Abstract
Vanillate and syringate are converted into protocatechuate (PCA) and 3-O-methylgallate (3MGA), respectively, by O-demethylases in Sphingomonas paucimobilis SYK-6. PCA is further degraded via the PCA 4,5-cleavage pathway, while 3MGA is degraded through multiple pathways in which PCA 4,5-dioxygenase (LigAB), 3MGA 3,4-dioxygenase (DesZ), and an unidentified 3MGA O-demethylase and gallate dioxygenase are participants. For this study, we isolated a 4.7-kb SmaI fragment that conferred on Escherichia coli the activity required for the conversion of vanillate to PCA. The nucleotide sequence of this fragment revealed an open reading frame of 1,413 bp (ligM), the deduced amino acid sequence of which showed 49% identity with that of the tetrahydrofolate (H4folate)-dependent syringate O-demethylase gene (desA). The metF and ligH genes, which are thought to be involved in H4folate-mediated C1 metabolism, were located just downstream of ligM. The crude LigM enzyme expressed in E. coli converted vanillate and 3MGA to PCA and gallate, respectively, with similar specific activities, and only in the presence of H4folate; however, syringate was not a substrate for LigM. The disruption of ligM led to significant growth retardation on both vanillate and syringate, indicating that ligM is involved in the catabolism of these substrates. The ability of the ligM mutant to transform vanillate was markedly decreased, and this mutant completely lost the 3MGA O-demethylase activity. A ligM desA double mutant completely lost the ability to transform vanillate, thus indicating that desA also contributes to vanillate degradation. All of these results indicate that ligM encodes vanillate/3MGA O-demethylase and plays an important role in the O demethylation of vanillate and 3MGA, respectively.
Lignin is a major component of woody plants and is the most abundant aromatic compound in nature. Therefore, the utilization of lignin is expected, although few practical uses for lignin have been established to date. One potential practical procedure employed for the utilization of lignin is the conversion of lignin into useful chemical materials by microbial lignin degradation enzyme systems.
Sphingomonas paucimobilis SYK-6 is able to grow on various lignin-derived biaryls as the sole source of carbon and energy; therefore, the enzyme systems in this strain are expected to convert lignin-derived compounds into valuable intermediate metabolites. Among the previously determined intermediate metabolites of lignin-derived compounds, 2-pyrone-4,6-dicarboxylate (PDC) has been found to be useful as a starting material for the synthesis of biodegradable polyamides, polyurethanes, and polyesters (27).
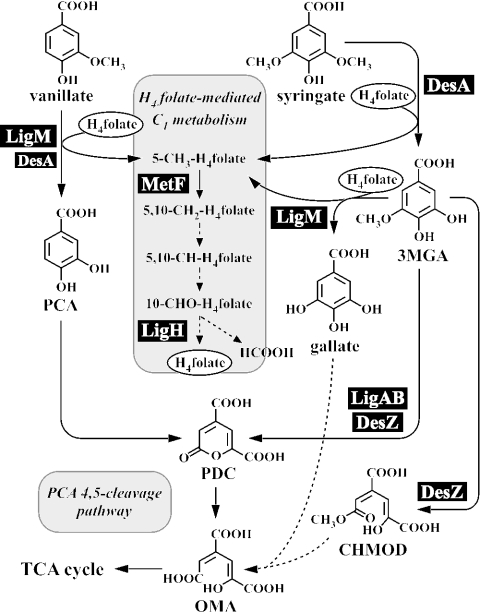
Vanillate and syringate have guaiacyl and syringyl moieties, respectively, which are well known as important chemicals derived from lignin. In SYK-6 cells, vanillate and syringate are converted to protocatechuate (PCA) and 3-O-methylgallate (3MGA), respectively, by tetrahydrofolate (H4folate)-dependent O-demethylases (Fig. 1) (20). PCA is further degraded through the PCA 4,5-cleavage pathway, and 3MGA is degraded via multiple pathways in which PCA 4,5-dioxygenase (LigAB), 3MGA 3,4-dioxygenase (DesZ), and an unidentified 3MGA O-demethylase are involved (11). Our investigations have characterized the structures and functions of all of the genes involved in the PCA 4,5-cleavage pathway (9, 10, 15, 17, 21) as well as those of the syringate O-demethylase gene (desA) (16). However, the details regarding each of the steps of O demethylation of vanillate and 3MGA remain largely unknown.
FIG. 1.
Proposed O demethylation system linked with H4folate-mediated C1 metabolism in S. paucimobilis SYK-6. The reactions indicated by dashed arrows have not been confirmed. Abbreviations: PCA, protocatechuate; 3MGA, 3-O-methylgallate; PDC, 2-pyrone-4,6-dicarboxylate; OMA, 4-oxalomesaconate; CHMOD, 4-carboxy-2-hydroxy-6-methoxy-6-oxohexa-2,4-dienoate; H4folate, tetrahydrofolate.
Two different aromatic demethylation systems have been documented to date. One of these enzyme systems is vanillate demethylase (VanA and VanB), which is a class IA oxygenase composed of an oxygenase and a reductase. This type of demethylase is involved in vanillate degradation by all of the vanillate-utilizing aerobic bacteria reported thus far, such as Pseudomonas and Acinetobacter (5, 6, 26, 31). The other enzyme system is the H4folate-dependent aromatic O-demethylase from anaerobic bacteria, including “Acetobacterium dehalogenans” (13), Acetobacterium woodii (2), and Moorella thermoacetica (18). In the vanillate degradation reaction of “A. dehalogenans,” a methyl transferase I catalyzes the transfer of the methyl moiety of vanillate to a corrinoid protein. A methyl transferase II catalyzes the subsequent transfer of the methyl group from the corrinoid protein to H4folate.
In the case of SYK-6, the O demethylation of vanillate and syringate is dependent on H4folate (20). The conversion of syringate to 3MGA is catalyzed by an H4folate-dependent O-demethylase, DesA. The deduced amino acid sequence of desA revealed approximately 26% identity with the aminomethyltransferase (GcvT) of Escherichia coli but showed no sequence similarity with the H4folate-dependent aromatic O-demethylase of anaerobic bacteria. DesA showed only weak activity with respect to the transformation of vanillate and 3MGA: the respective activities toward these compounds were only 3 and 0.4% of that of DesA toward syringate. A desA disruption mutant lost the ability to grow on syringate but retained the ability to grow on vanillate, indicating that an unidentified H4folate-dependent O-demethylase is involved in vanillate degradation. For this study, we isolated the vanillate O-demethylase gene and characterized its functions and roles in the metabolism of both vanillate and syringate.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used for this study are listed in Table 1. S. paucimobilis SYK-6 was grown in W minimal salt medium (22) containing a 10 mM concentration of vanillate or syringate or 0.2% yeast extract or in Luria-Bertani (LB) medium. The SYK-6 derivative strains DKLM and DKDA were grown in LB medium containing 50 mg of kanamycin (KAN)/liter, and DDAM was grown in LB containing 50 mg of KAN/liter and 300 mg of carbenicillin/liter.
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| S. paucimobilis | ||
| SYK-6 | Wild type; Nalr Smr | 12 |
| DKLM | SYK-6 derivative; ligM::kan; Nalr Smr Kmr | This study |
| DKDA | SYK-6 derivative; desA::kan; Nalr Smr Kmr | 16 |
| DDAM | DKDA derivative; ligM::bla desA::kan; Nalr Smr Kmr Cbr Nalr | This study |
| IAM12578 | 14 | |
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thiΔ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 32 |
| BL21 (DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3); T7 RNA polymerase gene under the control of the lacUV5 promoter | 29 |
| HB101 | supE44 hsd20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 3 |
| Plasmids | ||
| pUC18 and 19 | Cloning vectors; Apr | 32 |
| pBluescript II KS(+) and SK(+) | Cloning vectors; Apr | 28 |
| pET21a(+) | Expression vector, Apr T7 promoter | Novagen |
| pT7Blue | Cloning vector; Apr T7 promoter | Novagen |
| pVK100 | Broad-host-range cosmid vector; Kmr Tetr | 7 |
| pRK2013 | Kmr Tra+ Mob+ | 8 |
| pUC4K | Apr Kmr | 30 |
| pIK03 | KS(+) with a 1.3-kb EcoRV fragment carrying kan of pUC4K | 16 |
| pK19mobsacB | oriT sacB Kmr | 25 |
| pG-KJE7 | Carries groES, groEL, dnaK, dnaJ, and grpE genes under the control of derivative of araB promoter; pACYC184 replicon; Kmr | 19 |
| pVKS41-1 | pVK100 with approximately 20-kb SalI partially digested fragment of SYK-6 genomic DNA | This study |
| pBSM4.7 | KS(+) with a 4.7-kb SmaI fragment of pVKS41-1 | This study |
| pUB6.5 | pUC119 with a 6.5-kb BamHI fragment carrying metF and ligH | 20 |
| pTLM | pT7Blue with a 1.4-kb PCR amplified fragment carrying ligM | This study |
| pELM | pET21a(+) with a 1.4-kb NdeI-SacI fragment of pTLM | This study |
| pCSM4.7 | pUC19 with a 4.7-kb SmaI fragment of pVKS41-1 | This study |
| pCDLM | pCSM4.7 with an insertion of kan of pIK03 replacing 0.4-kb BglII fragment | This study |
| pDLM | pK19mobsacB with a 3.7-kb Eco47III fragment of pCDLM | This study |
| pCDALM | pCSM4.7 with an insertion of bla of pUC19 replacing 0.4-kb BglII fragment | This study |
| pDALM | pK19mobsacB with a 3.4-kb Eco47III fragment of pCDALM | This study |
Abbreviations: Nalr, Smr, Kmr, Apr, Cbr, and Tetr, resistance to nalidixic acid, streptomycin, kanamycin, ampicillin, carbenicillin, and tetracycline, respectively.
Chemicals.
Vanillate, syringate, vanillin, syringaldehyde, ferulic acid, and sinapinic acid were purchased from Tokyo Kasei Kogyo Co. (Tokyo, Japan). 3MGA was synthesized in a previous study (16). Tetrahydrofolate (H4folate) and 5-methyl-H4folate were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Cloning of the gene.
A partially SalI-digested gene library of SYK-6 constructed with pVK100 as the vector was introduced into S. paucimobilis IAM12578 by triparental mating (7). The resulting transconjugants were grown in LB medium containing 50 mg of KAN/liter for 3 days at 30°C. The cells were harvested, washed with a 0.9% NaCl solution, and incubated with 1 mM vanillate in 100 mM Tris-HCl buffer (pH 8.0) with shaking for 2 days at 30°C. After incubation, the cells were removed by centrifugation (15,000 × g for 1 min), and the supernatant was filtered. The amount of vanillate in the filtrates was analyzed by use of a high-pressure liquid chromatography (HPLC) system (HP1100 series; Agilent Technologies, Palo Alto, Calif.) equipped with a TSKgel ODS-80TM column (6 by 150 mm; Tosoh, Tokyo, Japan). For analysis of the conversion of substrates, the mobile phase was a mixture of water (79%), acetonitrile (20%), and acetic acid (1%), and the flow rate was 0.5 ml/min. Vanillate was detected at 260 nm, and the retention time of vanillate was 8.3 min.
Resting cell assay.
S. paucimobilis IAM12578 harboring pVKS41-1 was grown in LB medium containing 50 mg of KAN/liter. The cells were harvested by centrifugation (3,000 × g for 20 min), washed twice with a 0.9% NaCl solution, and suspended with the same solution. These cells were inoculated into 100 mM Tris-HCl buffer (pH 8.0) containing 1 mM vanillate or syringate to a turbidity at 600 nm of 0.8 and 3.0, respectively, and incubated with shaking for 24 h at 30°C. At selected times, 100-μl aliquots were centrifuged, filtered, and then analyzed by the method mentioned above.
DNA manipulations and nucleotide sequencing.
DNA manipulations were performed as described previously (1, 23). Nucleotide sequences were determined by the dideoxy termination method with a CEQ 2000XL genetic analysis system (Beckman Coulter, Inc., Fullerton, Calif.). A Sanger reaction (24) was carried out by use of a CEQ Dye Terminator cycle sequencing quick start kit (Beckman Coulter, Inc.). Sequence analysis was performed with the GeneWorks program (Intelligenetics, Inc., Mountain View, Calif.). Homology searches were done with Swiss-Prot/TrEMBL by use of the BLAST program and genomic BLAST. A pairwise alignment was performed with the EMBOSS alignment tool at the home page of the European Bioinformatics Institute (http://www.ebi.ac.uk/emboss/align).
Expression of ligM in E. coli and preparation of cell extracts.
The coding region of ligM was amplified by a PCR using Ex Taq polymerase (Takara Shuzo Co. Ltd., Kyoto, Japan), with pBSM4.7 as a template and with the ligMF primer (GGACTTAGCATATGTCGACACCTACC) and the ligMR primer (CAGAGCTCAGGCCGTGACG). The 1.4-kb PCR product was cloned into pT7Blue and sequenced. The 1.4-kb NdeI-SacI fragment of the resulting plasmid was inserted into pET21a(+) to generate pELM. E. coli BL21(DE3) harboring both pELM and pG-KJE7, which carries the dnaK-dnaJ-grpE and groEL-groES genes, was grown in LB medium containing 100 mg of ampicillin/liter and 25 mg of KAN/liter at 30°C. The expression of ligM was induced for 3 h by adding 1 mM isopropyl-β-d-thiogalactopyranoside when the turbidity of the culture at 600 nm reached 0.5, and at the same time, the expression of chaperones was induced by adding 10 mg of l-arabinose/ml. The cells were harvested by centrifugation at 15,000 × g for 1 min, suspended in 100 mM Tris-HCl buffer (pH 8.0), and washed once with the same buffer. Cells suspended in the buffer were sonicated, and the cell lysate was centrifuged at 15,000 × g for 5 min. The resulting supernatant was used as the cell extract. The protein concentration was determined by the method of Bradford (4). The expression of the gene was checked by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE). Gel staining was performed with Coomassie brilliant blue.
Enzyme assay.
The O-demethylase activities of the cell extracts toward vanillate, syringate, 3MGA, vanillin, syringaldehyde, sinapinic acid, and ferulic acid were determined by measuring the decrease in substrates by use of the HPLC system. The 1-ml assay mixture contained 100 mM Tris-HCl buffer (pH 8.0), 100 μM substrate, 1 mM H4folate, and the cell extract of E. coli BL21(DE3) harboring pELM and pG-KJE7 (300 μg of protein). Reactions were performed under anaerobic conditions at 30°C in an anaerobic box (Hirasawa Works Inc., Tokyo, Japan) that contained an atmosphere of 95% N2 and 5% H2 (<100 ppm of O2). A portion of the reaction mixture was taken at various sampling points and analyzed by HPLC. For analysis of the conversion of substrates, the mobile phase was a mixture of water (74%), acetonitrile (25%), and acetic acid (1%), and the flow rate was 1 ml/min. Compounds were detected at the following wavelengths: vanillate, 260 nm; syringate and 3MGA, 275 nm; vanillin, 294 nm; syringaldehyde and sinapinic acid, 324 nm; and ferulic acid, 308 nm. The retention times of vanillate, syringate, 3MGA, vanillin, syringaldehyde, sinapinic acid, and ferulic acid were 6.0, 5.9, 4.3, 9.1, 9.0, 8.5, and 9.0 min, respectively. One unit of enzyme activity was defined as the amount of enzyme that degraded 1 μmol of substrate per min at 30°C. Specific activities were expressed in units per milligram of protein. Each value reported is the average ± standard deviation of three independent experiments.
Identification of reaction products.
The 1-ml assay mixture contained 100 mM Tris-HCl buffer (pH 8.0), 500 μM vanillate or syringate, 1 mM H4folate, and the cell extract of E. coli BL21(DE3) harboring pELM and pG-KJE7 (1 mg of protein). The reaction was carried out at 30°C under anaerobic conditions and stopped by the addition of methanol (final concentration, 25%) after 10 min. The reaction mixture was acidified and extracted with ethyl acetate, and then the extract was trimethylsilylated (TMS) with the TMSI-H reagent (hexamethyldisilazane:trimethylchlorosilane:pyridine [2:1:10]; GL Science Inc., Tokyo, Japan) according to the procedure recommended by the manufacturer. The resultant TMS derivative was analyzed by gas chromatography-mass spectrometry (GC-MS) on a model 5971A apparatus with an Ultra-2 capillary column (50 m by 0.2 mm; Agilent Technologies). The column temperature was increased initially from 100 to 150°C and then from 150 to 300°C at rates of 20 and 3°C/min, respectively. The mobile phase was a helium gas, and the flow rate was 1.0 ml/min.
For identification of the one-carbon (C1) derivative of H4folate generated during O demethylation of vanillate catalyzed by LigM, electrospray ionization-MS (ESI-MS) was employed. The 1-ml assay mixture contained 100 mM Tris-HCl buffer (pH 8.0), 5 mM vanillate, 5 mM H4folate, and the cell extract of E. coli BL21(DE3) harboring pELM and pG-KJE7 (1 mg of protein). The reaction was carried out at 30°C under anaerobic conditions and stopped by the addition of methanol (final concentration, 25%) at 30 min. The reaction products were analyzed by ESI-MS (HP1100 series LC-MSD; Agilent Technologies). For this analysis, mass spectra were obtained by negative-mode ESI, with a needle voltage of −3.5 kV and a source temperature of 350°C. The mobile phase was a mixture of water (74%), acetonitrile (25%), and acetic acid (1%), and the flow rate was 1 ml/min.
Construction of insertion mutants of S. paucimobilis SYK-6.
The 4.7-kb SmaI fragment carrying ligM of pBSM4.7 was cloned into the SmaI site of pUC19 to generate pCSM4.7, and the 0.4-kb BglII fragment was deleted for ligM disruption. The 1.3-kb EcoRV fragment carrying the KAN resistance gene (kan) from pIK03 and the 1.0-kb BspHI fragment carrying the ampicillin resistance gene (bla) from pUC19 were inserted into the BglII site of the 4.3-kb SmaI fragment to construct pCDLM and pCDALM, respectively. pCDLM and pCDALM were digested with SmaI, and the inserts were cloned into pK19mobsacB to generate pDLM and pDALM, respectively.
pDLM and pDALM were introduced into SYK-6 and DKDA cells, respectively, by electroporation, and candidates for ligM mutants and ligM desA double mutants were screened by a method described in a previous study (17). Southern hybridization analysis was done to examine the disruption of ligM by use of a digoxigenin system (Roche Molecular Biochemicals, Mannheim, Germany). The total DNAs of candidates for ligM mutants and ligM desA double mutants were digested with SmaI. The 2.8-kb Eco47III fragment carrying ligM, the 1.3-kb EcoRV fragment carrying kan, and the 1.0-kb BspHI fragment carrying bla were labeled with the digoxigenin system and used as probes.
Preparation of cell extracts of SYK-6 and insertion mutants.
SYK-6 and its insertion mutants were grown in W medium containing 0.2% yeast extract. Cells grown on yeast extract until the turbidity of the culture at 600 nm reached 0.8 were harvested by centrifugation (3,000 × g for 20 min), washed twice with a 0.9% NaCl solution, and suspended with the same solution. To induce their O-demethylase activities, we inoculated these cells into W medium containing 10 mM vanillate or syringate to a turbidity at 600 nm of 0.5 and incubated them for 20 h. The vanillate, syringate, and 3MGA O-demethylase activities of the cell extracts (2 mg of protein/ml) were determined. Preparations of the cell extracts and the enzyme assay were essentially the same as those described above.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB186750.
RESULTS AND DISCUSSION
Isolation and nucleotide sequence of vanillate O-demethylase gene.
A gene library of SYK-6 constructed with the cosmid vector pVK100 was introduced into S. paucimobilis IAM12578, which is not able to degrade vanillate. One thousand transconjugants were screened for vanillate conversion activity by HPLC analysis, and a transconjugant that showed relevant activity was found. The cosmid pVKS41-1 was isolated from this positive clone and reintroduced into IAM12578. The resulting IAM12578 transformant harboring pVKS41-1 converted vanillate to PCA within 24 h but showed no activity toward syringate (data not shown). Because syringate O-demethylase (DesA) catalyzes the O demethylation of syringate to form 3MGA, the gene(s) included in pVKS41-1 appeared to differ from desA.
A subcloning experiment with pVKS41-1 revealed that pBSM4.7 containing a 4.7-kb SmaI fragment conferred the ability to convert vanillate on E. coli JM109. The nucleotide sequence of the 4.7-kb SmaI fragment was determined, and a 1,413-bp open reading frame (ORF) encoding a polypeptide with a molecular mass of 52,296 Da was found. The deduced amino acid sequence of this ORF showed 49 and 23% identity with DesA and H4folate-dependent aminomethyltransferase (GcvT), which is involved in glycine cleavage, respectively. These results suggested that this ORF encodes an H4folate-dependent vanillate O-demethylase, and this ORF was designated ligM. Note that the deduced amino acid sequence of ligM revealed 77, 56, and 51% identity with those of genes referred to as gcvT in the genomes of Novosphingobium aromaticivorans DSM12444, Agrobacterium tumefaciens C58, and Rubrobacter xylanophilus DSM9941, respectively. However, these sequences showed an identity of only approximately 20% with GcvT of E. coli.
Downstream of ligM, two ORFs, encoding 5,10-methylene-H4folate reductase (metF) and a putative 10-formyl-H4folate synthetase (ligH), both of which are involved in H4folate-mediated C1 metabolism, were identified. The tandem localization of ligM, metF, and ligH might suggest that these genes are transcribed in an operon. Further investigations will still be necessary in order to clarify the operon structure of these genes.
Upstream of ligM, the 729-bp orf2 and an incomplete ORF (orf1) were identified. The deduced amino acid sequences of orf1 and orf2 revealed 24 and 27% identity with those of formaldehyde dehydrogenase (AdhC) of E. coli K12 and alkyl salicylate esterase (SalE) of Acinetobacter sp. strain ADP1, respectively. On the basis of the functions of these genes, orf1 and orf2 seemed not to be involved in the O demethylation of vanillate.
LigM catalyzes the O demethylation of vanillate and 3MGA in the presence of H4folate.
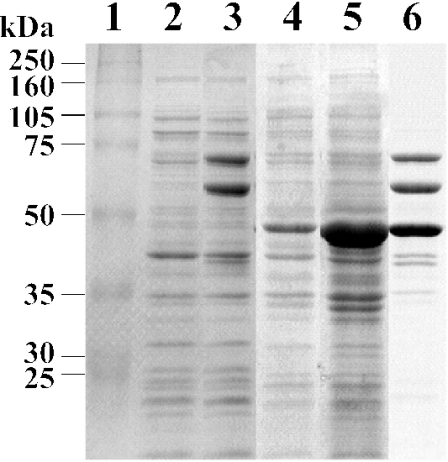
The 1.4-kb fragment carrying ligM was PCR amplified and cloned into pET21a(+) in order to generate pELM. The ligM gene was expressed in E. coli BL21(DE3) harboring pELM under the control of the T7 promoter. SDS-PAGE analysis indicated the production of a 47-kDa protein in an insoluble fraction, thus suggesting the formation of an inclusion body of LigM (Fig. 2, lanes 4 and 5). The size of this product was close to the value calculated from the deduced amino acid sequence of ligM (Mr, 52,296). To obtain the soluble form of LigM, we introduced pG-KJE7, carrying the dnaK-dnaJ-grpE and groEL-groES genes, into E. coli BL21(DE3) harboring pELM. The coexpression of these molecular chaperones had a marked effect on the production of LigM in its soluble form, presumably by facilitating correct folding (Fig. 2, lane 6).
FIG. 2.
SDS-PAGE analysis of LigM produced in E. coli BL21(DE3). Proteins (20 μg) were separated in an SDS-12% polyacrylamide gel and stained with Coomassie brilliant blue. Lanes: 1, molecular size markers; 2, crude extract of E. coli harboring pET21a(+); 3, crude extract of E. coli harboring pG-KJE7; 4, crude extract of E. coli harboring pELM; 5, SDS-solubilized cells of E. coli harboring pELM; 6, crude extract of E. coli harboring pELM and pG-KJE7. Molecular masses are given on the left.
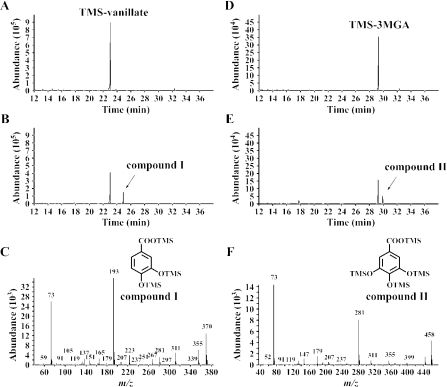
In order to examine whether ligM does indeed encode an H4folate-dependent vanillate O-demethylase, we measured the vanillate O-demethylase activity in the presence or absence of H4folate. The enzyme reactions were carried out under anaerobic conditions due to the lability of H4folate. GC-MS analysis showed that the crude LigM enzyme converted vanillate to PCA (compound I) only when H4folate was added to the reaction mixture (Fig. 3A to C). Interestingly, the crude LigM enzyme was able to convert 3MGA to gallate (compound II) (Fig. 3D to F), whereas syringate was not transformed. The transformation activities of the crude LigM enzyme with respect to vanillate and 3MGA, as measured in a 1-min reaction, were 125 ± 38 and 113 ± 35 mU/mg of protein, respectively. These results indicated that ligM encodes an H4folate-dependent vanillate/3MGA O-demethylase.
FIG. 3.
Identification of reaction products from vanillate and 3MGA catalyzed by LigM. The cell extract of E. coli BL21(DE3) harboring pELM and pG-KJE7 (1 mg of protein/ml) was incubated with 500 μM vanillate or 3MGA in the presence of 1 mM H4folate. (A and B) Gas chromatograms of the TMS derivative of the reaction product from vanillate at start and after 10 min of incubation, respectively. (D and E) Gas chromatograms of the TMS derivative of the reaction product from 3MGA at start and after 10 min of incubation, respectively. (C and F) Mass spectra of the TMS derivatives of compounds I and II, respectively.
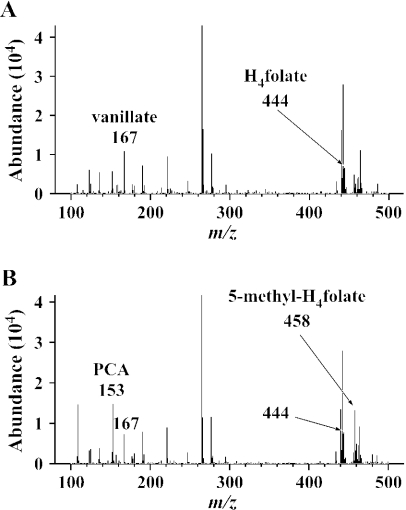
The reaction products from vanillate and H4folate catalyzed by the crude LigM enzyme were analyzed by negative-mode ESI-MS. The fragments at m/z 167 and 444, which correspond to the deprotonated molecular ions [(M − H)−] of vanillate and H4folate, respectively, were detected in the reaction mixture without LigM (Fig. 4A). In addition to these fragments, the generation of a fragment at m/z 153 (corresponding to [(M −H)−] of PCA) as well as the generation of a fragment at m/z 458 was observed in the reaction mixture incubated with LigM for 30 min (Fig. 4B). The product at m/z 458 was identified as 5-methyl-H4folate on the basis of a comparison of its molecular weight and the retention time of the authentic 5-methyl-H4folate analyzed by HPLC (16). This result strongly suggested that LigM catalyzes the transfer of the methyl moiety of vanillate to H4folate, thereby forming PCA and 5-methyl-H4folate. Moreover, during the O demethylation of 3MGA catalyzed by LigM, 5-methyl-H4folate appeared to be generated from H4folate.
FIG. 4.
Identification of C1-H4folate generated by O demethylation of vanillate catalyzed by LigM. The cell extract of E. coli BL21(DE3) harboring pELM and pG-KJE7 (1 mg of protein/ml) was incubated with 5 mM vanillate and H4folate. The results shown are negative-ion ESI-MS spectra of the reaction mixtures after 30 min of incubation without (A) or with (B) the enzyme.
Disruption of ligM in S. paucimobilis SYK-6.
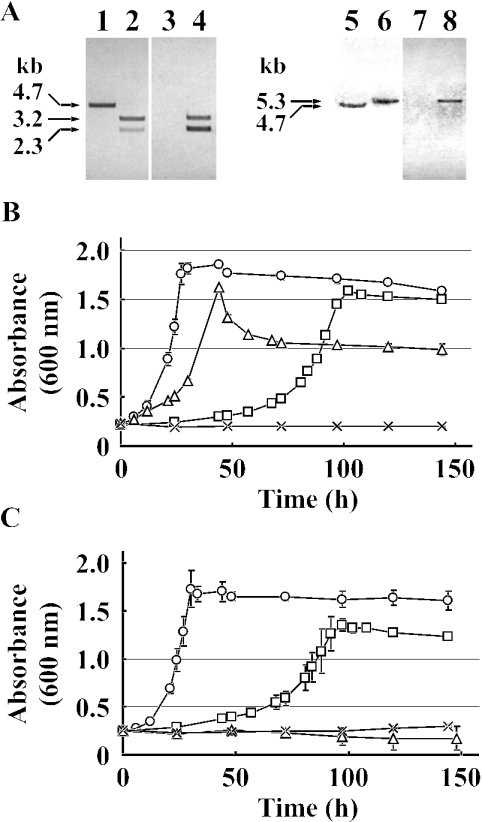
To examine the roles played by ligM in the catabolism of vanillate and syringate, we disrupted ligM in SYK-6 by a gene replacement technique using the ligM-disrupted plasmid pDLM, in which ligM was inactivated by insertion of the kan gene. Because DesA has only weak O demethylation activity with respect to vanillate (i.e., 3% of the specific activity toward syringate measured in a 1-min reaction), the ligM gene in a desA mutant (DKDA) constructed in a previous study (16) was disrupted with pDALM, in which ligM was inactivated by the insertion of bla. Southern hybridization analysis of the ligM mutants by the use of ligM, kan, and bla gene probes revealed that each ligM gene in SYK-6 and DKDA was inactivated by homologous recombination through the double crossover (Fig. 5A). The resultant ligM and ligM desA mutants were designated strains DKLM and DDAM, respectively.
FIG. 5.
Disruption of ligM and desA in SYK-6. (A) Southern hybridization analysis of insertion mutants. Lanes: 1, 3, 5, and 7, total DNAs of SYK-6 digested with SmaI; 2 and 4, total DNAs of DKLM digested with SmaI; 6 and 8, total DNAs of DDAM digested with SmaI. The 2.8-kb Eco47III fragment carrying ligM (lanes 1, 2, 5, and 6), the 1.3-kb EcoRV fragment carrying kan (lanes 3 and 4), and the 1.0-kb BspHI fragment carrying bla (lanes 7 and 8) were used as probes. (B and C) Growth on vanillate (B) and syringate (C) of SYK-6 (circles), DKLM (squares), DKDA (triangles), and DDAM (cross). These strains were grown in 10 ml of W medium containing 10 mM vanillate or syringate. Each value is the average ± standard deviation (error bars) of three independent experiments.
When DKLM was grown in W medium containing 10 mM vanillate, it took approximately 3.4 times longer for the DKLM cells than for the wild-type cells to enter stationary phase, but the DKLM cells retained the ability to grow on vanillate (Fig. 5B). On the other hand, DKDA cells showed weak growth retardation on vanillate, and the turbidity of the DKDA culture reached 61% of that of the wild-type culture. The disruption of both ligM and desA led to a growth defect on vanillate. These results indicated that ligM plays a crucial role in the growth of SYK-6 on vanillate and that desA is required for maximum growth on vanillate. DKDA and DDAM completely lost the ability to grow on syringate, in accord with the previous observation that the desA mutant no longer grew on syringate (Fig. 5C). Interestingly, a striking growth retardation of DKLM on syringate was observed (Fig. 5C). This result, considered together with the fact that LigM possesses activity toward 3MGA but not toward syringate, strongly suggested that ligM is indeed involved in 3MGA O demethylation. In a previous study, multiple pathways were proposed for 3MGA degradation, for which LigAB, DesZ, and 3MGA O-demethylase were identified as participants (11). The disruption of both ligB and desZ in SYK-6 resulted in the loss of dioxygenase-dependent 3MGA transformation activity, but the growth retardation of this mutant on syringate was not as significant as that of DKLM. The marked growth retardation of the ligM mutant on syringate suggests the predominance of O demethylation catalyzed by LigM in the multiple 3MGA degradation pathways.
O demethylation activities of ligM mutants toward vanillate, syringate, and 3MGA.
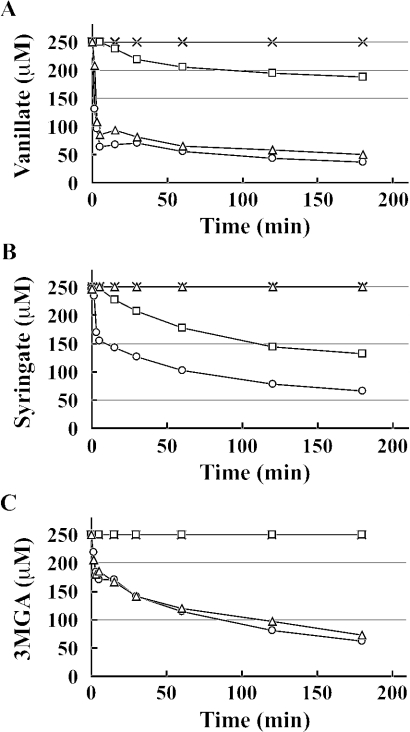
The O demethylation of vanillate, syringate, and 3MGA in cell extracts of the insertion mutants was examined in the presence of H4folate under anaerobic conditions, and HPLC was used to measure decreases in the amounts of substrate. In a previous study, we demonstrated that syringate O-demethylase activity increased approximately 10-fold when SYK-6 cells were incubated with syringate. The insertion mutants and SYK-6 were therefore first grown in W medium containing 0.2% yeast extract, and cells grown in this manner were then incubated with 10 mM vanillate (Fig. 6A) or syringate (Fig. 6B and C) for 20 h to induce the enzymes. The cell extract of SYK-6 incubated with vanillate showed approximately 20 times higher vanillate O-demethylase activity (27 mU/mg) than the corresponding extract incubated without vanillate. The vanillate conversion rate of the cell extract of DKLM incubated with vanillate was strikingly reduced, and the extract of DDAM cells no longer showed any such activity (Fig. 6A). These results indicated that only ligM and desA are involved in vanillate O demethylation, and furthermore, that ligM plays a major role in the O demethylation of vanillate. It was notable that the extremely low level of vanillate O-demethylase activity of DesA contributed to the maximum growth of SYK-6 cells on vanillate. The cell extract of DKLM incubated with syringate completely lost the ability to transform 3MGA under anaerobic conditions, whereby the ring cleavage of 3MGA by both LigAB and DesZ was inhibited; these results indicate that ligM encodes the essential O-demethylase for 3MGA (Fig. 6C). Unexpectedly, the ability of the cell extract of DKLM to transform syringate decreased, in contradiction with findings that syringate is not a substrate for LigM (Fig. 6B). The low level of relevant activity in DKLM cells may have been caused by a lack of desA induction. Thus, 3MGA O-demethylase activity might be necessary to produce an inducer of desA transcription. The finding that ligM disruption led to the observed significant decrease in DesA activity was indicative of the crucial role played by ligM in syringate catabolism.
FIG. 6.
O-demethylase activities of strains DKLM, DKDA, and DDAM. The time courses of degradation of vanillate (A), syringate (B), and 3MGA (C) by cell extracts (2 mg of protein/ml) of SYK-6 (circles), DKLM (squares), DKDA (triangles), and DDAM (cross) incubated with 10 mM vanillate (A) or syringate (B and C) are shown. Each cell extract was incubated with 250 μM vanillate, syringate, or 3MGA in the presence of 1 mM H4folate under anaerobic conditions. HPLC was used to monitor the time course of substrate removal.
Finally, it should be emphasized that ligM, metF, and ligH are tandemly localized in the SYK-6 genome. In a previous study, we demonstrated that PCA is not a growth substrate for SYK-6 cells, but we also showed that this strain is able to grow well on PCA in the presence of methionine. This previous finding suggested that the 5-methyl-H4folate generated by the O demethylation of vanillate is utilized for methionine biosynthesis. Therefore, the O demethylation of vanillate, syringate, and 3MGA catalyzed by LigM or DesA appears to be important for SYK-6 cells, not only as a degradation step for these compounds, but also to supply the 5-methyl-H4folate required for methionine biosynthesis. The resulting 5-methyl-H4folate might be metabolized through a C1 metabolic pathway in which metF and ligH are both participants (Fig. 1). For a clarification of the correlation between the O demethylation of lignin-derived compounds and C1 metabolism in SYK-6 cells, further investigations will be necessary, including analyses of the various functions of C1 metabolic genes and studies of the regulation of each of the O-demethylase and C1 metabolic genes.
Acknowledgments
We thank K. Nishihara and H. Yanagi for providing pG-KJE7.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Berman, M. H., and A. C. Frazer. 1992. Importance of tetrahydrofolate and ATP in the anaerobic O-demethylation reaction for phenylmethylethers. Appl. Environ. Microbiol. 58:925-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brunel, F., and J. Davison. 1988. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J. Bacteriol. 170:4924-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Civolani, C., P. Barghini, A. R. Roncetti, M. Ruzzi, and A. Schiesser. 2000. Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13. Appl. Environ. Microbiol. 66:2311-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara, H., E. Masai, Y. Katayama, and M. Fukuda. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6950-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara, H., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2003. Characterization of the 4-carboxy-4-hydroxy-2-oxoadipate aldolase gene and operon structure of the protocatechuate 4,5-cleavage pathway genes in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 185:41-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasai, D., E. Masai, K. Miyauchi, Y. Katayama, and M. Fukuda. 2004. Characterization of the 3-O-methylgallate dioxygenase gene and evidence of multiple 3-O-methylgallate catabolic pathways in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 186:4951-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katayama, Y., S. Nishikawa, M. Nakamura, K. Yano, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1987. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi 33:77-79. [Google Scholar]
- 13.Kaufmann, F., G. Wohlfarth, and G. Diekert. 1998. O-Demethylase from Acetobacterium dehalogenans substrate specificity and function of the participating proteins. Eur. J. Biochem. 253:706-711. [DOI] [PubMed] [Google Scholar]
- 14.Masai, E., Y. Katayama, S. Kawai, S. Nishikawa, M. Yamasaki, and N. Morohoshi. 1991. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J. Bacteriol. 173:7950-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masai, E., K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 2000. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6651-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masai, E., M. Sasaki, Y. Minakawa, T. Abe, T. Sonoki, K. Miyauchi, Y. Katayama, and M. Fukuda. 2004. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J. Bacteriol. 186:2757-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naidu, D., and S. W. Ragsdale. 2001. Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. J. Bacteriol. 183:3276-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishihara, K., M. Kanemori, M. Kitagawa, H. Yanagi, and T. Yura. 1998. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 64:1694-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishikawa, S., T. Sonoki, T. Kasahara, T. Obi, S. Kubota, S. Kawai, N. Morohoshi, and Y. Katayama. 1998. Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl. Environ. Microbiol. 64:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noda, Y., S. Nishikawa, K. Shiozuka, H. Kadokura, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 26.Segura, A., P. V. Bunz, D. A. D'Argenio, and L. N. Ornston. 1999. Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J. Bacteriol. 181:3494-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shigehara, K., C. Seki, N. Okamura, M. Matsumoto, Y. Katayama, and S. Nishikawa. 1998. Synthesis of polymers with 2H-pyran-2-one-4,6-dicarboxylate (PDC). Nuclei Polymer Preprints (Japan) 47:415. [Google Scholar]
- 28.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. λ ZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venturi, V., F. Zennaro, G. Degrassi, B. C. Okeke, and C. V. Bruschi. 1998. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 144:965-973. [DOI] [PubMed] [Google Scholar]
- 32.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]