Abstract
Colicin V (ColV) is a peptide antibiotic that kills sensitive cells by disrupting their membrane potential once it gains access to the inner membrane from the periplasmic face. Recently, we constructed a translocation suicide probe, RR-ColV, that is translocated into the periplasm via the TAT pathway and thus kills the host cells. In this study, we obtained an RR-ColV-resistant mutant by using random Tn10 transposition mutagenesis. Sequencing analysis revealed that the mutant carried a Tn10 insertion in the sdaC (also called dcrA) gene, which is involved in serine uptake and is required for C1 phage adsorption. ColV activity was detected both in the cytoplasm and in the periplasm of this mutant, indicating that RR-ColV was translocated into the periplasm but failed to interact with the inner membrane. The sdaC::Tn10 mutant was resistant only to ColV and remained sensitive to colicins Ia, E3, and A. Most importantly, the sdaC::Tn10 mutant was killed when ColV was anchored to the periplasmic face of the inner membrane by fusion to EtpM, a type II integral membrane protein. Taken together, these results suggest that the SdaC/DcrA protein serves as a specific inner membrane receptor for ColV.
Colicin V (ColV) is a peptide antibiotic secreted by some members of the Enterobacteriaceae to kill closely related bacterial cells, thereby reducing competition for essential nutrients. Four plasmid-borne genes (cvaA, cvaB, cvaC, and cvi) are involved in ColV synthesis, export, and immunity (11). ColV is encoded by the cvaC gene and synthesized as a 103-amino-acid primary translation product with a conserved double glycine leader peptide at its N terminus (13). It is secreted through a dedicated ABC exporter composed of three proteins—CvaA, CvaB, and TolC—and processed to an 88-amino-acid polypeptide (11, 14, 27, 32). The processing of ColV is concomitant with its secretion, dependent on the CvaA, CvaB, and TolC proteins, and requires membrane integrity (27, 33). Another chromosomal gene, cvpA is required for ColV production and secretion in Escherichia coli (8, 11). The function of the cvpA gene product is unknown (8). Mutants with mutations in the cir, tonB, and exbB genes are resistant to ColV, suggesting that these gene products are involved in ColV uptake (6). ColV kills sensitive cells by disrupting the membrane potential (30). Importantly, it is only bactericidal when it gains access to the inner membrane from the periplasmic face (16, 32). This might be explained by the fact that the active ColV contains a disulfide bond between the only cysteines at positions 91 and 102 of the precursor polypeptide (13). The cvi gene encodes the cognate immunity protein of 78 residues, with two transmembrane helices, which alone is sufficient to fully protect a cell from the bactericidal activity of ColV.
Recently, we replaced the natural double glycine leader peptide with the twin-arginine signal peptide (RR) of the E. coli trimethylamine N-oxide reductase (TorA). The chimeric RR-ColV is translocated into the periplasm via the twin arginine translocation (Tat) system and then kills the host cells by depolarization of the inner membrane (15, 16). Importantly, blockage of the RR-ColV translocation by mutations in the twin-arginine signal peptide or in the tat genes abolishes the toxic effect of RR-ColV. EtpM of E. coli O157:H7 is a component of type II secretion system and plays an important role in the pathogenesis of the enterohemorrhagic pathogen. It belongs to type II bitopic inner membrane proteins with a large C-terminal domain protruding into the periplasm (9, 22). When fused to the periplasmic domain of EtpM, the ColV moiety is translocated into the periplasm and the chimera EtpM-ColV preserves its bactericidal activity (10). In the present study, we exploit these translocation-suicide probes in order to isolate novel mutants resistant to the cytotoxicity of ColV. The insertion of the Tn10 mini-transposon into sdaC (also called dcrA) abolished the bactericidal effect of RR-ColV, despite the fact that ColV was accumulated in the periplasm. In contrast, the mutant is sensitive to the bactericidal activity of EtpM-ColV. These observations strongly suggest that SdaC/DcrA could be required for the ColV insertion into the inner membrane and probably functions as an inner membrane receptor of ColV.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The E. coli strains used in the present study included TG1 [Δ(lac-pro) supE thi hsdD5/F′ traD36 proA+B+ lacIq lacZΔM15; laboratory stock], MC4100A (F′ lac.U169 araD139 rpsL150 thi flbB5301 deoC7 ptsF25 relA1 ara+; laboratory stock), and the MC4100A derivatives B1LK0A (ΔtatC) (2), SC44 (tolC::Tn5), and WB591 (tonB). Plasmids pBAD24 (12), its derivative pRR-ColV (16), pEtpM167-ColV (10), and pCvi (16) were described previously.
The bacteria were routinely grown in Luria-Bertani (LB) medium or on LB plates. When required, glucose (0.2%), arabinose (0.2%), ampicillin (AMP; 100 μg/ml), chloramphenicol (25 μg/ml), tetracycline (TET; 10 μg/ml), trimethylamine N-oxide (TMAO; 1 mg/ml), and lactose (0.2%) were added. Precultures were grown from single colonies and used at 100-fold dilutions for inoculation of cultures.
To study the inhibitory effect of ColV on bacterial growth, a single colony carrying RR-ColV or its derivatives was inoculated in LB-AMP-glucose medium, and the culture was incubated at 37°C with shaking for about 2 h. The culture was then diluted to an A600 of 0.01 with LB-AMP-glucose or LB-AMP-arabinose medium, and growth was monitored by measuring the A600 at intervals with a Cary 50 spectrophotometer. Alternatively, cellular fractions were added to a culture or to the top soft agar (0.5% in water) poured on the surface of a plate containing indicator cells, and the A600 or inhibitory halo was measured, respectively, after incubation. After the protein samples were resolved on polyacrylamide gels, sensitive cells in soft-agar medium were poured on the gels in plates, and the bactericidal activity was indicated by the appearance of inhibition halos.
Construction of sdaC mutant.
For λ1098 phage mutagenesis, 3 ml of TG1 strain were inoculated at 1/100 in LB medium, followed by incubation at 37°C for 2 h without shaking and then for 1 h with a gentle shaking. Bacteria were recovered by centrifugation (10,000 × g for 5 min), and 3 ml of LB medium supplemented with 10 mM MgSO4. λ1098 phages were added to 1 ml of the bacteria suspension (2 × 108 bacteria) from the stock in a final ratio of one phage per bacterium. The suspension was incubated for 20 min without shaking and then for 90 min with shaking at 37°C. Aliquots of 100 μl or 200 μl of suspension were spread onto LB agar with 10 μg of TET/ml and incubated overnight at 37°C. To eliminate an eventual second mutation in a single cell, the insertions were transferred into TG1 or MC4100A by P1vir phage-mediated transduction as described previously (18).
Cloning of the sdaC::Tn10 mutation, the sdaC gene, and the sdaCB genes.
To identify the insertion sites, chromosomal DNA was prepared from the mutant strains, digested with PstI or SalI, and cloned into the same site of the vector pBluescript. For the complementation assays, the sdaC or sdaCB genes were amplified from TG1 by PCR with SdaCBNheIup (CTAGCTAGCATTCCTCCAGGAGAAATAGAT) and, respectively, sdaC-BamHI down (CGCGGATCCAAGCGGCGCGAAAGGACTTAG) or SdaCB-BamHIdown (CGCGGATCCGCTGGATGAGAAATCGGGAAG) as primers. The PCR products were digested by NheI and BamHI endonucleases and ligated in the XbaI and BamHI sites of the pUC19 vector.
Cellular fractionation.
Periplasmic and cytoplasmic fractions were prepared by lysozyme-EDTA-cold osmotic shock and ultracentrifugation as described previously (21, 23). The preparation of spheroplasts was described previously (21). The fractions were analyzed by immunoblot with anti-RBP or anti-GroEL sera as described previously (19, 20).
RESULTS AND DISCUSSION
Construction of ColV-resistant mutants.
Recently, a translocation suicide probe was constructed by fusing the twin-arginine signal peptide (RR) of the TorA with the mature moiety of the ColV (16). The synthesis of the resulting RR-ColV is under the tight control of the PBAD promoter, which is inducible by arabinose and repressed by glucose. In the wild-type strain grown with arabinose, RR-ColV is synthesized, exported into the periplasm, and kills the host cells (15, 16). In contrast, in the tatC mutant, RR-ColV accumulates in the cytoplasm and has no effect on cellular growth. At present, four functional tat genes (tatA, -B, -C, and -E) have been identified in E. coli (28). These gene products are integral membrane proteins, and the overproduction of the TatA, TatB, and TatC proteins allows the in vitro reconstitution of a functional TAT system (1, 29). However, it is unknown whether or not the TAT apparatus consists of other unidentified components. Therefore, we sought to use the suicide property of the RR-ColV translocation to identify novel tat gene(s). We reasoned that the translocation of the RR-ColV protein could be blocked upon mutation of the genes required for the TAT function. By selecting such mutants, new tat gene(s) might thus be identified. We used a random transposition mutagenesis mediated by a Tn10 mini-transposon to create a bank of mutants.
The E. coli TG1 wild-type strain was infected with λ1098 carrying the Tn10 mini-transposon, and TET-resistant colonies were selected on LB-TET plates (see Materials and Methods). About 20,000 colonies were pooled and transformed with pRR-ColV by electroporation. Seventy-five colonies were obtained on LB-TET-AMP plates supplemented with 0.2% arabinose. After reisolation on LB-AMP plates, the derivatives of the 75 colonies were analyzed for growth on LB-TET and LB-AMP-arabinose plates. Only 15 colonies grew on both media, and plasmids were prepared from these strains. Four were identical to the pRR-ColV plasmid, as revealed by endonuclease digestion profiles, and still conferred the capacity to kill the TG1 strain when they were introduced into the wild-type strain. Therefore, the insertion of the Tn10 transposon into the chromosome of the corresponding Tetr Ampr Arar colonies seemed to interrupt genes that might be required for RR-ColV translocation or ColV cytotoxicity. The mutations carried by the four Tn10 insertion derivatives of TG1 (TnTK, TnTL, TnTM, and TnTS) were transferred into the wild-type TG1 or MC4100A strains by P1 mediated transduction to eliminate eventual multiple insertions. The four TG1 derivatives maintained the Tetr Ampr Arar phenotype. Intriguingly, only two of four MC4100A derivatives, TnMK and TnMM acquiring, respectively, the mutations from TnTK and TnTM, displayed the Tetr Ampr Arar phenotype. In contrast, TnML and TnMS, transferred, respectively, with the mutations from TnTL and TnTS, lost resistance to RR-ColV expression. Therefore, either the chromosome of the TG1 strain contains a mutation that confers, together with the Tn10 insertion, the resistance to the RR-ColV or the chromosome of MC4100A carries a suppressor or regulator that abolishes the effect of the Tn10 insertion. We therefore studied TnTK, TnTM, and their MC4100A derivatives in the following experiments, since the phenotype of the two other mutants might be a result of multiple mutations.
TAT function was not affected by Tn10 insertion.
The Tn10 insertion-generated mutants exhibited an RR-ColV-resistant phenotype, suggesting that RR-ColV translocation might be affected. To test this hypothesis, the TAT functionality of the mutants was assessed. A standard assay is to test for anaerobic growth on TMAO minimal medium (2, 5, 24, 26). Because they have negligible levels of periplasmic TMAO reductase (TorA) and membrane-bound dimethyl sulfoxide reductase (DmsABC)activities, ΔtatC mutant cells are not capable of anaerobic growth with TMAO as the sole electron acceptor unless they are complemented in trans by a functional tatC gene (2). All mutants analyzed grew as well as the parental wild-type strain in minimal TMAO medium (data not shown). In addition, the translocation of three TAT substrates—TorA, RR-GFP, and hydrogenase-2—was not impaired (data not shown). Therefore, these mutations were unlikely to affect TAT function. Moreover, the sizes and endonuclease digestion profiles of the amplified chromosomal areas covering the tatABCD operon and the tatE gene of the mutants were identical to those of the parental wild-type strain (data not shown), thus ruling out the insertion of Tn10 in one of the known tat genes. Because a single ColV molecule is sufficient to kill a host cell, it would be very difficult to obtain mutants with a leaky TAT activity by this approach.
Identification of the mutated gene carried by TnTK and TnTM.
Growth behaviors on minimal TMAO media and intact translocation of TAT substrates suggested that the TAT system was not impaired in the TnTK and TnTM mutants. To identify sites of the Tn10 insertion in the TnTK and TnTM mutants, chromosomal DNA was prepared from these strains, digested with PstI or SalI, and cloned into the same site of the pBluescript vector. Ligation products were transformed into MC4100A, and plasmids carrying the Tn10 transposon were selected on LB-TET plates. The cloned fragments were amplified by using a pair of oligonucleotides that hybridized with the polylinker region of pBluescript and the left extreme end of the Tn10. Sequencing analysis of the amplified fragments revealed that Tn10 had inserted in the sdaC gene at codon 233 in both TnTK and TnTM.
The sdaC::Tn10 mutation does not affect RR-ColV translocation.
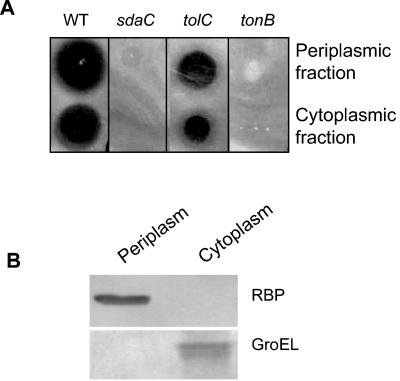
The sdaC gene is located at 63 min on the chromosome and forms an operon with the sdaB gene. The sdaC gene encodes an inner membrane threonine-insensitive serine transporter of 429 residues, with 11 predicted transmembrane helices (25). SdaC is also known as DcrA, which was previously shown to be required for the E. coli lytic phage C1 adsorption (17). To understand the in vivo resistance mechanism of the sdaC::Tn10 mutant toward RR-ColV, we analyzed the synthesis and the subcellular distribution of RR-ColV. Under natural conditions, ColV is secreted by the CvaAB-TolC system and infects sensitive cells via the TonB-ExbBD translocation machinery (25). Cytoplasmic and periplasmic fractions were prepared from the TnMK/pRR-ColV strain grown with arabinose, and the ColV activity was assessed by loading the fractions on a cellular lawn of wild-type strain and sdaC::Tn10, tolC, and tonB mutants. Inhibition halos were observed for both the wild-type strain and the tolC mutant incubated with the cytoplasmic fractions (Fig. 1). It has been repeatedly observed that RR-ColV is blocked in the cytoplasm of the tat mutants and that the periplasmic fractions obtained from the tat mutants lack the ColV bactericidal activity (15, 16). In marked contrast, the periplasmic fractions of the sdaC mutant expressing RR-ColV inhibited the growth of the wild type and the tolC mutant (Fig. 1A). The quality of cellular fractions was controlled by immunoblot analysis with antisera to RBP or GroEL as periplasmic and cytoplasmic fraction marker, respectively. Apparently, no contamination of the periplasm by cytoplasmic proteins was observed (Fig. 1B). Therefore, RR-ColV was synthesized, was exported into the periplasm, and preserved its potential bactericidal property in the sdaC::Tn10 mutant. Similar to the tonB mutant, no inhibition halos were observed for the sdaC::Tn10 mutant (Fig. 1A), suggesting the requirement of the SdaC protein for the bactericidal activity of ColV.
FIG. 1.
Cellular distribution of ColV activity expressed from pRR-ColV plasmid in TnMK. Periplasmic and cytoplasmic fractions were prepared from TnMK grown in the presence of arabinose. (A) Bactericidal activities were assessed by loading 5 μl of the fractions on the top of soft agar, which was prepared by mixing 1 ml of overnight test strain cultures of MC4100A (WT), TnMK (sdaC), SC44 (tolC), and WB591 (tonB) with 1 ml of 1% agar solution and poured on the surface of LB plates. Inhibition halos were recorded after overnight incubation of plates at 37°C. (B) The quality of the periplasmic and cytoplasmic fractions was analyzed by immunoblotting with anti-RBP (periplasmic marker) and anti-GroEL (cytoplasmic marker).
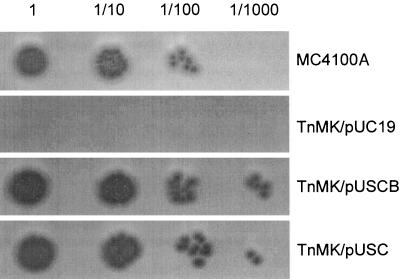
To further confirm that the resistance of the TnMK mutant to ColV was the direct consequence of the sdaC gene interruption, we cloned the sdaCB operon and the sdaC gene (see Materials and Methods) and analyzed the sensitivity of MC4100A, TnMK/pUC19, TnMK/pUSCB [(sdaCB)+], and TnMK/pUSC (sdaC+) to the killing effect of ColV added from outside of the cells. No inhibition halo was observed for TnMK/pUC19 under all concentrations of the ColV added (Fig. 2). In contrast, the same mutant carrying pUSCB was sensitive to ColV. In addition, inhibition was observed for the strain TnMK/pUSC, which indicated that the sdaC gene alone was sufficient to complement the sdaC::Tn10 mutation (Fig. 2). Inhibition was observed for TnMK/pUSCB and TnMK/pUSC at a ColV concentration 10-fold lower than that for the wild-type strain. Therefore, the expression of sdaC gene from a multicopy plasmid seems to increase the sensitivity of TnMK cells to the killing effect of ColV compared to the wild-type parental strain. Taken together, these data revealed that the sdaC::Tn10 mutation impairs neither RR-ColV synthesis nor its translocation but rather confers resistance to ColV.
FIG. 2.
The ColV-resistant phenotype of the TnMK mutant is the direct consequence of the sdaC mutation. Portions (5 μl) of either undiluted or 10-fold-serial-diluted ColV solution prepared from the cytoplasmic fraction of the tatC mutant expressing RR-ColV were loaded on the soft agar containing MC4100A, TnMK/pUC19, TnMK/pUSCB, or TnMK/pUSC prepared as described in Fig. 1.
Specificity of SdaC toward other colicins.
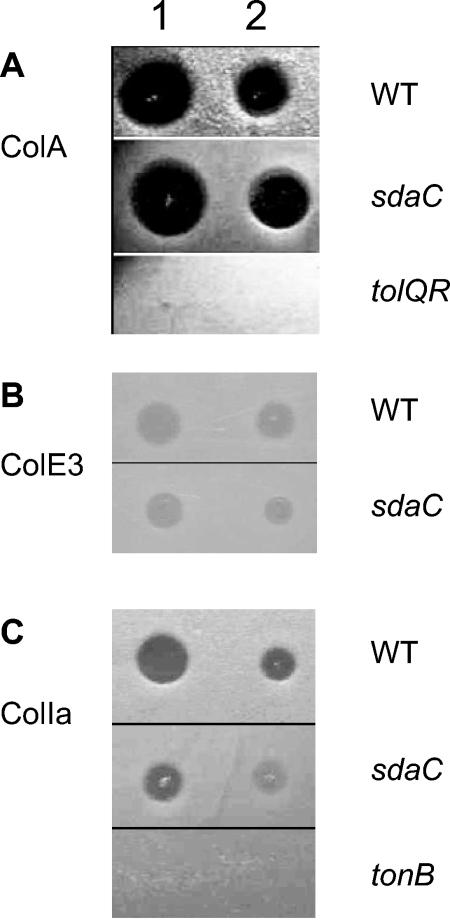
To be bactericidal, most colicins must cross the outer membrane and inner membrane of sensitive cells (7). ColV needs only to cross the outer membrane and forms a pore in the inner membrane. Despite a wide variety of lethal activities, colicins pass through the outer membrane via the porin OmpF or the iron receptor FepA, BtuB, Cir, or FhuA and requires the participation of TolA, TolB, TolC, or TonB (3, 4, 7). These results showed that SdaC is required for the bactericidal activity of ColV. To assess whether SdaC is also required for the lethal activities of other colicins, we analyzed the killing effect of pore-forming colicins ColA, ColIa, and RNase ColE3 by growth inhibition assays. Like ColV, ColIa uses Cir and TonB-ExbBD to kill sensitive cells, whereas ColA and ColE3 require BtuB and TolABQR-OmpF (4). As shown in Fig. 3, the addition of solutions containing any one of these other colicins fully inhibited the growth of the sdaC mutant as observed for the wild-type strain. Therefore, SdaC seems to be specifically required for ColV lethal action.
FIG. 3.
Sensitivity of the sdaC mutant to different colicins. Portions (10 μl) either undiluted (lane 1) or 10-fold diluted (lane 2) of ColA (A), ColE3 (B), or ColIa (C) were loaded on cellular lawns of the wild-type strain MC4100A, the sdaC mutant TnMK, or the tolQR or tonB mutant. The growth inhibition effect was analyzed after overnight incubation at 37°C.
SdaC may function as a specific ColV inner membrane receptor.
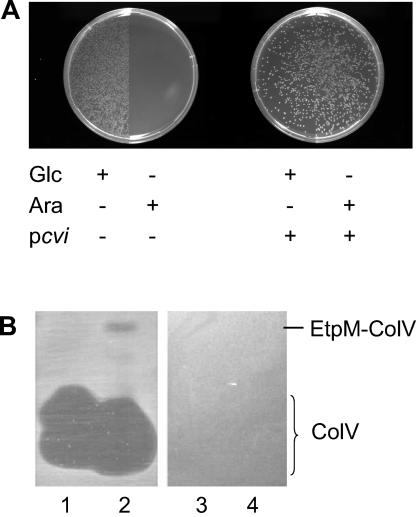
Since the sdaC::Tn10 mutant produced an active RR-ColV, which accumulated in the periplasm but could not kill host cells, SdaC might function as a receptor to bring the ColV to the membrane or might be required for the pore-forming activity of ColV. Recently, we constructed an EtpM-ColV chimera by fusing ColV to the C terminus of the EtpM protein (10). EtpM belongs to the GspM protein family and is a bitopic type II inner membrane protein with a large C-terminal domain protruding into the periplasm (9, 22). The ColV moiety is anchored to the inner membrane on the periplasmic side by EtpM, and membrane insertion of EtpM-ColV depends on YidC (10). In direct contrast to RR-ColV, the sdaC::Tn10 mutant was killed when expression of EtpM-ColV was induced by arabinose (Fig. 4A). In addition, coexpression of the Cvi immunity protein fully protected the sdaC::Tn10 mutant from the killing effect (Fig. 4A). Therefore, SdaC seems to be dispensable for the ColV bactericidal activity when ColV is brought into close contact to the inner membrane.
FIG. 4.
Sensitivity of TnMK to endogenously produced or exogenously added EtpM-ColV fusion. (A) CFU of TnMK/pEtpM167-ColV grown on an LB-AMP plate supplemented with glucose (Glc +) or arabinose (Ara +) without (pcvi −) or with (pcvi +) coexpression of ColV immunity protein. (B) Crude extracts containing ColV (lanes 1 and 3) or ColV and EtpM-ColV (lanes 2 and 4) were resolved on 0.1% sodium dodecyl sulfate-20% polyacrylamide gels. The gels were then incubated for 30 min in a 50% methanol-10% acetic acid solution, washed twice with 250 ml of sterile water, and placed onto LB plates. Soft agar containing the wild-type MC4100A strain (lanes 1 and 2) or the sdaC mutant (lanes 3 and 4) was poured on the top, and the petri dishes were incubated overnight at 37°C. The migration positions of ColV and EtpM-ColV are indicated.
The sensitivity of the sdaC mutant to exogenously added ColV and EtpM-ColV was analyzed. Crude extracts containing ColV or EtpM-ColV were separated on sodium dodecyl sulfate-polyacrylamide gels, test cells were poured onto the surface of the gels in soft-agar overlays, and bactericidal activity was indicated by the inhibition of growth (see Materials and Methods). As shown in Fig. 4B, growth of the wild-type parental strain was inhibited by both ColV and EtpM-ColV. In contrast, the sdaC::Tn10 mutant was resistant to both forms of the ColV (Fig. 4B). Therefore, the sdaC::Tn10 mutant was resistant to the EtpM-ColV when it was added from the outside, even though it was killed by the EtpM-ColV produced by itself. The sensitivity of the sdaC mutant to the bactericidal activity of EtpM-ColV might result from an indirect effect of an overexpression of the membrane EtpM moiety. These results could also support a new concept: SdaC should function as a specific receptor used by ColV to anchor to the inner membrane, which is an essential step to kill cells by disrupting their membrane potential.
In general, colicins are composed of three domains that apparently perform different functions (4). For the pore-forming colicins (ColA, ColE1, ColIa, ColN, and ColB), the central region forms unique structures that bind to outer membrane receptor proteins and the N terminus is required for the transport of the toxin through the outer membranes. The C terminus creates voltage-gated pores in the cytoplasmic membrane (31). The bactericidal activity of pore-forming colicins requires the conversion of their channel domain from a water-soluble state to a hydrophobic integral transmembrane channel; this involves massive conformational changes (31). These structural changes are mediated by the outer membrane receptor system (BtuB/TolC and BtuB/OmpF) and the Tol or Ton protein network that provides trans-envelope transport for the N-terminal translocation and C-terminal channel domains of the colicin. ColV does not contain the three apparent domains and does not belong to the bacteriocin family in a strict sense. However, it shares some similarities with the pore-forming colicins with respect to TonB/ExbBD dependency and membrane depolarization activity from the periplasmic side. It has been suggested that ColV inserts into the inner membrane spontaneously, without the assistance of other proteins, once it gains access to the inner membrane from the periplasmic side. In the present study, we showed for the first time that the ColV bactericidal activity seems to require the inner membrane protein SdaC. Since the sdaC mutant remained sensitive to other pore-forming colicins analyzed, the sdaC mutation should not affect Tol-Ton network function that is generally required for colicins to cross the outer membrane. Because EtpM-ColV was capable of killing the sdaC mutant when EtpM-ColV is anchored in the membrane, SdaC might be a specific receptor for ColV to gain intimate contact with the inner membrane, a step essential for the pore-forming procedure. Because such an in vivo interaction cannot be shown directly (due to the killing effect of ColV), further experiments could be driven by using ColV derivatives that cannot disrupt the inner membrane potential but are still able to interact with the membrane. Nevertheless, these preliminary data may open the door to the understanding of the membrane insertion mechanism of the pore-forming colicins.
Acknowledgments
We are grateful for the valuable discussions and suggestions of A. Chanal, V. Douet, K. Gouffi, B. Ize, and C.-L. Santini. We thank D. Cavard and D. Duche for the colicins and valuable suggestions and V. Méjean for advice regarding Tn10 mutagenesis.
REFERENCES
- 1.Alami, M., D. Trescher, L.-F. Wu, and M. Muller. 2002. Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 277:20499-20503. [DOI] [PubMed] [Google Scholar]
- 2.Bogsch, E., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson, and T. Palmer. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003-18006. [DOI] [PubMed] [Google Scholar]
- 3.Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Benedetti, and R. Lloubes. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84:413-421. [DOI] [PubMed] [Google Scholar]
- 4.Cao, Z., and P. E. Klebba. 2002. Mechanisms of colicin binding and transport through outer membrane porins. Biochimie 84:399-412. [DOI] [PubMed] [Google Scholar]
- 5.Chanal, A., C.-L. Santini, and L.-F. Wu. 1998. Potential receptor function of three homologous components, TatA, TatB and TatE, of the twin-arginine signal sequence-dependent metalloenzyme translocation pathway in Escherichia coli. Mol. Microbiol. 30:674-676. [DOI] [PubMed] [Google Scholar]
- 6.Chehade, H., and V. Braun. 1988. Iron-regulated synthesis and uptake of colicin V. FEMS Microbiol. Lett. 52:177-182. [Google Scholar]
- 7.de Zamaroczy, M., and R. H. Buckingham. 2002. Importation of nuclease colicins into Escherichia coli cells: endoproteolytic cleavage and its prevention by the immunity protein. Biochimie 84:423-432. [DOI] [PubMed] [Google Scholar]
- 8.Fath, M. J., H. K. Mahanty, and R. Kolter. 1989. Characterization of a purF operon mutation which affects colicin V production. J. Bacteriol. 171:3158-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 10.Gerard, F., N. Pradel, C. Ye, B. Ize, L. Yi, J. Xu, R. E. Dalbey, and L.-F. Wu. 2004. Putative membrane assembly of EtpM-colicin V chimeras. Biochimie 86:283-286. [DOI] [PubMed] [Google Scholar]
- 11.Gilson, L., H. K. Mahanty, and R. Kolter. 1990. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J. 9:3875-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havarstein, L. S., H. Holo, and I. F. Nes. 1994. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by gram-positive bacteria. Microbiology 140:2383-2389. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, J., X. Zhong, and P. C. Tai. 1997. Interactions of dedicated export membrane proteins of the colicin V secretion system: CvaA, a member of the membrane fusion protein family, interacts with CvaB and TolC. J. Bacteriol. 179:6264-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ize, B., F. Gerard, and L.-F. Wu. 2002. In vivo assessment of the Tat signal peptide specificity in Escherichia coli. Arch. Microbiol. 178:548-553. [DOI] [PubMed] [Google Scholar]
- 16.Ize, B., F. Gérard, M. Zhang, A. Chanal, R. Voulhoux, T. Palmer, A. Filloux, and L.-F. Wu. 2002. In vivo dissection of the Tat translocation pathway in Escherichia coli. J. Mol. Biol. 317:327-335. [DOI] [PubMed] [Google Scholar]
- 17.Likhacheva, N. A., V. V. Samsonov, and S. P. Sineoky. 1996. Genetic control of the resistance to phage C1 of Escherichia coli K-12. J. Bacteriol. 178:5309-5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Pradel, N., C. L. Santini, C. Y. Ye, L. Fevat, F. Gerard, M. Alami, and L.-F. Wu. 2003. Influence of tat mutations on the ribose-binding protein translocation in Escherichia coli. Biochem. Biophys. Res. Commun. 306:786-791. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigue, A., N. Batia, M. Müller, O. Fayet, R. Böhm, M. A. Mandrand-Berthelot, and L.-F. Wu. 1996. Involvement of the GroE chaperonins in the nickel-dependent anaerobic biosynthesis of NiFe-hydrogenases of Escherichia coli. J. Bacteriol. 178:4453-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigue, A., D. H. Boxer, M. A. Mandrand-Berthelot, and L.-F. Wu. 1996. Requirement for nickel of the transmembrane translocation of NiFe- hydrogenase 2 in Escherichia coli. FEBS Lett. 392:81-86. [DOI] [PubMed] [Google Scholar]
- 22.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 23.Santini, C. L., B. Ize, A. Chanal, M. Müller, G. Giordano, and L.-F. Wu. 1998. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17:101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao, Z., R. T. Lin, and E. B. Newman. 1994. Sequencing and characterization of the sdaC gene and identification of the sdaCB operon in Escherichia coli K-12. Eur. J. Biochem. 222:901-907. [DOI] [PubMed] [Google Scholar]
- 26.Weiner, J. H., P. T. Bilous, G. M. Shaw, S. P. Lubitz, L. Frost, G. H. Thomas, J. Cole, and R. J. Turner. 1998. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93:93-101. [DOI] [PubMed] [Google Scholar]
- 27.Wu, K.-H., and P. C. Tai. 2004. Cys32 and His105 are the critical residues for the calcium-dependent cysteine proteolytic activity of CvaB, an ATP-binding cassette transporter. J. Biol. Chem. 279:901-909. [DOI] [PubMed] [Google Scholar]
- 28.Wu, L.-F., B. Ize, A. Chanal, Y. Quentin, and G. Fichant. 2000. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J. Mol. Microbiol. Biotechnol. 2:179-189. [PubMed] [Google Scholar]
- 29.Yahr, T. L., and W. T. Wickner. 2001. Functional reconstitution of bacterial Tat translocation in vitro. EMBO. J. 20:2472-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, C. C., and J. Konisky. 1984. Colicin V-treated Escherichia coli does not generate membrane potential. J. Bacteriol. 158:757-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zakharov, S. D., and W. A. Cramer. 2002. Insertion intermediates of pore-forming colicins in membrane two-dimensional space. Biochimie 84:465-475. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, L. H., M. J. Fath, H. K. Mahanty, P. C. Tai, and R. Kolter. 1995. Genetic analysis of the colicin V secretion pathway. Genetics 141:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong, X., R. Kolter, and P. C. Tai. 1996. Processing of colicin V-1, a secretable marker protein of a bacterial ATP binding cassette export system, requires membrane integrity, energy, and cytosolic factors. J. Biol. Chem. 271:28057-28063. [DOI] [PubMed] [Google Scholar]