Abstract
In contrast to previous findings, we demonstrate that the dissimilatory (bi)sulfite reductase genes (dsrAB) of Desulfobacula toluolica were vertically inherited. Furthermore, Desulfobacterium anilini and strain mXyS1 were identified, by dsrAB sequencing of 17 reference strains, as members of the donor lineage for those gram-positive Desulfotomaculum species which laterally acquired dsrAB.
Dissimilatory (bi)sulfite reductase catalyzes the energy-generating step during the anaerobic respiration of sulfite or sulfate and thus represents a key enzyme of all sulfite- and sulfate-reducing prokaryotes (11, 22, 34). Recently, the genes encoding the alpha- and beta-subunits of this enzyme (dsrAB) have been used to infer the evolutionary history of dissimilatory (bi)sulfite reductases. For this purpose, a dsrAB database containing 75 entries for described sulfate-reducing prokaryotes (SRPs) (representing all known major evolutionary lineages of this guild) and four for sulfite-reducing microorganisms has been established (9, 12, 14-17, 23, 24, 28, 32, 33). Comparison of 16S rRNA- and DsrAB-based phylogenetic trees revealed congruent topologies for many SRP lineages, suggesting an ancient origin of dissimilatory (bi)sulfite reductase (33). This finding is consistent with isotopic evidence for biological sulfate reduction at 3.47 Gyr ago (31). However, we now recognize that the distribution of dsrAB among sulfate-reducing species reflects a combination of divergence through speciation (vertical descent) and acquisition via lateral gene transfer from distantly related prokaryotes (15). The archaeal SRPs of the genus Archaeoglobus, the deep-branching thermophilic SRPs of the genus Thermodesulfobacterium, as well as a large number of thermophilic gram-positive Desulfotomaculum species, possess laterally-acquired (bi)sulfite reductases. In addition, the deltaproteobacterial SRP Desulfobacula toluolica was postulated to have laterally acquired its (bi)sulfite reductase relatively recently, since its dsrAB genes differed significantly from those of its close relatives, including Desulfobacter latus, which have vertically transmitted (bi)sulfite reductase genes. In the DsrAB tree, the putative DsrAB sequence of D. toluolica formed a well-supported monophyletic cluster with the laterally acquired DsrAB sequences of Desulfotomaculum species. Therefore, it was speculated that D. toluolica and the Desulfotomaculum species received their dsrAB from a common but so far unidentified deltaproteobacterial donor lineage (15). However, no information on transfer mechanism or donor lineages is available for any of the recognized dsrAB lateral gene transfer events.
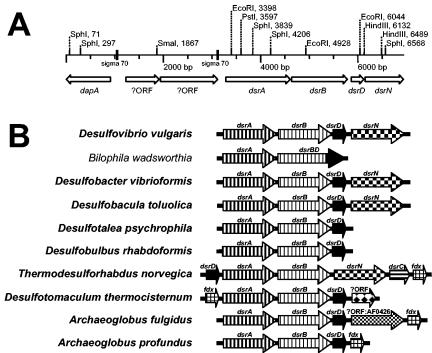
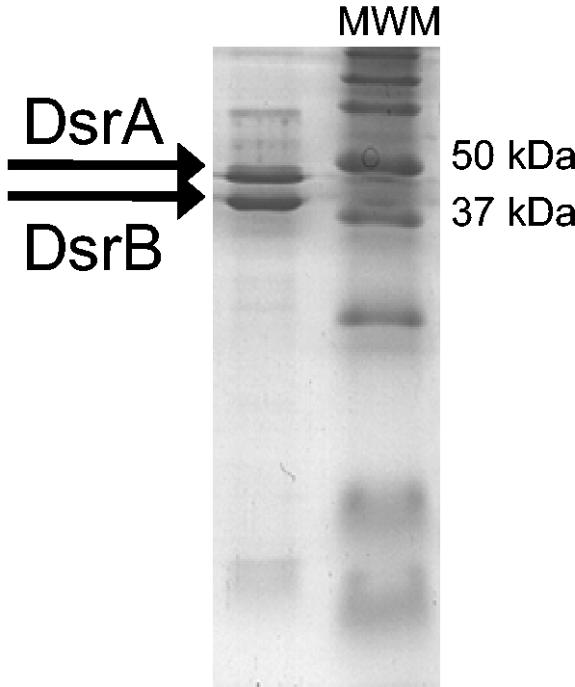
In an attempt to determine which additional genes might have been cotransferred with the dsrAB genes of D. toluolica and to reveal genetic traces indicative of the responsible transfer mechanism, the dsr operon (and flanking regions) of this SRP was sequenced. In a first step, a digoxigenin (DIG)-labeled 152-bp polynucleotide probe targeting dsrA was generated from D. toluolica DNA by using the primers DsrA415F (5′-TATCARGATGAGCTKCATCGYCC-3′) and DsrA542R(5′-ACYGCDTCCTGATCAATVCGGATAT-3′) and thePCR DIG probe synthesis kit (Roche, Mannheim, Germany). Using this probe, it was demonstrated by Southern hybridization of DNA restriction fragments from D. toluolica at low stringency that this organism contains a single dsrA in its genome (data not shown). After cloning of genomic DNA of D. toluolica into a lambda vector, phages containing the dsrA gene were identified by plaque hybridization with the polynucleotide probe (29). An 8.9-kb insert of D. toluolica DNA in a phage clone which hybridized with the dsrA polynucleotide probe was sequenced by primer walking (GenBank accession no. AJ457136). Gene sequence comparison revealed that this fragment contained the dsr operon consisting of the genes dsrA, dsrB, dsrD, and dsrN (Fig. 1A). The dsrABDN operon structure of D. toluolica has previously been detected in other deltaproteobacterial SRPs (Fig. 1B). Surprisingly, comparative sequence analysis of dsrA and dsrB of D. toluolica revealed that these genes were clearly different (less than 66% nucleic acid similarity) from the dsrAB gene fragments of this organism which were previously published (15). Sequence analysis of the target sites of the PCR primers used for D. toluolica dsrAB gene fragment amplification by Klein et al. (15) revealed that dsrA has three mismatches with primer DSR1F and dsrB has one mismatch with primer DSR4R. Therefore, the dsrAB gene fragment of D. toluolica could not be amplified with these primers (data not shown). Thus, the dsrAB sequence which was previously reported (15) most likely originated from a laboratory contamination and the dsr operon sequence reported in the present paper is the actual dsr sequence of D. toluolica. The newly determined dsrAB sequence of D. toluolica phylogenetically clusters together with dsrAB sequences of the genus Desulfobacter (Fig. 2) independently from the treeing method applied (15, 19). Since this affiliation is consistent with the respective 16S rRNA gene tree topology, D. toluolica contains a vertically transmitted dsr operon. Two additional experiments were undertaken to further support this finding. First, the sequence of a 1.9-kb dsrAB PCR fragment of Desulfobacula phenolica, the closest known relative of D. toluolica, was determined and found to be almost identical (97.5% and 99.5% dsrA and dsrB nucleic acid similarity, respectively) to the respective gene sequences of D. toluolica (Fig. 2). Second, the DsrA and DsrB enzyme subunits were purified from cell extracts (7) of D. toluolica (Fig. 3) and N-terminal sequencing (6) of the DsrA subunit (N-terminal sequencing of DsrB failed) revealed 100% accordance with the respective amino acid stretch (AKHETPFL) predicted from the dsrA sequence. All predicted N-terminal amino acid sequences of DsrA from other SRPs differ in at least one amino acid from this sequence.
FIG. 1.
(A) Schematic map showing the genetic organization of a dsr operon-containing genomic fragment of D. toluolica. Restriction sites of common endonucleases and sequence motifs similar to Escherichia coli sigma 70 promoters are shown. The following abbreviations represent fully sequenced open reading frames: dsrA and dsrB, alpha and beta subunits, respectively, of the dissimilatory (bi)sulfite reductase; dsrD, dissimilatory (bi)sulfite reductase D; which has a possible role in the regulation of dsr gene transcription (21); dsrN, putative siroheme amidase; dapA, dihydrodipicolinate synthase; ?ORF, unidentified open reading frame. (B) Genetic organization of all known dsr operons from SRPs and Bilophila wadsworthia (18). Prokaryotes which are able to use sulfate as an electron acceptor for anaerobic respiration are indicated in boldface type. Open reading frames: dsrC, gamma subunit of the dissimilatory (bi)sulfite reductase; fdx, ferredoxin. Accession numbers: Desulfovibrio vulgaris, AE017285; B. wadsworthia, AF269147; Desulfobacter vibrioformis, AJ250472; D. toluolica, AJ457136; D. psychrophila, NC_006138; Desulfoblulbus rhabdoformis, AJ250473; Thermodesulforhabdus norvegica, AJ277293; Desulfotomaculum thermocisternum, AF074396; A. fulgidus, NC_000917; and Archaeoglobus profundus, AF071499.
FIG. 2.
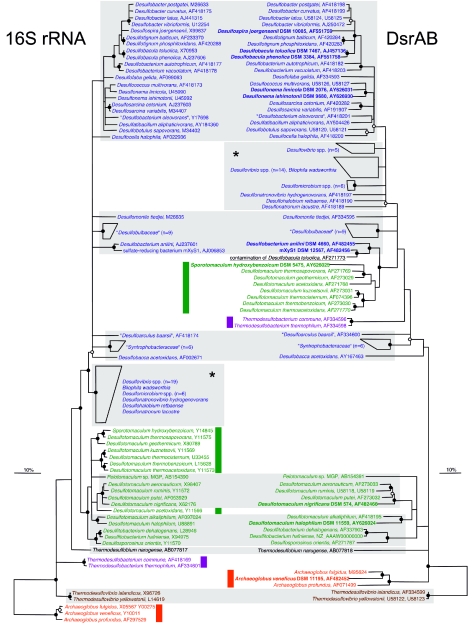
Comparison of 16S rRNA- and DsrAB-based phylogenetic consensus trees. Sequences determined in this study are in boldface. 16S rRNA phylogenetic analyses were performed on alignment positions conserved in at least 50% of all bacteria. Alignment regions of insertions and deletions were omitted in DsrAB amino acid sequence analyses. Polytomic nodes connect branches for which a relative order could not be determined unambiguously by using distance matrix, maximum-parsimony, and maximum-likelihood methods. Filled circles indicate branch points highly supported by maximum-parsimony bootstrap analysis (>90% in 1,000 resamplings). Open circles at nodes indicate 75 to 90%, while nodes without circles showed <75% bootstrap support. The bars represent 10% sequence divergence as estimated from maximum-likelihood and distance matrix analysis for the 16S rRNA and DsrAB trees, respectively. “Deltaproteobacteria,” low G+C gram-positive bacteria (Firmicutes), Thermodesulfobacterium species, Thermodesulfovibrio species, Thermodesulfobium narugense, and Archaeoglobus species are depicted in blue, green, violet, brown, black, and red, respectively. The wrong dsrAB sequence of D. toluolica published by Klein et al. (15) is underlined. Colored bars indicate species which harbor laterally acquired dsrAB genes. Consistent groups between both trees are shaded gray. Note that the apparently inconsistent positions of SRP groups that are labeled by an asterisk are not well resolved in the respective trees and thus cannot be interpreted as indicators of lateral gene transfer events. The strain A. veneficus SNP6 (DSM 11195) (containing plasmid XY), had been deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) by K. O. Stetter, Lehrstuhl für Mikrobiologie, Universität Regensburg, Regensburg, Germany. An ungrouped version of this figure (supplementary web Fig. 1) can be downloaded from our web site (http://www.microbial-ecology.net/supplements.asp) together with the respective ARB dsrAB database.
FIG. 3.
Denaturing polyacrylamide gel electrophoresis analysis of the dissimilatory (bi)sulfite reductase purified from D. toluolica. The predicted sizes of DsrA and DsrB according to the determined gene sequences are 49.9 and 42.5 kDa, respectively. MWM, molecular weight marker.
The sequence of D. toluolica extends the number of available complete dsrAB sequences from reference cultures to 11. These sequences can be used to validate the suitability of the commonly used PCR primers for dsrAB amplification from SRP pure cultures and from environmental or clinical samples (Table 1). With the exception of the dsrAB of D. toluolica and Desulfotalea psychrophila (28), the primers DSR1F and DSR4R (33) and their recently published variants (19) perfectly match the available complete dsrAB genes. For improved coverage of SRPs in future experiments, it is recommended that primer pairs DSR1Fc-DSR4Rd and DSR1Fd-DSR4Re, targeting dsrAB of D. toluolica and D. psychrophila, respectively, are added to the primer variant mixture (Table 1). Furthermore, PCR annealing stringency should be kept low in environmental dsrAB diversity surveys (e.g., references 1-5, 8, 13, 20, 25, and 26) because it is likely that additional sequence variants in the dsrAB PCR primer binding sites exist.
TABLE 1.
DSR1F and DSR4R primer binding sites as recognized from completely sequenced dsrAB genes of SRPs and B. wadsworthia
| dsrAB-containing prokaryotes | dsrA-targeted forward primer binding site (5′→3′)a | Perfectly matching forward primer (5′→3′) | Forward primer reference | dsrB-targeted reverse primer binding site (5′→3′)a | Perfectly matching reverse primer (5′→3′) | Reverse primer reference |
|---|---|---|---|---|---|---|
| D. vulgaris AE017285 | ACCCACTGGAAGCACG | DSR1F: ACSCACTGGAAGCACG | 33 | TGCGGTAACTGCTACAC | DSR4R: GTGT AGCAGTTACCGCA | 33 |
| D. desulfuricans AJ249777 | ACCCATTGGAAACACG | DSR1Fa: ACCCAYTGGAAACACG | 19 | TGCGGAAACTGCTACAC | DSR4Rc: GTGTAGCAGTTK CCGCA | 19 |
| B. wadsworthia AF269147 | ACGCACTGGAAGCACG | DSR1F: ACSCACTGGAAGCACG | 33 | TGCGGTAACTGCTACAC | DSR4R: GTGTAGCAGTTACCGCA | 33 |
| D. vibrioformis AJ250472 | ACCCACTGGAAACACG | DSR1Fa: ACCCAYTGGAAACACG | 19 | TGCGGTAACTGTTACAC | DSR4Rb: GTGTAACAGTTAC CGCA | 19 |
| D. toluolica AJ457136 | ACCCATTGGAAACATG | DSR1Fc: ACCCATTGGAAACATG | This study | TGTGGTAACTGCTACAC | DSR4Rd: GTGTAGCAG TTACCACA | This study |
| D. psychrophila NC_006138 | ACTCACTGGAAGCACG | DSR1Fd: ACTCACTGGAAGCACG | This study | TGTGGTAACTGTTACAC | DSR4Re:GTGTA ACAGTTACCACA | This study |
| D. rhabdoformis AJ250473 | ACCCATTGGAAACACG | DSR1Fa: ACCCAYTGGAAACACG | 19 | TGCGGTAACTGCTACAC | DSR4R: GTGTAGCAGTTACCG CA | 33 |
| T. norvegica AJ277293 | GGCCACTGGAAGCACG | DSR1Fb: GGCCACTGGAAGCACG | 19 | TGCGGAAACTGCTACAC | DSR4Rc: GTGTAGCAGTTKCCGCA | 19 |
| D. thermocisternum AF074396 | ACCCACTGGAAACACG | DSR1Fa: ACCCAYTGGAAACACG | 19 | TGCGGCAACTGCTACAC | DSR4Rc: GTGTAGCAGTT KCCGCA | 19 |
| A. fulgidus NC_000917 | ACGCACTGGAAGCACG | DSR1F: ACSCACTGGAAGCACG | 33 | TGCGGTAACTGCTACAC | DSR4R: GTGTAGCAGTTACCGCA | 33 |
| A. profundus AF071499 | ACGCACTGGAAGCACG | DSR1F: ACSCACTGGAAGCACG | 33 | TGTGGAAACTGTTACAC | DSR4Ra: GTGTAACAGTTTCCACA | 19 |
Highly conserved nucleic acid positions are in boldface.
In the DsrAB tree, none of the SRPs which received the genes for this enzyme by lateral transfer group with species possessing vertically transmitted enzyme genes. This suggests that the dsrAB donor lineages have yet to be described or, alternatively, are no longer extant. In order to more fully describe the evolutionary history of the dissimilatory (bi)sulfite reductase, PCR-amplified dsrAB gene fragments (19, 33) of 15 SRPs and the syntrophic gram-positive bacterium Sporotomaculum hydroxybenzoicum were cloned, sequenced, and phylogenetically analyzed (Fig. 2). The identity of the analyzed reference cultures was confirmed by comparative 16S rRNA gene sequence analysis (27).
Interestingly, the DsrAB sequences of the deltaproteobacterial SRPs Desulfobacterium anilini (30) and strain mXyS1 (10) formed a well-supported monophyletic branch with the laterally acquired sulfite reductases of the gram-positive Desulfotomaculum species (Fig. 2). This affiliation, which is consistently inferred by different treeing methods, suggests that D. anilini and strain mXyS1 either acquired their (bi)sulfite reductase genes from the same unknown donor lineage as the gram-positive SRPs or that these two organisms are members of the dsrAB lineage, which served as donor for the gram-positive SRPs. Since D. anilini and strain mXyS1, which are marine mesophilic bacteria, form an independent lineage within the “Deltaproteobacteria” in the 16S rRNA and DsrAB trees (with the exception of the gram-positive bacteria with the laterally acquired dsrAB) (Fig. 2), this lineage is the most parsimonious dsrAB donor candidate for the gram-positive SRPs.
As S. hydroxybenzoicum forms a monophyletic branch in the 16S rRNA tree together with Desulfotomaculum species known to have received deltaproteobacterial dsrAB, it was not unexpected that S. hydroxybenzoicum also contains laterally acquired dsrAB. Similarly, Archaeoglobus veneficus possesses, like the other two species of this genus, laterally transferred dsrAB. The phylogenetic affiliations of the 12 remaining novel dsrAB sequences were found to be largely congruent with the respective 16S rRNA phylogeny of the organisms (Fig. 2). This observation further supports our current perception that the dissimilatory (bi)sulfite reductase is an ancient enzyme whose evolutionary history is largely consistent with vertical transmission but has also been influenced by periodic lateral gene transfer events. To avoid publication of incorrectly assigned dsrAB sequences in the future, we recommend careful checking of the purity of reference cultures. Furthermore, PCR-independent control experiments (e.g., Southern hybridization with genomic DNA using a dsrAB-targeted highly specific oligonucleotide probe) should be implemented for those reference cultures whose DsrAB sequences do not phylogenetically cluster with DsrAB sequences of recognized close relatives (as inferred from phylogenetic 16S rRNA trees).
Acknowledgments
We thank Natuschka Lee for database maintenance, Gerda Harms for valuable discussion, and Michael Taylor for critical review of the manuscript. Christian Baranyi, Stephan Duller, and Sibylle Schadhauser are acknowledged for their excellent technical assistance.
This research was supported by grants of the bmb+f (01 LC 0021 subproject 2 in the framework of the BIOLOG program) and of the DFG (in the framework of the project “Degradation of marine pollutants by cyanobacterial mats—an interdisciplinary approach”) to M.W. and by a Marie Curie Intra-European Fellowship (VENTSULFURMICDIV) within the 6th European Community Framework Programme to A.L. D.A.S. was supported by grant DEB-0213186 from the U.S. National Science Foundation.
REFERENCES
- 1.Baker, B. J., D. P. Moser, B. J. MacGregor, S. Fishbain, M. Wagner, N. K. Fry, B. Jackson, N. Speolstra, S. Loos, K. Takai, B. S. Lollar, J. Fredrickson, D. Balkwill, T. C. Onstott, C. F. Wimpee, and D. A. Stahl. 2003. Related assemblages of sulphate-reducing bacteria associated with ultradeep gold mines of South Africa and deep basalt aquifers of Washington State. Environ. Microbiol. 5:267-277. [DOI] [PubMed] [Google Scholar]
- 2.Castro, H., K. R. Reddy, and A. Ogram. 2002. Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68:6129-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, Y.-J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubilier, N., C. Mulders, T. Ferdelman, D. de Beer, A. Pernthaler, M. Klein, M. Wagner, C. Erseus, F. Thiermann, J. Krieger, O. Giere, and R. Amann. 2001. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411:298-302. [DOI] [PubMed] [Google Scholar]
- 6.Eckerskor, C., and F. Lottspeich. 1989. Internal amino acid sequence analysis of proteins separated by gel electrophoresis after tryptic digestion in polyacrylamide matrix. Chromatographia 28:92-94. [Google Scholar]
- 7.Fauque, G., A. R. Lino, M. Czechowski, L. Kang, D. V. DerVartanian, J. J. Moura, J. LeGall, and I. Moura. 1990. Purification and characterization of bisulfite reductase (desulfofuscidin) from Desulfovibrio thermophilus and its complexes with exogenous ligands. Biochim. Biophys. Acta 1040:112-118. [DOI] [PubMed] [Google Scholar]
- 8.Fishbain, S., J. G. Dillon, H. L. Gough, and D. A. Stahl. 2003. Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl. Environ. Microbiol. 69:3663-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich, M. W. 2002. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J. Bacteriol. 184:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harms, G., K. Zengler, R. Rabus, F. Aeckersberg, D. Minz, R. Rosselló-Mora, and F. Widdel. 1999. Anaerobic oxidation of o-xylene, m-xylene, and homologous alkylbenzenes by new types of sulfate-reducing bacteria. Appl. Environ. Microbiol. 65:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatchikian, E. C., and J. G. Zeikus. 1983. Characterization of a new type of dissimilatory sulfite reductase present in Thermodesulfobacterium commune. J. Bacteriol. 153:1211-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hipp, W. M., A. S. Pott, N. Thum-Schmitz, I. Faath, C. Dahl, and H. G. Trüper. 1997. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology 143:2891-2902. [DOI] [PubMed] [Google Scholar]
- 13.Joulian, C., N. B. Ramsing, and K. Ingvorsen. 2001. Congruent phylogenies of most common small-subunit rRNA and dissimilatory sulfite reductase gene sequences retrieved from estuarine sediments. Appl. Environ. Microbiol. 67:3314-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karkhoff-Schweizer, R. R., D. P. W. Huber, and G. Voordouw. 1995. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl. Environ. Microbiol. 61:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen, O., T. Lien, and N. K. Birkeland. 2000. Characterization of the desulforubidin operons from Desulfobacter vibrioformis and Desulfobulbus rhabdoformis. FEMS Microbiol. Lett. 186:41-46. [DOI] [PubMed] [Google Scholar]
- 17.Larsen, O., T. Lien, and N. K. Birkeland. 2001. A novel organization of the dissimilatory sulfite reductase operon of Thermodesulforhabdus norvegica verified by RT-PCR. FEMS Microbiol. Lett. 203:81-85. [DOI] [PubMed] [Google Scholar]
- 18.Laue, H., M. Friedrich, J. Ruff, and A. M. Cook. 2001. Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 183:1727-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loy, A., K. Küsel, A. Lehner, H. L. Drake, and M. Wagner. 2004. Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70:6998-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuno, N., G. Voordouw, K. Miki, A. Sarai, and Y. Higuchi. 2003. Crystal structure of dissimilatory sulfite reductase D (DsrD) protein-possible interaction with B- and Z-DNA by its winged-helix motif. Structure 11:1133-1140. [DOI] [PubMed] [Google Scholar]
- 22.Molitor, M., C. Dahl, I. Molitor, U. Schäfer, N. Speich, R. Huber, R. Deutzmann, and H. G. Trüper. 1998. A dissimilatory sirohaem-sulfite-reductase-type protein from the hyperthermophilic archaeon Pyrobaculum islandicum. Microbiology 144:529-541. [DOI] [PubMed] [Google Scholar]
- 23.Mori, K., H. Kim, T. Kakegawa, and S. Hanada. 2003. A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extremophiles 7:283-290. [DOI] [PubMed] [Google Scholar]
- 24.Morse, R., G. R. Gibson, and M. D. Collins. 2000. Secondary structure analysis of the dissimilatory sulphite reductase in Desulfovibrio desulfuricans. Lett. Appl. Microbiol. 30:375-378. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa, T., J.-I. Ishibashi, A. Maruyama, T. Yamanaka, Y. Morimoto, H. Kimura, T. Urabe, and M. Fukui. 2004. Analysis of dissimilatory sulfite reductase and 16S rRNA gene fragments from deep-sea hydrothermal sites of the Suiyo Seamount, Izu-Bonin Arc, Western Pacific. Appl. Environ. Microbiol. 70:393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Jimenez, J. R., L. Y. Young, and L. J. Kerkhof. 2001. Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrAB) genes. FEMS Microbiol. Ecol. 35:145-150. [DOI] [PubMed] [Google Scholar]
- 27.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Röser, and H. P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 28.Rabus, R., A. Ruepp, T. Frickey, T. Rattei, B. Fartmann, M. Stark, M. Bauer, A. Zibat, T. Lombardot, I. Becker, J. Amann, K. Gellner, H. Teeling, W. D. Leuschner, F. O. Glöckner, A. N. Lupas, R. Amann, and H. P. Klenk. 2004. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 6:887-902. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schnell, S., F. Bak, and N. Pfennig. 1989. Anaerobic degradation of aniline and dihydroxybenzenes by newly isolated sulfate-reducing bacteria and description of Desulfobacterium anilini. Arch. Microbiol. 152:556-563. [DOI] [PubMed] [Google Scholar]
- 31.Shen, Y., R. Buick, and D. E. Canfield. 2001. Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature 410:77-81. [DOI] [PubMed] [Google Scholar]
- 32.Stahl, D. A., S. Fishbain, M. Klein, B. J. Baker, and M. Wagner. 2002. Origins and diversification of sulfate-respiring microorganisms. Antonie Leeuwenhoek 81:189-195. [DOI] [PubMed] [Google Scholar]
- 33.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfe, B. M., S. M. Lui, and J. A. Cowan. 1994. Desulfoviridin, a multimeric-dissimilatory sulfite reductase from Desulfovibrio vulgaris (Hildenborough). Purification, characterization, kinetics and EPR studies. Eur. J. Biochem. 223:79-89. [DOI] [PubMed] [Google Scholar]