Abstract
A total of 64 type, reference, clinical, health food, and stock isolates of microaerophilic Lactobacillus species were examined by restriction fragment length polymorphisms. Of particular interest were members of six of the eight species most commonly recovered from the vaginas of healthy premenopausal women, namely, Lactobacillus jensenii, L. casei, L. rhamnosus, L. acidophilus, L. plantarum, and L. fermentum. Six main groupings were identified on the basis of ribotyping. This technique was able to classify fresh isolates to the species level. In the case of the ribotype A grouping for L. rhamnosus, differences between strains were evident by chromosome typing (chromotyping). Many isolates did not possess plasmids. Six L. rhamnosus strains isolated from four different health food products appeared to be identical to L. rhamnosus ATCC 21052. The molecular typing system is useful for identifying and differentiating Lactobacillus isolates. Studies of strains of potential importance to the urogenital flora should include molecular characterization as a means of comparing genetic traits with those of strains whose characteristics associated with colonization and antagonism against pathogens have been defined.
Lactobacilli colonizing human tissues have long been considered important for the maintenance of a healthy gastrointestinal tract (4) and urogenital tract (20, 21, 22, 25). The disruption of the lactobacillus flora has been associated with many urogenital infections, and as these afflict over 150 million women worldwide each year, this area of study is an important one. Indeed, many patients resort to taking health food products containing lactobacilli as a means of trying to maintain a healthy intestinal (and in some cases vaginal) flora.
The typing of lactobacilli has generally been conducted by cell and colony morphology and biochemical tests. These techniques type bacteria based on their ability to ferment sugars and produce acids such as lactic acid and acetic acid (9). Unfortunately, these typing methods are not completely accurate, and strains which show intermediate characteristics are frequently encountered (9, 33).
Many studies emphasize that the classification of lactobacilli is unsatisfactory and does not reflect the real phylogenetic relatedness of different strains and species (6, 19, 30). Several new genetic and chemotaxonomic approaches have been used during the last 14 years with an aim of improving the classification and identification of lactobacilli: for example, analysis of plasmid content (18), sodium dodecyl sulfate-polyacrylamide gel electrophoresis patterns of whole-cell protein (19) and of total soluble cell protein (8), sequencing of rRNA (5, 6, 19), restriction endonuclease fingerprinting (14, 30), and DNA-DNA hybridization (6, 17, 19, 27). All of these approaches have improved the taxonomic knowledge of the generic and suprageneric relationships of lactobacilli. However, no analysis of urogenital or health food isolates of lactobacilli by molecular typing has been reported.
Plasmid typing of Lactobacillus strains has been suggested as a taxonomic tool in a number of reports (27, 31), but there is evidence to suggest that it is not very effective (2, 12, 15, 28), since plasmids can be absent or unstable.
Chromosome typing (restriction endonuclease fingerprinting of chromosomal DNA) (chromotyping) has been applied to the discrimination of strains of lactobacilli and has been found more specific and reproducible than plasmid content analysis (14). However, one of the disadvantages of chromotyping is that comparing electrophoretic patterns consisting of up to 100 bands is difficult. Hence, chromotyping alone may not be widely used for typing large numbers of strains.
A sensitive but rapid method for species differentiation involves the use of ribotyping (1, 27), which combines Southern hybridization of chromosomal DNA fingerprints with the use of Escherichia coli rRNA probes, thereby discriminating between various species and individual strains of lactobacilli. The method involves the separation and identification of the rRNA genes present within the bacterial genome, which exist in various copy numbers (10, 13), making it possible to delineate species based on differences in the restriction fragment length polymorphisms of the rRNA genes. Of particular importance is the fact that specific regions of the rRNA genes have remained well conserved because of their functional importance, thus allowing the detection of a broad range of bacteria with 16S and 23S rRNA of E. coli as probes.
The aim of this study was to develop a method to test the efficacy of ribotyping of Lactobacillus type and reference strains and to use this method to characterize a number of clinical, health food, and laboratory isolates.
MATERIALS AND METHODS
Bacteria.
American Type Culture Collection (ATCC) and National Collection of Food Bacteria (NCFB) bacterial strains were used along with clinical and stock cultures studied previously by our group (Table 1). The type and reference strains had been identified to the species level by ATCC and NCFB prior to 1993, and the clinical and stock isolates had been identified to the species level by standard, authorized procedures prior to 1991. The nomenclature used here for the type and reference strains was that given by ATCC and NCFB prior to 1993, and that used for the clinical and stock isolates was the same as that reported in publications by our group (23, 24).
TABLE 1.
Molecular typing of Lactobacillus isolates with respect to ATCC typed strains
| Original nomenclature | Ribotype with BclI | Chromotype with BclI |
|---|---|---|
| L. rhamnosus ATCC 7469T | A | 1.1 |
| L. rhamnosus ATCC 11443 | A | 1.1 |
| L. casei subsp. rhamnosus 81 | A | 1.1 |
| L. casei subsp. rhamnosus 76 | A | 1.2 |
| L. rhamnosus ATCC 8530 | A | 1.3 |
| L. casei subsp. rhamnosus GR-1 | A | 1.4 |
| L. casei subsp. rhamnosus 36 | A | 1.4 |
| Vaginal isolate w | A | 1.4 |
| Vaginal isolate g | A | 1.4 |
| L. rhamnosus NCFB 2964 | A | 1.5 |
| L. casei subsp. rhamnosus RC-17 | A | 1.6 |
| L. fermentum A60 | A | 1.6 |
| L. plantarum RC-6 | A | 1.7 |
| L. rhamnosus ATCC 21052 | A | 2.1 |
| C1-A | A | 2.1 |
| C3-A | A | 2.1 |
| C3-B | A | 2.1 |
| C5-A | A | 2.1 |
| C7-A | A | 2.1 |
| C7-B | A | 2.1 |
| L. casei subsp. casei ATCC 393 | B | 10.1 |
| L. casei subsp. casei ATCC 4940 | B1 | 11.1 |
| L. paracasei subsp. paracasei ATCC 25302T | C | 20.1 |
| L. casei subsp. casei ATCC 334 | C | 20.2 |
| L. casei subsp. casei ATCC 4007 | C1 | 21.1 |
| L. casei subsp. casei ATCC 4913 | C2 | 22.1 |
| L. casei subsp. casei 55 | C2 | 23.1 |
| L. paracasei subsp. paracasei ATCC 25303 | C2 | 24.1 |
| L. casei subsp. rhamnosus 43 | C3 | 25.1 |
| L. casei 8 | C3 | 25.1 |
| L. casei 65 | C3 | 25.1 |
| L. plantarum ATCC 14917T | D | 30.1 |
| L. plantarum UH 2153 | D | 30.1 |
| L. plantarum ATCC 10012 | D | 30.2 |
| L. plantarum ATCC 11974 | D1 | 31.1 |
| L. plantarum ATCC 14431 | D2 | 32.1 |
| L. plantarum ATCC 8014 | D3 | 33.1 |
| L. plantarum 260 | D3 | 33.1 |
| L. plantarum RC-20 | D4 | 34.1 |
| L. acidophilus 75 | D5 | 34.2 |
| US2-A | D6 | 35.1 |
| US2-B | D6 | 35.1 |
| US2-C | D6 | 35.1 |
| US3-A | D7 | 35.2 |
| US3-B | D7 | 35.2 |
| US6-A | D8 | 36.1 |
| US6-B | D8 | 36.1 |
| US6-C | D8 | 36.1 |
| C4-A | D8 | 36.1 |
| C4-B | D8 | 36.1 |
| US8-A | D9 | 37.1 |
| L. fermentum ATCC 14931T | E | 40.1 |
| L. fermentum ATCC 23271 | E1 | 40.2 |
| L. fermentum ATCC 8289 | E2 | 40.3 |
| L. fermentum ATCC 9338 | E2 | 40.3 |
| L. fermentum ATCC 11739 | E3 | 41.1 |
| L. fermentum ATCC 14932 | E4 | 42.1 |
| L. fermentum B-54 | E5 | 43.1 |
| L. acidophilus RC-14 | E5 | 43.1 |
| L. acidophilus RC-14 (subcultured 20 times) | E5 | 43.1 |
| L. acidophilus ATCC 4356T | F | 50.1 |
| US4 | F | 50.2 |
| L. acidophilus T-13 | F1 | 51.1 |
| L. jensenii ATCC 25258T | G | 60.1 |
Nine health food products were examined. In each, five capsules were opened and the contents were cultured in 40 to 80 ml of MRS broth anaerobically for 48 h. The dominant strains of lactobacilli which grew under these conditions were then processed for further study. The products were as follows: C1, from Rhone Poulenc, Montreal, Quebec, Canada, containing Lactobacillus species; C3, from Pharma Plus Drugmarts, Toronto, Ontario, Canada, containing Lactobacillus acidophilus, L. bulgaricus, and Streptococcus thermophilus; C4 Fermalac Vaginal, from Rougier Inc., Chambley, Province Quebec, Canada, containing L. acidophilus, L. bulgaricus, and S. thermophilus; C5, from Gahler Enterprises, Burnaby, British Columbia, Canada, containing L. acidophilus and L. bifidus; C7, from Nu-Life Nutritional Product, Toronto, Ontario, Canada, containing L. acidophilus (L. rhamnosus A) and L. acidophilus ATCC; US2 Probiotic Acidophilus for Life, from Nutrition Now Inc., Vancouver, Wash., containing L. acidophilus, L. plantarum, L. bulgaricus, L. casei, S. thermophilus, S. faecium, Bifidobacterium bifidum, and B. infantis; US3 Lactobacillus acidophilus, from Source Naturals Inc., Scotts Valley, Calif., containing L. bifidus, L. brevis, L. caucasicus, L. salivarius, L. casei, L. bulgaricus, L. youghurtii, S. faecalis, and S. thermophilus; US4, from Solaray Inc., Ogden, Utah, containing L. bulgaricus and L. acidophilus; US6, from Wakunaga of America Co., Mission Viejo, Calif., containing L. acidophilus and B. bifidum; and US8, from America’s Finest, lactospore flora containing L. sporogenes.
Cell lysis and genomic DNA isolation.
Genomic DNA isolation was conducted by a method modified from that of Bialkowska-Hobrzanska et al. (2). All strains were used after a maximum of three subcultures. Cultures were grown in 40 to 80 ml of MRS broth containing 1% glycine at 37°C in 5% CO2 for 24 to 72 h to mid-log growth, harvested (6,000 × g, 10 min, 4°C), and washed once and resuspended in extraction buffer (50 mM Tris-Cl [pH 8.0], 50 mM EDTA, 50 mM NaCl) to a final concentration of 109 cells/ml. Mutanolysin (Boehringer-Mannheim Canada, Laval, Quebec, Canada) and lysozyme (Boerhinger-Mannheim Canada) were added to final concentrations of 400 U/ml and 10 mg/ml, respectively, and the suspension was incubated at 55°C for 8 to 16 h. Following incubation, RNase A (200 μg/ml; Boehringer-Mannheim Canada) and sodium dodecyl sulfate (1%) were added, and the suspension was incubated for an additional 30 min at 55°C. Proteinase K (200 μg/ml; Boehringer-Mannheim Canada) was added, and the suspension was incubated for 1 h at 50°C and then for 10 min at 65°C. Cell components were removed with three phenol-chloroform (1:1) extractions and three ether extractions and then dialyzed for 3 to 12 h against a buffer containing 10 mM Tris-Cl (pH 7.6) and 1 mM EDTA three times. Following dialysis, the DNA samples were separated (1.5 h, 100 V) on an 0.8% agarose mini horizontal slab gel (Bio-Rad Laboratories, Mississauga, Ontario, Canada) in a buffer containing 0.089 M Tris-borate and 0.002 M EDTA to test for DNA quality and quantity. For plasmid profiling and chromotyping analyses, 3 to 5 μg of DNA was used.
Plasmid profiling.
The cleared-lysate procedure of total DNA purification described in the preceeding paragraph was used throughout the study for plasmid profile analysis, and a supercoiled DNA ladder (Gibco BRL Life Technologies, Burlington, Ontario, Canada) was used as a size marker.
Chromotyping.
The chromotyping analysis consisted of digestion of 3 to 5 μg of total genomic DNA with one of two selected restriction enzymes. Previous work (3a) with a variety of restriction enzymes showed that the enzymes BclI and DraI produce Lactobacillus strain-discriminating DNA banding patterns. Total genomic DNA was digested with at least 10 U of either BclI or DraI (Boehringer-Mannheim Canada or Promega Corp., Madison, Wis., respectively) per μg of DNA for 5 h under conditions recommended by the supplier, after which time the activity of the enzyme BclI or DraI was stopped by heating at 65°C for 10 min or 75°C for 15 min, respectively. The size marker used for chromosomal DNA was a 1-kb DNA ladder (Gibco BRL Life Technologies). The DNA fragments were separated on an 0.8% agarose horizontal slab gel as described above for plasmid profile analysis. The DNA banding patterns were visually analyzed to determine chromotype profiles.
Ribotyping.
Ribotyping was conducted by a method modified from that of Bialkowska-Hobrzanska et al. (1, 3, 27). The restriction fragments were transferred to a nitrocellulose membrane (Schleicher & Schuell, Inc., Keene, N.H.) as described by Southern (29). Then, rRNA (16S plus 23S) from E. coli (Boehringer-Mannheim Canada) was dephosphorylated with calf intestinal phosphatase and labelled with [γ-32P]dATP (ICN Pharmaceuticals Canada, Montreal, Quebec, Canada) by use of polynucleotide kinase. The specific activity of rRNA was 108 cpm/μg. Prehybridization, hybridization, and autoradiography were carried out as described previously (3). The sizes of the DNA fragments hybridizing with labelled rRNA were determined by comparison with [α-32P]dCTP and the labelled 1-kb DNA ladder. Distinct ribotype patterns were defined on the basis of hybridization bands and differences in the numbers and sizes of rRNA gene restriction fragments.
RESULTS
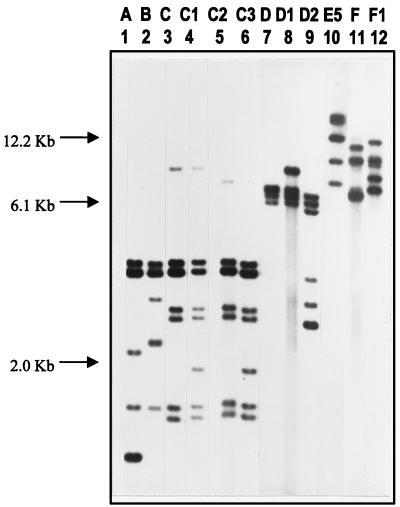
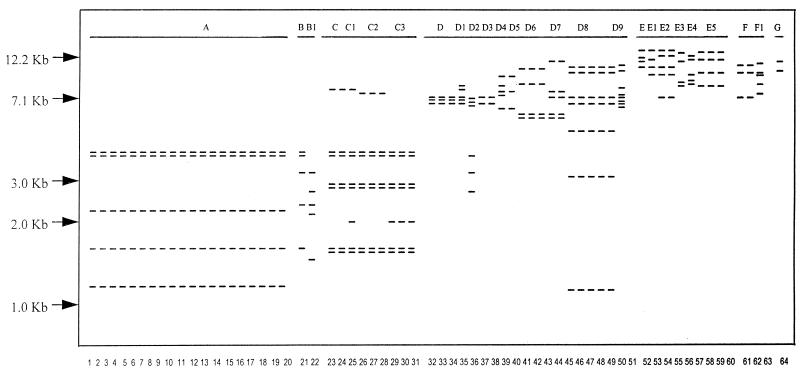
The molecular typing results are presented in Table 1. The chromotyping results are shown in Fig. 2, and the ribotyping results are shown in Fig. 2 and 3. The numbering system was created to differentiate strains; for example, chromotyping grouped each species as 1 to 9, 10 to 19, 20 to 29, and so forth. When strain banding patterns were very close but not exactly the same, a decimal point number was used to differentiate them (e.g., 1.1 versus 1.2). A similar system was used for ribotyping, except that differences were depicted only by a number added to a letter. For example, within each group, a difference in minor bands resulted in a strain being termed D2 (32.1) relative to type strain D (30.1).
FIG. 2.
(A) Chromotyping. Agarose gel electrophoresis of total DNA after digestion with BclI. (B) Ribotyping. Autoradiogram of Southern blot of BclI-digested DNA after hybridization with 32P-labelled rRNA from E. coli. Lanes 1 to 8, health food strains of Lactobacillus (C3-B, C4-B, C7-B, US2-B, US2-C, US3-B, US4, and US6-C); lane 9, L. fermentum ATCC 11739; lane 10, L. fermentum ATCC 8289; lane 11, clinical strain of Lactobacillus (vaginal isolate w); lane 12, L. acidophilus ATCC 4356T; lane 13, clinical strain of Lactobacillus (strain 36).
FIG. 3.
Autoradiogram of Southern blot of BclI-digested DNA after hybridization with 32P-labelled rRNA from E. coli. Lanes 1 to 12 represent ribotype groups A to F. Lane 1, L. rhamnosus ATCC 7469T; lane 2, L. casei subsp. casei ATCC 393; lane 3, L. paracasei subsp. paracasei ATCC 25302T; lane 4, L. casei subsp. casei ATCC 4007; lane 5, L. paracasei subsp. paracasei ATCC 25303; lane 6, L. casei 8; lane 7, L. plantarum ATCC 14917T; lane 8, L. plantarum ATCC 11974; lane 9, L. plantarum ATCC 14431; lane 10, L. fermentum B-54; lane 11, L. acidophilus ATCC 4356T; lane 12, L. acidophilus T-13.
Six main ribotypes were identified for the 64 strains tested, coinciding with six of the eight key species found in the healthy vagina. While L. jensenii is very commonly found in the vagina (26), none of the fresh or health food strains tested here showed patterns similar to that of ATCC 25258T. The roots and break points for the ribotype patterns were exclusive for group A L. rhamnosus strains, but it is acknowledged that in other cases, patterns with some deviation were grouped together. The subgrouping was based upon strains sharing similarly sized band regions; thus, the rRNA genes were closely associated (Fig. 1). For example, strains D2, D4, and D5 had banding patterns closest to that of group D, and the API50 CHL system (API Systems, La Balme les Grottes, France; species identification in accordance with the Virginia Polytechnic Institute Manual) identified them as L. plantarum (results not shown), so they were placed in group D. Ribotyping was able to classify fresh isolates to the species level, such as vaginal isolates w and g as L. rhamnosus. In the case of ribotype A grouping, differences between strains were evident by chromotyping.
FIG. 1.
Schematic representation of the species-specific ribotypes obtained after digestion of total DNA from Lactobacillus isolates with BclI and hybridization with 32P-labelled rRNA from E. coli. Lanes 1 to 64 represent strains, and groups A to G represent ribotypes described in Table 1.
Many isolates did not have plasmids, and there was no correlation between plasmid content and species identification.
L. rhamnosus contains five reference strains of this species. Seven strains previously studied for bacterial interference against uropathogens, L. casei subsp. rhamnosus GR-1, 81, 76, 36, and RC-17, L. fermentum A60, and L. plantarum RC-6, were found to have ribotype A characteristics and are hereby reclassified as L. rhamnosus GR-1, 81, 76, 36, RC-17, A60, and RC-6, respectively.
Other reclassified strains included L. paracasei subsp. paracasei 8, 65, 55, and 43, L. plantarum 75, and L. fermentum RC-14. It should be noted that after 20 subcultures, L. fermentum RC-14 had stable molecular types (Fig. 1). Two vaginal isolates from healthy women, termed w and g, and strain 36 showed patterns identical to that of L. rhamnosus GR-1 (Fig. 2), shown to colonize the human vagina (22).
Of the organisms which we isolated from the nine health food products, six isolates from four Canadian products (C1-A, C3-A, C3-B, C5-A, C7-A, and C7-B) had the same banding patterns and profiles as L. rhamnosus ATCC 21052. Nine isolates from four U.S. health food products (US2-A, US2-B, US2-C, US3-A, US3-B, US6-A, US6-B, US6-C, and US8-A) had molecular patterns consistent with that of L. plantarum, while only US4 was an L. acidophilus strain, similar to L. acidophilus ATCC 4356T (Fig. 1 and 2). The Canadian strains C4-A and C4-B showed the same pattern as U.S. strains US6-A, US6-B, and US6-C.
DISCUSSION
Ribotyping proved to be a more effective and practical method than chromotyping for Lactobacillus species identification and strain discrimination. The species examined represented six of those most commonly recovered from the vaginas of healthy premenopausal women (26). Ribotyping relied on the same assumptions as chromotyping in that as strains diverge, they acquire random mutations which result in unique restriction sites in the genomic DNA. Here, the analyses of strains and comparison to ATCC and NCFB type strains with BclI showed that species-specific ribotypes do exist. The plasmid typing patterns were found to be unreliable for species identification, and 39 of the strains had no plasmids, a common finding for strains isolated from the vaginas of healthy women (26).
An examination of the ribotypes of the 19 fresh clinical and laboratory isolates studied previously by us (16, 23, 24) indicated that 13 of the isolates should be reassigned and renamed.
Chromotyping discriminated strains, but because of the complexity of the patterns produced (Fig. 2), we do not view it as being an effective method for species determination. Ribotyping produces fewer bands, which makes species identification easier; however, ribotyping does not have the precision that is necessary for completely accurate strain discrimination, so we use ribotyping for species discrimination. These characteristics may explain the finding that L. paracasei subsp. paracasei ATCC 25302T and ATCC 25303 have a ribotype which is highly related to that of L. casei subsp. casei ATCC 334, ATCC 4007, and ATCC 4913, even though there is apparently a 15% DNA divergence between them (5).
One important outcome of the analyses is the better characterization of organisms which are believed to be potentially useful in colonizing humans and reducing the risk of disease. However, the profile data per se are insufficient to determine which DNA patterns correspond to properties necessary for a probiotic effect and which strains would not be expected to be useful. Still, a comparative examination can be made between the results obtained here and those reported previously for hydrophobicity (7, 16, 24), adhesion to epithelial cells (23), resistance to nonoxynol-9 (32), inhibition of pathogenic attachment and growth (23), and ability to colonize the vagina (23). This comparison shows that strains within each ribotype possess so-called protective properties, whether they be high-level adherence and/or inhibition of pathogens. What is not yet clear is how to identify lactobacilli which are of no use as probiotic organisms.
We found some clones which differed only in the acquisition or loss of plasmids, such as hydrophobic strains L. fermentum B-54 and RC-14, which showed identical patterns, and L. rhamnosus RC-17 and A60, whose plasmid patterns differed from each other. Two isolates from the US6 health food product were identical apart from one having apparently lost its plasmids. The same plasmid profile was never found in more than one species.
It is interesting to note that apparently identical pairs of Lactobacillus strains were isolated in very different settings: L. fermentum B-54 is a poultry isolate sent to us from Ottawa, and RC-14 was recovered from the vagina of a woman in Toronto; L. plantarum ATCC 14917T was purchased by us from ATCC, and UH 2153 was isolated from the vagina of a woman in London (Ontario); and L. rhamnosus A60 is a poultry isolate sent to us from Ottawa, and RC-17 was isolated from the vagina of a woman in Toronto. Vaginal isolates L. rhamnosus w and g appear identical, yet they were isolated from two women in London (Ontario); L. rhamnosus GR-1 was isolated from the urethra of a woman in Kingston (Ontario) in 1981, and 36 was isolated from the vagina of a woman in Toronto in 1989.
The ability to use ribotyping to identify a strain with some degree of accuracy could be used to monitor its colonization of, for example, the vagina. This technique has been used successfully to show that L. rhamnosus GR-1 is able to colonize the vagina of a patient prone to urogenital infections (22). Having stated that, some strains appear to have identical ribotype and chromotype patterns; therefore, more specific identification methods, such as the use of repetitive sequence PCR probes, would be preferred.
A previous report stated that health food products can be unreliable in their content (11). This statement cannot be confirmed here, and we are not claiming that the products are either unreliable or mislabelled. Also, it is possible that some predominant strains present in the products were not recovered by our harvesting procedures. Still, it was interesting to note the inclusion of the same ribotype, chromotype, and plasmid profiles of L. rhamnosus ATCC 21052 in four different health food products. No efforts were made to match the species listed on the labels with the isolates recovered; however, L. rhamnosus was recovered from three of the Canadian health food products (C1, C3, and C5) but was not listed on the labels. None of the health food products indicated the origin of the organisms or the reason for their inclusion, and none claimed to be able to reduce the risk of urinary tract infection. Of the U.S. health food products, only US2 was labelled to contain L. plantarum, yet this species was also recovered from US3, US6, and US8.
In summary, the data illustrate that Lactobacillus species can be discriminated effectively on the basis of their ribotypes, constituting a specific and reproducible method for determining bacterial type. The technique may be useful for identifying species present in functional food products.
ACKNOWLEDGMENTS
The assistance of Diane Jaskot and the support of St. Joseph’s Health Centre and the Lawson Research Institute are appreciated.
We thank the Faculty of Medicine at the University of Western Ontario for a grant-in-aid.
REFERENCES
- 1.Bialkowska-Hobrzanska H, Harry V, Jaskot D, Hammerberg O. Typing of coagulase-negative staphylococci by Southern hybridization of chromosomal DNA fingerprints using an rRNA gene probe. Eur J Clin Microbiol Infect Dis. 1990;9:588–594. doi: 10.1007/BF01967213. [DOI] [PubMed] [Google Scholar]
- 2.Bialkowska-Hobrzanska H, Jaskot D, Hammerberg O. Evaluation of restriction endonuclease fingerprinting of chromosomal DNA and plasmid profile analysis for characterization of multiresistant coagulase-negative staphylococci in bacteremic neonates. J Clin Microbiol. 1990;28:269–275. doi: 10.1128/jcm.28.2.269-275.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bialkowska-Hobrzanska H, Jaskot D, Millsap K, Reid G. Program and abstracts of the 34th Interscience Conference on Antimicrobial Agents and Chemotherapy. 1994. Species identification of lactobacilli by restriction fragment length polymorphism analysis of rRNA genes, abstr. D-9. [Google Scholar]
- 3a.Bialkowska-Hobrzanska, H. Unpublished data.
- 4.Bibel D J. Elie Metchnikoff’s bacillus of long life. ASM News. 1988;54:661–665. [Google Scholar]
- 5.Collins M D, Phillips B A, Zanoni P. Deoxyribonucleic acid homology studies of Lactobacillus casei, Lactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. Int J Syst Bacteriol. 1989;39:105–108. [Google Scholar]
- 6.Collins M D, Rodrigues U, Ash C, Aguire M, Farrow J A E, Martinez-Murcia A, Phillips B A, Williams A M, Wallbanks S. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16s rRNA. FEMS Microbiol Lett. 1991;77:5–12. [Google Scholar]
- 7.Cuperus P, van der Mei H C, Reid G, Khoury A H, Bruce A W, Rouxhet P G, Busscher H J. The effect of serial passaging of lactobacilli in liquid medium on their physicochemical and structural surface characteristics. Cells Mater. 1992;2:271–280. [Google Scholar]
- 8.Dicks L M T, Van Vuuren H J J. Relatedness of heterofermentative lactobacillus species revealed by numerical analysis of total cell protein patterns. Int J Syst Bacteriol. 1987;37:437–440. [Google Scholar]
- 9.Gasser F. Electrophoretic characterization of lactic dehydrogenases in the genus Lactobacillus. J Gen Microbiol. 1970;62:223–229. doi: 10.1099/00221287-62-2-223. [DOI] [PubMed] [Google Scholar]
- 10.Grimont F, Grimont P A D. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986;137b:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 11.Hughes V L, Hillier S L. Microbiologic characteristics of Lactobacillus products used for colonization of the vagina. Obstet Gynecol. 1990;75:244–248. [PubMed] [Google Scholar]
- 12.Larsen J L, Olsen J E. Occurrence of plasmids in Danish isolates of Vibrio anguillarum serovars O1 and O2 and association of plasmids with phenotypic characteristics. Appl Environ Microbiol. 1991;57:2158–2163. doi: 10.1128/aem.57.8.2158-2163.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindahl L, Zengel J M. Ribosomal genes in Escherichia coli. Annu Rev Genet. 1986;20:297–326. doi: 10.1146/annurev.ge.20.120186.001501. [DOI] [PubMed] [Google Scholar]
- 14.Manachini P L, Parini C. DNA restriction endonuclease cleavage patterns, DNA sequence similarity and phenotypical characteristics in some strains of Lactobacillus helveticus and Lactobacillus jugurti. Antonie Leeuwenhoek. 1983;49:143–152. doi: 10.1007/BF00393672. [DOI] [PubMed] [Google Scholar]
- 15.Mickelson M N, Brown R W. Physiological characteristics of Streptococcus dysgalactiae and Streptococcus uberis and the effect of lactoperoxidase complex on their growth in chemically-defined medium and milk. J Dairy Sci. 1985;68:1095–1102. doi: 10.3168/jds.S0022-0302(85)80934-6. [DOI] [PubMed] [Google Scholar]
- 16.Millsap K, Reid G, van der Mei H C, Busscher H J. Cluster analysis of genotypically characterized Lactobacillus species based on physicochemical cell surface properties and their adhesion to hexadecane. Can J Microbiol. 1996;43:284–291. [Google Scholar]
- 17.Miteva V I, Abadjieva A N, Stefanova T T. M13 DNA fingerprinting, a new tool for classification and identification of Lactobacillus spp. J Appl Bacteriol. 1992;73:349–354. doi: 10.1111/j.1365-2672.1992.tb04988.x. [DOI] [PubMed] [Google Scholar]
- 18.Nes I F. Plasmid profiles of ten strains of Lactobacillus plantarum. FEMS Microbiol Lett. 1984;21:359–361. [Google Scholar]
- 19.Pot B, Hertel C, Ludwig W, Descheemaeker P, Kersters K, Schleifer K H. Identification and classification of Lactobacillus acidophilus, L. gasseri and L. johnsonii strains by SDS-PAGE and rRNA-targeted oligonucleotide probe hybridization. J Gen Microbiol. 1993;139:513–517. doi: 10.1099/00221287-139-3-513. [DOI] [PubMed] [Google Scholar]
- 20.Redondo-Lopez V, Cook R L, Sobel J D. Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev Infect Dis. 1990;12:856–872. doi: 10.1093/clinids/12.5.856. [DOI] [PubMed] [Google Scholar]
- 21.Reid G, Bruce A W. The role of the urogenital flora in probiotics. In: Lappin-Scott H, editor. Bacterial biofilms. Cambridge, England: Cambridge University Press; 1995. pp. 274–281. [Google Scholar]
- 22.Reid G, Bruce A W, Taylor M. Instillation of Lactobacillus and stimulation of indigenous organisms to prevent recurrence of urinary tract infections. Microecol Ther. 1995;23:32–45. [Google Scholar]
- 23.Reid G, Cook R L, Bruce A W. Examination of strains of lactobacilli for properties which may influence bacterial interference in the urinary tract. J Urol. 1987;138:330–335. doi: 10.1016/s0022-5347(17)43137-5. [DOI] [PubMed] [Google Scholar]
- 24.Reid G, Cuperus P L, Bruce A W, Tomeczek L, van der Mei H C, Khoury A H, Busscher H J. Comparison of contact angles and adhesion to hexadecane of urogenital, dairy, and poultry lactobacilli: effect of serial culture passages. Appl Environ Microbiol. 1992;58:1549–1553. doi: 10.1128/aem.58.5.1549-1553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid G, Bruce A W, McGroarty J A, Cheng K-J, Costerton J W. Is there a role for lactobacilli in prevention of urogenital and intestinal infections? Clin Microbiol Rev. 1990;3:335–344. doi: 10.1128/cmr.3.4.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid G, McGroarty J A, Tomeczek L, Bruce A W. Identification and plasmid profiles of Lactobacillus species from the vagina of 100 healthy women. FEMS Immunol Med Microbiol. 1996;15:23–26. doi: 10.1111/j.1574-695X.1996.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 27.Rodtong S, Tannock G W. Differentiation of Lactobacillus strains by ribotyping. Appl Environ Microbiol. 1993;59:3480–3484. doi: 10.1128/aem.59.10.3480-3484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skov M N, Pedersen K, Larsen J L. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl Environ Microbiol. 1995;61:1540–1545. doi: 10.1128/aem.61.4.1540-1545.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 30.Stahl M, Molin G, Persson A, Ahrne S, Stahl S. Restriction endonuclease patterns and multivariable analysis as a classification tool for Lactobacillus spp. Int J Syst Bacteriol. 1990;40:189–193. [Google Scholar]
- 31.Tannock G W, Fuller R, Pedersen K. Lactobacillus succession in the piglet digestive tract demonstrated by plasmid profiling. Appl Environ Microbiol. 1990;56:1310–1316. doi: 10.1128/aem.56.5.1310-1316.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomeczek L, Reid G, Cuperus P L, McGroarty J A, van der Mei H C, Bruce A W, Khoury A H, Busscher H J. Correlation between hydrophobicity and resistance to nonoxynol-9 and vancomycin for urogenital isolates of lactobacilli. FEMS Microbiol Lett. 1992;94:101–104. doi: 10.1111/j.1574-6968.1992.tb05297.x. [DOI] [PubMed] [Google Scholar]
- 33.Williams R A D, Sadler S A. Electrophoresis of glucose-6-phosphate dehydrogenase, cell wall composition and the taxonomy of heterofermentative lactobacilli. J Gen Microbiol. 1971;65:351–358. doi: 10.1099/00221287-65-3-351. [DOI] [PubMed] [Google Scholar]