h of development (3). To provide access to the developmental phase of M. xanthus, the promoter that drives lacI in pLOJ6 was changed to a pilA promoter in pLOJ28 (PpilA-lacI), which was active during development (24). DK12604 had pLOJ28 integrated into the chromosomal Mx8 attB site of DK12602, and its genomic structure was verified by PCR. Expression from PpilA-lacO-RBS-lacZ during fruiting body development is shown in Fig. 4. Without IPTG the initial expression from PpilA-lacO-RBS-lacZ in DK12604 was 604 ± 165 U of β-galactosidase, and the value increased to 1,137 ± 206 U after 24 h of development. This pattern of expression revealed that LacI was not able to fully repress expression from the constructed promoter during development, as was observed during vegetative growth. Nevertheless, when PpilA-lacO-RBS-lacZ in DK12604 was induced with 1 mM IPTG, the expression increased to 3,473 ± 466 U after 24 h, and overall there was 5.8-fold induction (Fig. 4). The final derepressed or fully induced level was similar to that of DK12602, which had no repressor and whose activity increased from 1,900 U at zero time to 3,000 U at 24 h due to the developmental increase in pilR expression (6). These data show that it is possible to induce PpilA-lacO-RBS-lacZ with 1 mM IPTG to the maximum activity by 24 h of development.
FIG. 4.
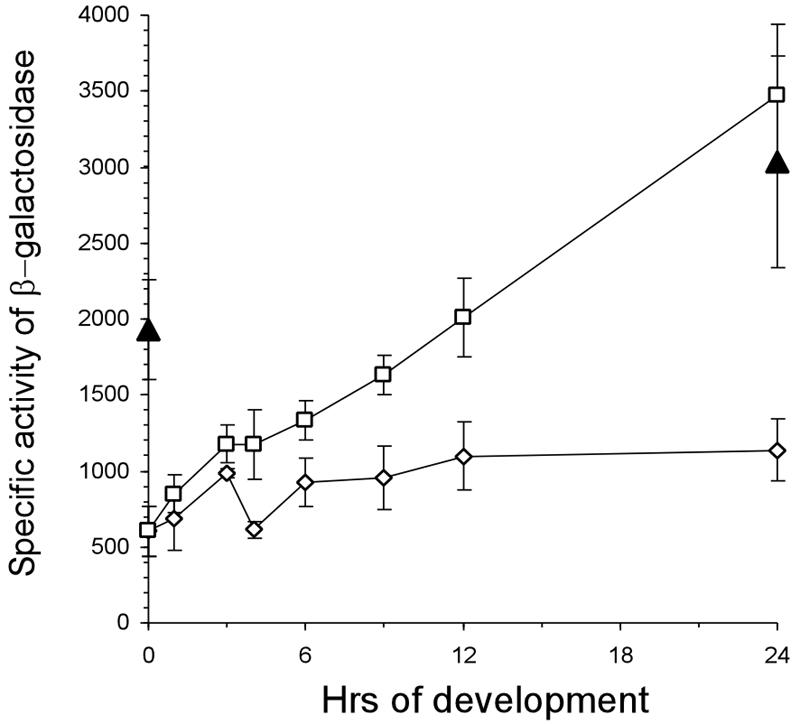
IPTG induction of lacZ from PpilA-lacO-RBS-lacZ during development: specific activity of β-galactosidase expressed in DK12602 (PpilA-lacO-RBS-lacZ) and DK12604 (PpilA-lacO-RBS-lacZ, PpilA-lacI) during development. Specific activities of β-galactosidase were determined in cells after exposure to starvation in MC7 buffer with or without 1 mM IPTG for different times. Symbols: ▴, DK12602; □, DK12604 with IPTG; ⋄, DK12604 without IPTG. The specific activities of β-galactosidase are expressed in nanomoles of o-nitrophenol produced from o-nitrophenol-β-d-galactoside per minute per milligram of protein. Each value is the average of the values from two independent experiments, and the error bars indicate standard deviations.
Dependence of pilus assembly on pilin concentration.
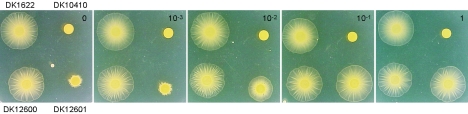
The IPTG-dependent induction of pilin in DK12601 offered the possibility of investigating the relationship between total pilin concentration and the pilus activity for S-motile swarming. To assess S motility, the swarm areas after 48 h were measured for DK1622, DK10410, DK12600, and DK12601 with IPTG concentrations ranging from 10−3 to 1 mM (Fig. 5). Swarming was quantified by determining the area of swarming beyond the inoculation zone, as described above. Strain DK12600 (PpilA-lacO-RBS-pilA), which had no lac repressor, swarmed to the same extent as DK1622 (wild type) at all concentrations of IPTG (Fig. 5). However, with the repressor, the PpilA-lacO-RBS-pilA lacI strain DK12601 swarmed at a rate that was only 6% of the rate of DK1622 without IPTG, and with 10−3 mM IPTG there were only a few short motility flares evident at the periphery of the swarm (Fig. 5). With 10−2 mM IPTG, however, the morphology of the DK12601 swarm was similar to that of the DK1622 swarm, but the area was only 40% of the area of the DK1622 swarm. With 10−1 and 1 mM IPTG, concentrations at which full induction of pilin was observed (Fig. 3C and D), DK12601 swarmed as well as DK1622. Evidently, S motility and the swarm expansion rate do increase in proportion to the pilin concentration.
FIG. 5.
Dose-dependent induction of S motility in DK12601. Strain DK12601 is DK10410::PpilA-lacO-RBS-pilA lacI. Strains DK1622 (wild type), DK10410 (ΔpilA), and DK12600 (DK10410::PpilA-lacO-RBS-pilA) were included as positive and negative controls. A total of 2.5 × 107 cells were spotted on 0.5× CTT-0.4% agar plates and incubated at 32°C. The concentration of IPTG used (millimolar) is indicated in the top right corner of each panel. The swarming was inspected after 48 h. The results are representative of the results of three independent experiments.
To assess the functional dependence of S motility on the amount of pilin, the relative swarm rates shown in Fig. 5 were plotted against the relative pilin levels shown in Fig. 3D for the same IPTG concentrations (Fig. 6). To ensure that the plot covered the full range of PilR-PilS-regulated pilin expression, a pilin-overproducing strain, DK10415 (ΔpilS), was added to the experiment. This strain had an internal deletion of 165 amino acids in the pilS gene and produced ∼40% more pilin than DK1622 produced (Fig. 3C) (22, 24). As shown in Fig. 6, the dependence of the swarm rate on the amount of pilin was not linear. Low concentrations of pilin supported negligible amounts of swarming. When the DK12601 strain produced ∼50% of the wild-type pilin level, it swarmed at only 6% of the rate of DK1622.
FIG. 6.
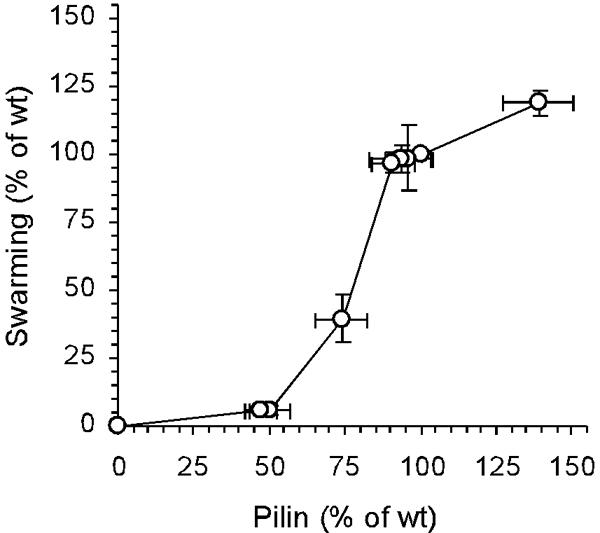
Relationship between the amount of pilin synthesized and S motility. The levels of S motility at different IPTG concentrations, as shown in Fig. 5, are plotted as a function of the levels of pilin at the corresponding IPTG concentrations shown in Fig. 3C and D. The values are expressed relative to wild-type S motility and wild-type pilin production. Each value is the average of the values from three independent experiments, and the error bars indicate standard deviations. wt, wild type.
Swarming depends on pilus retraction; pilT mutants that have pili do not swarm (25). To verify that the disproportion between pilin and the swarm rate is due to a disproportion with the amount of pili, the latter was measured in two different ways. The first way was by agglutination of piliated cells, and the results are shown in Fig. 7. The assay was based on clumping of piliated cells in suspension under carefully selected conditions in which cells without pili do not clump (25). The ΔpilA strain, DK10410, which had no pili, had not agglutinated after 120 min (OD550, 100% ± 3%). this finding illustrated the pilus specificity of this assay. Uninduced strain DK12601 behaved like the ΔpilA mutant; it had an OD550 of 91% ± 9% and remained in suspension at 120 min. However, fully IPTG-induced strain DK12601 agglutinated like DK1622 after 120 min (OD550, 9% ± 3%).
FIG. 7.
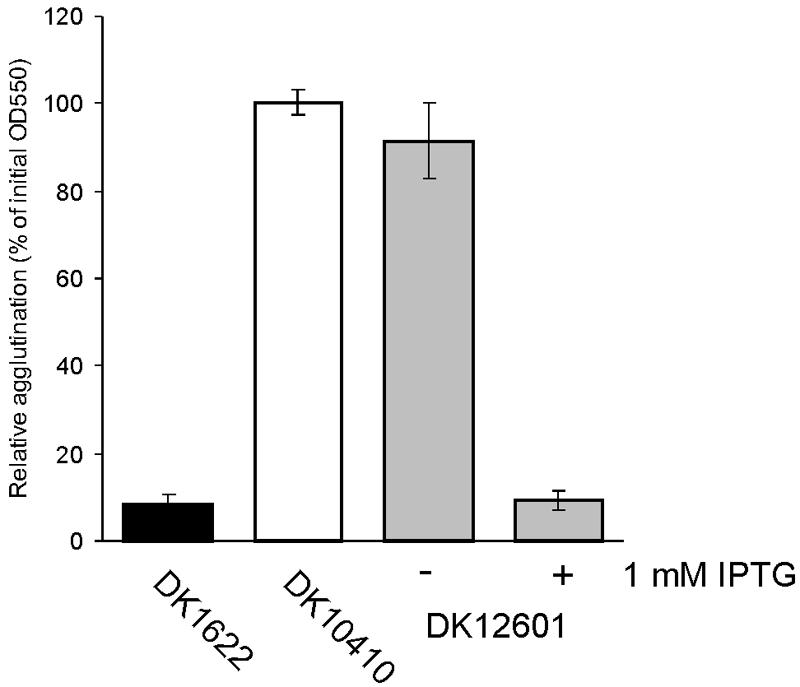
Cell-cell agglutination assay. Cells of DK1622 (wild type), DK10410 (ΔpilA), and DK12601 (DK10410::PpilA-lacO-RBS-pilA lacI) were grown at 32°C overnight in CTT medium with or without 1 mM IPTG, and the OD550 was adjusted to 0.6 with CTT broth. The absorbance after 120 min was determined relative to the initial absorbance for each strain. Solid bar, DK1622; open bar, DK10410; shaded bars, DK12601. Each value is the average of the values from three independent experiments, and the error bars indicate standard deviations.
In a second assay we measured the total mass of pilin assembled into pili by shearing the pili from cells and then separating pilus fragments from cells by differential centrifugation as described by Wall et al. (19). The mass of the shear-sensitive pilin was quantified on immunoblots with anti-PilA antibodies, as described above for Fig. 3C and D. Figure 8 shows that DK12601 incubated without IPTG released after shearing 14% ± 5% of the mass of pilin that was produced by the wild type even though DK12601 produced ∼50% as much total pilin as the wild type produced under these conditions. This low level of assembled pilin agrees with the low level of agglutination (Fig. 7) and explains the low rate of S-motile swarming. This consistency justifies the three assays used to quantify pili. Figure 6 shows that the level of total pilin must be ≥50% of the maximum level for efficient pilus assembly in M. xanthus.
FIG. 8.
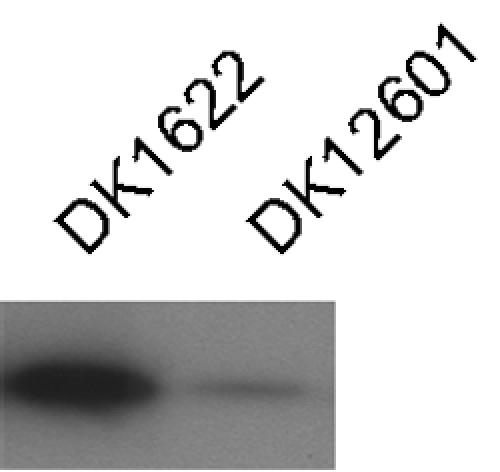
Level of piliation in DK1622 (wild type) and DK12601 (DK10410::PpilA-lacO-RBS-pilA lacI) measured by sensitivity to shearing. Material removed from cells by shearing was purified from DK1622 and DK12601 cultures after incubation at 32°C on 0.5×CTT agar plates for 12 h. Material purified from 100 μg of cells was loaded in each lane and reacted with anti-PilA antibody. The results are representative of the results of three independent experiments.
DISCUSSION
Here we describe the construction of a regulated promoter for M. xanthus that functions during growth and fruiting body development. For this study, we adapted the IPTG-inducible lac repressor lacI and the lac operator from E. coli to regulate expression from the M. xanthus native pilA promoter. Constructed in this way, the inducible pilA promoter was shown to reach the same maximum level of expression as the native pilA promoter in wild-type cells. The synthesized promoter was shown to depend on the NtrC-like regulator PilR in the same way that the native promoter depends on it. The data indicate that the synthesized promoter uses the transcriptional control apparatus in M. xanthus in the same way that the native pilA promoter uses it. When both pilA and lacZ were used as reporters, induction was observed to initiate during growth within 1 h after addition of 1 mM IPTG and to reach full expression by 8 h. IPTG appears to enter M. xanthus cells readily since a response was detected at concentrations below 10−2 mM. However, in the absence of IPTG induction, the Lac repressor allowed almost 50% of the maximum expression during growth. During development, induction of lacZ by 1 mM IPTG began within the first 2 h, and full expression was reached after 24 h, when fruiting body development was complete and spore maturation was under way. Although the uninduced level of expression was similar to the level during growth, 5.8-fold induction was observed.
A second lac operator was added in an attempt to decrease the uninduced level of expression from the synthetic promoter. The uninduced level of expression from the promoter with two lac operators was less than the uninduced level of expression from the promoter with one operator, although expression was not abolished (data not shown). The relatively high level of uninduced expression from the promoter may limit its use, but since the level of expression from the promoter can reach 3,500 U of β-galactosidase activity after 24 h of development, it should be capable of reaching the maximum expression levels of many genes of M. xanthus (11) and thus be suitable for untimely expression of those genes during development. Because the expression from the promoter varies with the IPTG concentration, this technique offers a new tool for examining thresholds in some organelle assembly processes and possibly some regulatory circuits in M. xanthus.
By using the IPTG-induced promoter, S motility was shown to have a threshold of pilin expression below which neither production of shear-sensitive pilin, pilus-dependent cell agglutination, nor S-motile swarming was observed. At the threshold, an M. xanthus culture producing 50% the wild-type level of total pilin produces only 14% the wild-type level of shear-sensitive pilin. Evidently, in M. xanthus pilin assembly limits pilus production and S motility. This limitation may be related to the high frequency of reversal of gliding direction observed in M. xanthus. When developing cells are moving in traveling waves, they reverse their gliding direction regularly every 8 min (20), and growing cells reverse on average after roughly 10 min (1). Reversal implies loss of pili at the leading pole of the cell and commissioning of new pilus assembly at the opposite pole because 95% of cells have pili only at one pole (7, 8). Since one cell cycle in growing cells is more than 200 min long, Myxococcus needs to assemble new pili 20 or more times per cycle, as well as once for each new cell that is born. Developing M. xanthus cells have a similar need because they reverse even more frequently.
Frequent reversals have several regulatory correlates. In the first place, the pilA promoter in M. xanthus has a much higher potential expression rate than the average gene when β-galactosidase reporter activities are compared (24; this study). The coupling of a high expression level and a threshold for assembly suggests that the expression of pilA is negatively regulated by the accumulation of pilin at a cell end, which may be sensed by the PilS-PilR two-component system. The expression of the pilA promoter in a ΔpilA or ΔpilS mutant strain has been found to be three- to fourfold greater than that in the wild type (Fig. 3 and 6) (24). When the absence of unassembled pilin is sensed, PilS increases the activity of PilR, and PilR increases pilA expression, as shown by experiments reported here. The conclusion that unassembled pilin is sensed is supported by the fact that other mutants having defects in pilus assembly or pilus retraction, such as pilB, pilC, tgl, and pilT mutants, which do accumulate pilin, have been observed to express pilA at approximately wild-type levels. Also suggestive is the fact that PilR expression increases threefold during development compared with the expression during growth (6). We concluded that the increase in PilR prepares cells for the frequent reversals of gliding direction that they have to make during fruiting body development.
Once a newly commissioned cell end senses with PilS that it has no pilin, the pilA gene is expressed at a high rate. A high level of expression continues until the pilin at the newly commissioned cell end has assembled into pili, and the cell is prepared to glide in the reverse direction. M. xanthus pili extend forward 1 to 2 cell lengths (9). The length of the pili helps to explain why the pilA promoter is very strong. In addition to PilS and PilR, the PilS2-PilR2 two-component system plays a significant, albeit unknown, role in pilus assembly because disrupting the pilR2 gene knocks out S-motile swarming (2).
Acknowledgments
We thank Lars Jelsbak for helpful discussions and critical reading of the manuscript.
This work was supported by Public Health Service grant GM23441 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Blackhart, B. D., and D. Zusman. 1985. Frizzy genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. USA 82:8767-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caberoy, N. B., R. D. Welch, J. S. Jakobsen, S. C. Slater, and A. G. Garza. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao, T., D. Biran, G. J. Velicer, and L. Kroos. 2002. Identification of the Ω4514 regulatory region, a developmental promoter of Myxococcus xanthus that is transcribed in vitro by the major vegetative RNA polymerase. J. Bacteriol. 184:3348-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in M. xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 5.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in M. xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 6.Jakobsen, J. S., L. Jelsbak, L. Jelsbak, R. Welch, C. Cummings, B. Goldman, E. Stark, S. C. Slater, and D. Kaiser. 2004. σ54 Enhancer binding proteins and Myxococcus xanthus fruiting body development. J. Bacteriol. 186:4361-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaiser, D., and C. Crosby. 1983. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motility 3:227-245. [Google Scholar]
- 10.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 11.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 12.Letouvet-Pawlak, B., C. Monnier, S. Barray, D. A. Hodgson, and J. F. Guespin-Michel. 1990. Comparison of β-galactosidase production by two inducible promoters in Myxococcus xanthus. Res. Microbiol. 141:425-435. [DOI] [PubMed] [Google Scholar]
- 13.Li, S., B. U. Lee, and L. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 14.Li, Y., H. Sun, X. Ma, A. Lu, R. Lux, D. Zusman, and W. Shi. 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merz, A. J., and K. T. Forest. 2002. Bacterial surface motility: slime trails, grappling hooks and nozzles. Curr. Biol. 12:R297-R303. [DOI] [PubMed] [Google Scholar]
- 16.Shi, W., and D. R. Zusman. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA 90:3378-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spratt, B. G., P. J. Hedge, S. teHessen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 18.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 19.Wall, D., S. S. Wu, and D. Kaiser. 1998. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J. Bacteriol. 180:759-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch, R., and D. Kaiser. 2001. Cell behavior in traveling wave patterns of myxobacteria. Proc. Natl. Acad. Sci. USA 98:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12:1-20. [DOI] [PubMed] [Google Scholar]
- 22.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 23.Wu, S. S., and D. Kaiser. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J. Bacteriol. 178:5817-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]