Abstract
Motivation
High-quality computational structural models are now precomputed and available for nearly every protein in UniProt. However, the best way to leverage these models to predict which pairs of proteins interact in a high-throughput manner is not immediately clear. The recent Foldseek method of van Kempen et al. encodes the structural information of distances and angles along the protein backbone into a linear string of the same length as the protein string, using tokens from a 21-letter discretized structural alphabet (3Di).
Results
We show that using both the amino acid sequence and the 3Di sequence generated by Foldseek as inputs to our recent deep-learning method, Topsy-Turvy, substantially improves the performance of predicting protein–protein interactions cross-species. Thus TT3D (Topsy-Turvy 3D) presents a way to reuse all the computational effort going into producing high-quality structural models from sequence, while being sufficiently lightweight so that high-quality binary protein–protein interaction predictions across all protein pairs can be made genome-wide.
Availability and Implementation
TT3D is available at https://github.com/samsledje/D-SCRIPT. An archived version of the code at time of submission can be found at https://zenodo.org/records/10037674.
1 Introduction
Experimental protein–protein interaction (PPI) data remain sparse in most model organisms and even more so in other species. Recent deep learning methods that predict PPIs solely from sequence seek to address this limitation. In prior work, we introduced D-SCRIPT (Sledzieski et al. 2021) and Topsy-Turvy (Singh et al. 2022), two deep learning methods that rapidly predict whether two proteins will physically bind in the cell using only protein sequence information. We call these methods lightweight deep-learning methods, since they are computationally efficient enough to be run genome-wide. These methods can be contrasted with classical PPI docking methods (Porter et al. 2019) that require different inputs (namely the 3D structures of the proteins), and also produce different outputs (in addition to predicting if the proteins bind, they also model how they bind).
The advent of large deep learning methods for structure prediction like OmegaFold (Wu et al. 2022), AlphaFold2 (Jumper et al. 2021), ESMFold (Lin et al. 2023), and RoseTTAFold (Baek et al. 2021), however, mean that high-quality 3D protein structural models can now be produced when only protein sequence is available as input. While these methods are too expensive to run from scratch at genome-wide scale, thanks to large community-wide efforts, there is no longer a need to run them from scratch: high quality computational structural models are now being made publicly available for nearly every protein in UniProt (Varadi et al. 2022, Burley et al. 2023). In this work, we ask how this wealth of computational work and high-quality predicted structural information can be re-used to improve lightweight deep-learning methods that rapidly predict whether two proteins will physically bind in the cell. One potential approach is to run computational fold-and-dock methods such as AlphaFold-Multimer (Evans et al. 2021, Zhu et al. 2023), or full complex structure prediction (Weissenow et al. 2022, Burke et al. 2023, Harini et al. 2023, Lensink et al. 2023). While these approaches are powerful for a small set of candidate pairs, they are still too computationally expensive to scale genome-wide, e.g. to create a full predicted PPI atlas for a nonmodel organism.
However, the wide availability of protein structure prediction methods has also coincided with breakthroughs in compact representation of protein structure and structure search. One such example is Foldseek (van Kempen et al. 2023), which uses a vector-quantized variational autoencoder (VQ-VAE) (Van Den Oord et al. 2017) to encode a protein structure as a sequence of discrete embedding vectors, each of which is then mapped onto a set of characters which called the 3D interaction alphabet (3Di). This process maps the 3D space of protein structure into a single dimensional 3Di sequence, which can then be used with fast sequence search tools such as BLAST (Altschul et al. 1997) or MMseqs2 (Steinegger and Söding 2017) to identify structurally similar proteins (Barrio-Hernandez et al. 2023).
Here, we introduce Topsy-Turvy 3D (TT3D), which builds off of prior work in sequence-based PPI prediction (Singh et al. 2022) to incorporate structure by jointly modeling both amino acid sequence and 3Di sequence. We demonstrate that TT3D is able to take advantage of the compact representation of protein structure to improve the accuracy of PPI prediction in a cross-species context. In an era where high-quality predictions of protein structure are readily available for many proteins, we expect that TT3D can be easily substituted into pipelines which use lightweight sequence-only deep learning prediction methods to make high-quality predictions, while remaining fast enough to be applied at genome scale.
2 Materials and methods
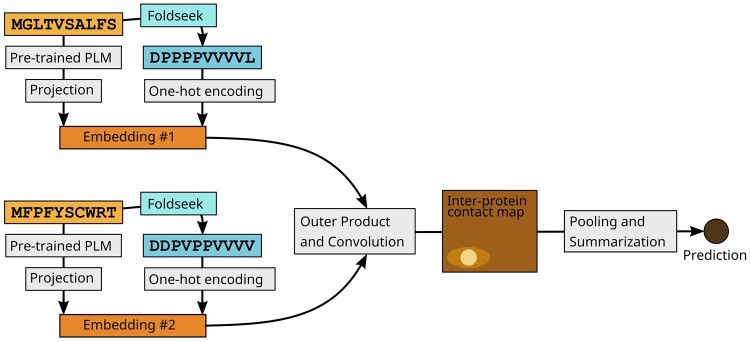
TT3D augments the inputs to the basic Topsy-Turvy architecture with encodings of the Foldseek-generated 3Di sequence (see Fig. 1) (van Kempen et al. 2023). In Topsy-Turvy, the amino acid sequence is numerically encoded using the Bepler & Berger protein language model (Bepler and Berger 2019, 2021) as , which is then reduced in dimension via a multi-layer perceptron to a projection .
Figure 1.
TT3D model architecture. TT3D follows the structure and training procedures of Topsy-Turvy, but with an augmented protein embedding. We concatenate a one-hot encoding of the Foldseek 3Di (van Kempen et al. 2023) string to the protein language model (PLM)-based embedding before passing this representation into the convolutional portion of the architecture.
In TT3D, we additionally convert the protein sequence x to a 3Di sequence using Foldseek. If a crystal structure is available for the protein, Foldseek can be directly applied. If the sequence is not available in the PDB, we query for an exact match for it in AlphaFoldDB (Varadi et al. 2022), and this structure is then used for extraction of the 3Di sequence by Foldseek. If no such hit can be found, we conservatively add an uninformative all-X 3Di sequence. We represent y with a one-hot encoding, yielding . We then concatenate the embeddings from the language model and from Foldseek, resulting in a joint embedding . Given two protein sequences , we combine embeddings as in D-SCRIPT and Topsy-Turvy (Sledzieski et al. 2021, Singh et al. 2022) to predict a probability of interaction. The Topsy-Turvy loss function is used to train the model using back-propagation.
3 TT3D outperforms state-of-the-art deep learning-based methods
We evaluate TT3D in the same cross-species setting where D-SCRIPT and Topsy-Turvy were originally tested. Following (Sledzieski et al. 2021), TT3D was trained and validated on known human PPI from the STRING database (Szklarczyk et al. 2021), filtered for experimentally determined physical binding interactions.
Then, the best model trained on human PPIs was tested on known interactions from other model organisms such as mouse (Mus musculus), fly (Drosophila melanogaster), roundworm (Caenorhabditis elegans), Escherichia coli, and brewer’s yeast (Saccharomyces cerevisiae), also from STRING. Sequences were clustered with human sequences at 40% similarity using CD-HIT (Li and Godzik 2006) and those with high similarity to proteins in the training set were removed. We measure model performance using the area under the precision–recall curve (AUPR). The test sets were constructed to have a 1:10 ratio of positives to negatives, so a random method would have an AUPR of .
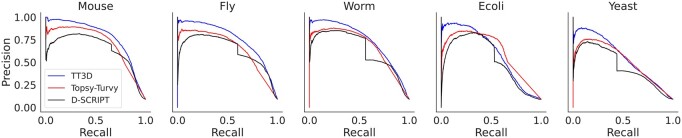
We compare TT3D to D-SCRIPT and Topsy-Turvy, neither of which incorporate structural information, and find that augmenting the Topsy-Turvy model with the encoded 3Di Foldseek sequence improves its PPI predictions. We also test against simple sequence and structure homology-based approaches. In Fig. 2, we show precision–recall curves for each of the three deep learning methods on the five benchmark test sets. TT3D performs significantly better than the other methods for all organisms that we tested on. In addition to overall performance, early precision (i.e. precision at low recall) is important, because often only a small number of highly predicted interactions are selected for downstream experimental prediction. We find that the precision values at low recall are closer to 1 for TT3D, which indicates that its top predictions are much more accurate than both D-SCRIPT and Topsy-Turvy.
Figure 2.
Precision–recall curves for TT3D, Topsy-Turvy, and D-SCRIPT. Our experiments in organisms: Mouse, Fly, Roundworm, E.coli, and Brewer’s Yeast show TT3D significantly outperforming the other methods while predicting unknown PPI interactions.
4 Comparing TT3D’s performance with simple sequence and structure homology transfer approaches
Sequence or structure-based homology approaches can also be used to transfer PPI annotations across species. We benchmarked TT3D against two such approaches, one based on Ensembl-provided sequence homology (Smedley et al. 2009), and the other based on structural homology inferred using a pipeline of AlphaFoldDB, Foldseek, and MMseqs2 (see Supplementary Materials Online). We note two major challenges with such annotation transfer approaches. First, due to the bias in how candidate PPIs were chosen for assays, just knowing that a pair of target proteins have human homologs turns out to be a surprisingly good predictor of their interaction, achieving precision (recall) of 0.2065 (0.658) and 0.2313 (0.4442) in fly and yeast, respectively. Second, such an approach does not provide a probability of an interaction, so neither an average precision nor precision–recall curve can be computed. Nonetheless, we compared TT3D to these approaches by generating the exhaustive set of fly (or yeast) PPI candidates by considering all possible transfers of human PPIs and scored these against ground-truth PPIs. TT3D outperformed both sequence and structure-based annotation transfer, achieving about 5× and 17× greater precision in fly than sequence and structure-based approaches, respectively (see Supplementary Materials for detailed precision–recall metrics).
5 Availability and implementation
For inference with TT3D, as well as with Topsy-Turvy and D-SCRIPT, we make available a web interface at https://cb.csail.mit.edu/cb/dscript/. This interface is implemented with Gradio (Abid et al. 2019) and hosted on HuggingFace spaces, and allows the user to upload a .fasta formatted file with sequences and a .tsv file with candidate protein pairs, and get back predictions for the desired model. This interface additionally leverages 3Di sequences from (Heinzinger et al. 2023).
For model training or larger-scale inference from the command line, TT3D is implemented in Python 3 as part of the dscript package for predicting PPIs, which is available from the PIP package repository (pip install dscript) or on GitHub at https://github.com/samsledje/D-SCRIPT. Model training and inference was performed on a machine with a 112-core Intel Xeon Gold 6258R CPU and using a single NVIDIA A100 GPU. TT3D is trained for a maximum of 10 epochs, and the best performing model in cross-validation is used for making predictions. We make the trained model available to download at https://d-script.readthedocs.io/en/stable/, where it can be used to make new predictions with the dscript predict command.
TT3D requires that Foldseek (van Kempen et al. 2023) be installed and that 3Di sequences be generated for protein sequences in the training or inference set. Structures in .pdb format must be available for all sequences, either natively or generated by a structure-prediction method such as OmegaFold (Wu et al. 2022), AlphaFold2 (Jumper et al. 2021), or RoseTTAFold (Baek et al. 2021). Foldseek can be downloaded and build from source on Github at https://github.com/steineggerlab/foldseek. For convenience, we provide the command dscript extract-3Di, which uses the user’s installed Foldseek to translate a set of structures into a .fasta file containing 3Di sequences.
To run TT3D, users should run the command dscript train – allow_foldseek, where – allow_foldseek is an optional command that runs the training iterations in “Foldseek” mode. While running in this mode, the user should provide the corresponding 3Di sequences in .fasta format using the – foldseek_fasta argument.
Supplementary Material
Contributor Information
Samuel Sledzieski, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139, United States.
Kapil Devkota, Department of Computer Science, Tufts University, 177 College Avenue, Medford, MA 02155, United States.
Rohit Singh, Department of Biostatistics & Bioinformatics, Duke University, Durham, NC 27705, United States; Department of Cell Biology, Duke University, Durham, NC 27705, United States.
Lenore Cowen, Department of Computer Science, Tufts University, 177 College Avenue, Medford, MA 02155, United States.
Bonnie Berger, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139, United States; Department of Mathematics, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, United States.
Author contributions
All authors conceived the research, performed the experiments, analyzed the results, and wrote the manuscript. S.S., K.D., and R.S. wrote the code.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
This work was supported by the National Science Foundation Graduate Research Fellowship under Grant No. 2141064 [to S.S.], the National Science Foundation [Grant CCF-1934553 to L.C.], and the National Institutes of Health [Grant R35GM141861 to B.B.].
References
- Abid A, Abdalla A, Abid A. et al. Gradio: Hassle-free sharing and testing of ML models in the wild. arXiv, arXiv:1906.02569, 2019, preprint: not peer reviewed.
- Altschul SF, Madden TL, Schäffer AA. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, DiMaio F, Anishchenko I. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021;373:871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio-Hernandez I, Yeo J, Jänes J. et al. Clustering predicted structures at the scale of the known protein universe. bioRxiv, 10.1101/2023.03.09.531927, 2023, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- Bepler T, Berger B. Learning protein sequence embeddings using information from structure. In: International Conference on Learning Representations (ICLR). arXiv, arXiv: 1902.08661, 2019, preprint: not peer reviewed.
- Bepler T, Berger B.. Learning the protein language: evolution, structure, and function. Cell Syst 2021;12:654–69.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DF, Bryant P, Barrio-Hernandez I. et al. Towards a structurally resolved human protein interaction network. Nat Struct Mol Biol 2023;30:216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Bhikadiya C, Bi C. et al. RCSB protein data bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res 2023;51:D488–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R, O’Neill M, Pritzel A. et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv, 10.1101/2021.10.04.463034, 2021, preprint: not peer reviewed. [DOI]
- Harini K, Christoffer C, Gromiha MM. et al. Pairwise and multi-chain protein docking enhanced using LZerD web server. In: Shahid M. (Ed.) Protein–Protein Interactions: Methods in Molecular Biology 2690. New York, NY: Springer; Nature, 2023, 355–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzinger M, Weissenow K, Sanchez JG. et al. ProstT5: Bilingual language model for protein sequence and structure. bioRxiv, 10.1101/2023.07.23.550085, 2023, preprint: not peer reviewed. [DOI]
- Jumper J, Evans R, Pritzel A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lensink M, Brysbaert G, Raouraoua N. et al. Impact of AlphaFold on structure prediction of protein complexes: the CASP15-CAPRI experiment. Authorea 2023. 10.22541/au.168888815.53957253/v1 [DOI] [PMC free article] [PubMed]
- Li W, Godzik A.. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006;22:1658–9. [DOI] [PubMed] [Google Scholar]
- Lin Z, Akin H, Rao R. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023;379:1123–30. [DOI] [PubMed] [Google Scholar]
- Porter KA, Desta I, Kozakov D. et al. What method to use for protein–protein docking? Curr Opin Struct Biol 2019;55:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Devkota K, Sledzieski S. et al. Topsy-Turvy: integrating a global view into sequence-based PPI prediction. Bioinformatics 2022;38:i264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledzieski S, Singh R, Cowen L. et al. D-SCRIPT translates genome to phenome with sequence-based, structure-aware, genome-scale predictions of protein–protein interactions. Cell Syst 2021;12:969–82.e6. Focus on RECOMB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B. et al. BioMart–biological queries made easy. BMC Genomics 2009;10:22–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinegger M, Söding J.. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 2017;35:1026–8. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Nastou KC. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021;49:D605–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Oord A, Vinyals O, Kavukcuoglu K.. Neural discrete representation learning. Adv Neural Inf Process Syst 2017;30: 6306–15. [Google Scholar]
- van Kempen M, Kim SS, Tumescheit C. et al. Fast and accurate protein structure search with Foldseek. Nat Biotechnol 2023. 10.1038/s41587-023-01773-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 2022;50:D439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenow K, Heinzinger M, Rost B.. Protein language-model embeddings for fast, accurate, and alignment-free protein structure prediction. Structure 2022;30:1169–77.e4. [DOI] [PubMed] [Google Scholar]
- Wu R, Ding F, Wang R. et al. High-resolution de novo structure prediction from primary sequence. bioRxiv, 10.1101/2022.07.21.500999, 2022, preprint: not peer reviewed. [DOI]
- Zhu W, Shenoy A, Kundrotas P. et al. Evaluation of AlphaFold-Multimer prediction on multi-chain protein complexes. Bioinformatics 2023;39:btad424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.