Abstract
Rok is a repressor of the transcriptional activator ComK and is therefore an important regulator of competence in Bacillus subtilis (T. T. Hoa, P. Tortosa, M. Albano, and D. Dubnau, Mol. Microbiol. 43:15-26, 2002). To address the wider role of Rok in the physiology of B. subtilis, we have used a combination of transcriptional profiling, gel shift experiments, and the analysis of lacZ fusions. We demonstrate that Rok is a repressor of a family of genes that specify membrane-localized and secreted proteins, including a number of genes that encode products with antibiotic activity. We present evidence for the recent introduction of rok into the B. subtilis-Bacillus licheniformis-Bacilllus amyloliquefaciens group by horizontal transmission.
ComK is the major transcription factor driving the development of competence for transformation in Bacillus subtilis (37). rok encodes a direct repressor of comK transcription (18) and thereby plays an important, but incompletely understood, role in the complex regulation that takes place at the promoter of comK. Five proteins are known to bind to PcomK (reviewed in reference 9): ComK itself, DegU, Rok, AbrB, and CodY, the last three of which exert negative effects.
The use of a rok-lacZ fusion construct revealed no major change in rok transcription during growth in the population as a whole (18). Given this, it is surprising that the regulation of rok transcription is complex; rok is repressed by Rok itself, by ComK, and by the transition state regulators SinR and AbrB, although only ComK and Rok have been shown to bind directly to Prok (18). As cultures approach the end of exponential growth, the concentrations of active AbrB and SinR decrease (reviewed in references 31 and 32), leading to the expectation that rok transcription would increase. However, since ComK acts negatively on Prok, the increased synthesis of ComK in competent cells after the cessation of exponential growth would tend to place a limit on increased rok transcription. In addition, negative autoregulation at Prok would be expected to maintain a constant time-averaged level of rok expression. Transient changes in the concentration of Rok may play a role in the timing of competence expression, in the selection of which cells will develop competence, and in limiting the final level of comK expression in the competent subpopulation. Although transient changes have not been detected, it may be that our experiments lacked sufficient time resolution to detect these fluctuations. A major unanswered question concerns the possibility that the activity of Rok is somehow regulated, perhaps responding to the presence of an unknown corepressor.
The origin of rok is also interesting, since it has orthologs among sequenced organisms only in Bacillus licheniformis (68% identical [http://63.198.8.200/]) and Bacillus amyloliquefaciens (83% identical [R. Borriss, personal communication]). No orthologs are detected using either BLAST or Psi-BLAST (http://www.ncbi.nlm.nih.gov/BLAST/), even in other bacilli, such as B. anthracis, B. halodurans, and B. cereus (reference 18 and unpublished data). Both rok and its convergently transcribed immediate downstream neighbor yknT are missing from these related species.
To extend our understanding of the role of Rok, we identified additional genes regulated by this protein and asked whether Rok was capable of acting positively or only as a repressor. By transcriptional profiling and the use of lacZ fusions, we have identified at least seven gene clusters that are negatively regulated by rok, including several involved in the production of bacteriocin-like antibiotics, and we have shown that this repression is caused by direct binding of Rok to target DNA sequences.
MATERIALS AND METHODS
Strains and general procedures.
The B. subtilis strains used in this study are derived from strain 168 and most are isogenic with BD630 (his leu met). Selective and growth media, the growth of strains to competence, and transformation were described or referenced by Albano et al. (1) and by Tortosa et al. (35). Strains are listed in Table 1. Molecular biological methods were essentially as described by Sambrook et al. (30). β-Galactosidase assays were carried out as described previously (35) by using cultures grown in competence medium (1), which is a minimal salts medium supplemented with glucose, casein hydrolysate, and yeast extract.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BD630 | his leu met | Laboratory stock |
| BD2955 | his leu met rok::miniTn10 (Spcr) | 18 |
| BD3196 | his leu met rok (Kanr) | This work |
| ORB3147 | trpC2 pheA1 alb::pMUE1 (Ermr) | 40 |
| ORB3162 | trpC2 pheA1 SPβc2del2::Tn917::pTK-sboΔEB (Ermr Cmr) | 40 |
| BD2121 | his leu met comK (Kanr) | 38 |
| BD3128 | his leu met comK rok::miniTn10 (Kanr Spcr) | This work |
| BD3702a | his leu met albA-lacZ | This work |
| BD3703a | his leu met albA-lacZ rok::miniTn10 (Kanr Spcr) | This work |
| BD3704b | his leu met sboA-lacZ (Ermr Cmr) | This work |
| BD3705b | his leu met sboA-lacZ rok::miniTn10 (Ermr Cmr Spcr) | This work |
| BD3761 | his leu met yxaJ-lacZ | This work |
| BD3762 | his leu met yxaJ-lacZ rok::miniTn10 (Ermr Spcr) | This work |
| BD3763 | his leu met yybM-lacZ (Ermr) | This work |
| BD3764 | his leu met yybM-lacZ rok (Ermr Kanr) | This work |
| BD3767 | his leu met sunT-lacZ | This work |
| BD3768 | his leu met sunT-lacZ rok::miniTn10 (Ermr Spcr) | This work |
| BD3814 | his leu met sdpA-lacZ | This work |
| BD3815 | his leu met sdpA-lacZ rok (Ermr Kanr) | This work |
| BD3821 | his leu met yjcN-lacZ | This work |
| BD3822 | his leu met yjcN-lacZ rok (Ermr Kanr) | This work |
| BD3765 | his leu met yydl-lacZ | This work |
| BD3766 | his leu met yydl-lacZ rok (Ermr Kanr) | This work |
The alb-lacZ construct was obtained from ORB3147.
The sboA-lacZ construct was obtained from ORB3162.
Construction of lacZ fusions.
To generate fusion constructs of lacZ to sunT, yxaJ, yybM, yydI, yjcN, and sdpA, internal fragments of these genes were isolated by PCR by using the primers listed in Table 2. The fragments were cleaved with HindIII and BamHI and inserted into pMUTIN2 (36), which was also cleaved with these two enzymes. The resulting recombinant plasmids were used to transform BD630 and BD2955 with selection for erythromycin resistance and were thereby integrated in the chromosomes of these strains by single reciprocal recombination (Table 1).
TABLE 2.
Primers used in this study
| Primer | Target fragment | Sequence | Reference |
|---|---|---|---|
| psboA1 | PsboA | 5′-CCTCATAAAAAGCATTTCCT-3′ | This study |
| psboA2 | PsboA | 5′-AATTGAATCCTCCCTTTTTT-3′ | This study |
| psunA1 | PsunA | 5′-ATCCATAATAGTCAATTTTA-3′ | This study |
| psunA2 | PsunA | 5′-TTGTAAAACCTCCCCATTTG-3′ | This study |
| pyjcN-F | PyjcN | 5′-CTTTATTATGTAGTGGCTGCA-3′ | This study |
| pyjcN-R | PyjcN | 5′-TCATTTTTTCCTCCGATACCTT-3′ | This study |
| pK-F | PcomK | 5′-AATCTATCGACATATCCTGCAA-3′ | This study |
| pKplusR-R | PcomK | 5′-TTATACTAATAATCTATCATCTGTTT-3′ | This study |
| rokint1 | rok CSa | 5′-CGGGATCCCGCTTGCGGTTAGAA-3′ | This study |
| rokint2 | rok CSa | 5′-GGAATTCCGAAGCTCCGAGCTCT-3′ | This study |
| psdpF | PsdpA | 5′-CCCAAGCTTAGTTCCCAAATTCAATTCTG-3′ | This study |
| psdpR | PsdpA | 5′-GGAATTCTAATAGGAAACATATAGTCATTAC-3′ | This study |
| pyydF-F | PyydF | 5′-ATATTGATGTTTTACTTTTCAT-3′ | This study |
| pyydF-R | PyydF | 5′-CATATTATCCCTCCTCCT-3′ | This study |
| pyybN-FL | PyybN | 5′-CATGCAATTTAGTGATCCAA-3′ | This study |
| pyybN-R | PyybN | 5′-CATAATTTTACAATCCTTTCA-3′ | This study |
| pKOH-R | PcomK | 5′-TTCTGACTCATATTATGGCCT-3′ | This study |
| pykuWpa | Prok | 5′-CGGGATCCGCTTCTCTTTCATTAAACAT-3′ | 18 |
| prokps | Prok | 5′-CGGAATTCGATGTTTTTCCTCAATTTTAG-3′ | 18 |
| pcomG1 | PcomG | 5′-CCGGAATTCATGGTGACCATGTCTGCT-3′ | 33 |
| pcomG2 | PcomG | 5′-CGCGGATCCCTCTCCTTTCAACGC-3′ | 33 |
| pskfA-F | PskfA | 5′-CAAGCCGTACGAACGGACTG-3′ | This study |
| pskfA-R | PskfA | 5′-TGGATACGACCTCTTTGCCC-3′ | This study |
| pyydH-F | PyydH | 5′-GAAAAGTTAAATTCCAATTTGC-3′ | This study |
| pyydH-R | PyydH | 5′-TCATATTTTCCATCTTCATACTT-3′ | This study |
| pbhlA-F | PbhlA | 5′-GGCACAGTTAAGCTTGG-3′ | This study |
| pbhlA-R | PbhlA | 5′-GTTCACCACCTTAGTACC-3′ | This study |
| pykuJ-F | PykuJ | 5′-TAATTATGTGCATGACATTCAAAAAAAG-3′ | This study |
| pykuJ-R | PykuJ | 5′-TGCAGCCGTGTGATGATACC-3′ | This study |
| pyxaJ-F | PyxaJ | 5′-TACATGCAATATGGTATGGTG-3′ | This study |
| pyxaJ-R | PyxaJ | 5′-GTCATGATCGCCACGCTATT-3′ | This study |
| sunT-F | sunTb | 5′-GCCAAGCTTTGGGGATAAGGAAGGCT-3′ | This study |
| sunT-R | sunTb | 5′-CGGGATCCACCCACAACGAACAAGGA-3′ | This study |
| yxaJ-F | yxaJb | 5′-GCCAAGCTTAGCTGGCATGTGGTCA-3′ | This study |
| yxaJ-R | yxab | 5′-CGGGATCCGCCACAAACCAAATGACG-3′ | This study |
| yybM-F | yybMb | 5′-GCCAAGCTTTCTGGCACTTCCGT-3′ | This study |
| yybM-R | yybMb | 5′-CGGGATCCAAGGCTGTTATCAGGGA-3′ | This study |
| yydl-F | yydlb | 5′-GCCAAGCTTAGCAAAAGAATCGGCAG-3′ | This study |
| yydl-R | yydlb | 5′-CGGGATCCATCTTCTCGGGGTTCTC-3′ | This study |
| yjcN-F | yjcNb | 5′-CCCAAGCTTCTCCTGTTGTCTTTACAGCTTCTTCGG-3′ | This study |
| yjcN-R | yjcNb | 5′-CGCGGATCCCCCTGTGCCACAACTTACTTCGTATTC-3′ | This study |
| sdpA-F | sdpAb | 5′-CCCAAGCTTGAGGTTGAGCAGGACTACTATC-3′ | This study |
| sdpA-R | sdpAb | 5′-CGCGGATCCCTATTGCCTGAACTCTTCCTTC-3′ | This study |
Coding sequence.
The target sequences from this genes was inserted into pMUTIN2 to create a fusion to lacZ.
DNA microarrays and transcriptional profiling.
We purchased a B. subtilis oligonucleotide library, manufactured by Sigma Genosys and designed by Compugen. The library consisted of a collection of 4,128 oligonucleotides (65-mers) representing 4,106 B. subtilis genes, 10 control oligonucleotides (from Escherichia coli and Brome mosaic virus), and 12 random oligonucleotides. The oligonucleotides were designed to represent the B. subtilis genes as found in the genome data release R16.1 (26 April 2001) at the SubtiList website (http://genolist.pasteur.fr//SubtiList/). A single oligonucleotide was made for each gene. The oligonucleotides were spotted onto poly-l-lysine-coated glass slides at a concentration of 25 μM, and approximately 0.7 nl, containing ∼17.5 fmol, was delivered per spot. Each gene was represented once per slide. The oligonucleotides were spotted, and the hybridized arrays were scanned in the Center for Applied Genomics facility maintained at the Public Health Research Institute.
BD2121 (comK rok+) and BD3128 (comK rok::miniTn10) were grown in competence medium (1), and samples were harvested for RNA isolation during exponential growth (corresponding to a reading in a Klett-Summerson colorimeter of about 70) and at T2, defined as 2 h after the cultures departed from exponential growth. Samples were chilled rapidly by diluting them into an ice slurry of 0.3 M NaCl and 0.03 M Na citrate and collected by centrifugation at 4°C. Total cell RNA was isolated as described previously (5), from at least four independently grown cultures of the two strains. The RNA samples were analyzed on agarose gels to assess quality before being used in the preparation of cDNA.
cDNA was prepared from the RNA samples as previously described (5), by reverse transcription of 25 μg of total RNA to incorporate aminoallyl-dUTP into first-strand cDNA. The cDNA products were subsequently labeled by direct coupling either to Alexa Fluor 555 or Alexa Fluor 647 amine-reactive dyes (Molecular Probes), as described by the manufacturer. Labeled probes were mixed and purified using Qiaquick PCR spin columns; reciprocal labeling (dye swapping) of cDNA was done for all experiments.
Probes were purified for hybridization to microarrays essentially as described elsewhere (5) without filtration and were then boiled for 2 min, cooled to room temperature, and applied to the microarrays under coverglasses. The microarrays were placed in HybChambers (GeneMachines, San Carlos, Calif.) and incubated at 68°C overnight. The arrays were washed and dried as described previously (5). The slides were scanned, and images were produced with a GenePix 4000A microarray scanner (Axon Instruments).
Raw data files produced by GenePix (GPR files) were exported to Excel and prepared for analysis with CyberT software (24), as suggested by the program's authors (http://visitor.ics.uci.edu/genex/cybert/). The data were normalized by global scaling as described by Baldi and Hatfield (4). Each intensity value for each gene for a given wavelength was divided by the sum of the intensities for all genes at that wavelength. The resulting values were used to calculate red/green ratios, and the ratios were natural log transformed, as required by the software. A Bayesian paired expression value estimate was calculated for each gene according to the methods described in the CyberT website. Parameters for Bayesian standard deviation estimation were as suggested by the authors (sliding window size, 101; confidence value, 10).
Expression and purification of Rok-His6.
An overnight culture of E. coli strain ED428 (18) was diluted 1:50 into fresh TY medium supplemented with 100 μg of ampicillin/ml and 25 μg of kanamycin/ml and grown at 37°C with vigorous shaking. At an A600 of 0.6, expression of the recombinant protein was induced with 1 mM isopropyl thio-β-d-galactoside. Growth was continued for 4 h, and cells from 1 liter of culture were collected by centrifugation (10 min, 8,000 rpm, 4°C) in an Avanti J-20 XP centrifuge (Beckmann Coulter). The pellet was washed with 50 ml of buffer A (50 mM NaHPO4, 300 mM NaCl, 10 mM imidazole, 3.5% glycerol, 1 mM β-mercaptoethanol; pH 8.0) and stored at −80°C for future use. The pellet was resuspended in 5 ml of buffer A and supplemented with Complete Mini protease inhibitor (Roche), and cells were disrupted by sonication. Cellular debris was removed by centrifugation (30 min, 25,000 rpm, 4°C), and the supernatant fraction was loaded onto a Superflow Ni-nitrilotriacetic acid resin column (QIAGEN) equilibrated with buffer A. The column was washed with buffer B (identical to buffer A, but with 20 mM imidazole). The protein was eluted from the column by using a linear gradient of 20 to 500 mM imidazole, and 0.5-ml fractions were collected. Fractions were checked for protein content and purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and for DNA content by ethidium bromide staining of agarose gels. Fractions containing the protein were pooled and loaded onto a MonoQ column (Amersham Pharmacia) and equilibrated with buffer C (20 mM Tris, 1 mM EDTA; pH 8.0) to remove DNA. Elution was carried out using a linear gradient of 0 to 2 M KCl. Fractions containing protein, but negligible amounts of DNA, were used in subsequent experiments. Protein was quantified using the RC/DC protein determination kit (Bio-Rad).
Gel retardation.
Gel retardations were performed essentially as described previously (14). Fragments for use as probes were amplified with Expand (Roche) or Pwo (Roche) DNA polymerase using the primers from Table 2. The PCR products were end labeled with T4 polynucleotide kinase and [γ-32P]ATP. Protein and probe were mixed on ice and subsequently incubated for 20 min at 37°C. Samples were loaded onto a 6% nondenaturing polyacrylamide gel prepared with 1× TAE (40 mM Tris acetate [pH 8.0], 2 mM EDTA) and run in a 0.5-to-2.0× gradient of TAE at 100 V for 45 to 60 min in a MiniProtean electrophoresis system (Bio-Rad). Gels were dried in a vacuum dryer (model 583; Bio-Rad), and signals were recorded using phosphoscreens and a Cyclone PhosphorImager (Packard).
RESULTS
Transcriptional profiling.
In order to further define the set of genes regulated by rok, we used transcriptional profiling by microarray, with a chip containing synthetic oligonucleotides representing all of the annotated open reading frames in the sequenced B. subtilis genome. RNA was isolated from a strain with an inactivated rok gene and from an isogenic strain with intact rok. Since Rok is known to be a repressor of comK, which in turn regulates many genes (5, 13, 27), both strains used as sources of RNA for this experiment also carried a comK null mutation, to simplify analysis of the results. RNA was isolated at two stages during growth in competence medium (1): from exponentially growing cultures, and 2 h after the departure from exponential growth (defined as T2). Results of these transcriptional profiling experiments can be viewed at the website http://www.phri.org/research/res_pidubnau.asp and are summarized in Table 3 and in Fig. 1. The genes selected for inclusion in Table 3 satisfied three criteria: at least a 1.8-fold difference between the comK rok and comK RNA samples, Bayesian P values less than 0.01 calculated using the program Cyber T (24), and at least six successful independent measurements. If either the T2 or exponential samples satisfied these criteria, the gene was listed in Table 3. Twenty of 39 genes listed in Table 3 satisfied these criteria at both growth stages. Four satisfied the criteria only during exponential growth, while 15 genes satisfied the criteria only at T2, since they exhibited less than a 1.8-fold difference during exponential growth. This bias suggests that rok may be more active in stationary phase or, more likely, that the genes it controls may tend to be those that are expressed after the cessation of exponential growth, even in the absence of Rok. Two additional genes (sdpA and ykuK), which fall short of the criteria, are also listed in Table 3 because they are adjacent to sdpB and sdpC on the one hand and to ykuJ and ykzF on the other. Although the listed genes were candidates for negative regulation by rok, no positively regulated genes were detected.
TABLE 3.
Transcriptional profiling of rok mutantsa
| Gene | Ratio (T2)b | P value (T2)c | Ratio (Exp.)b | P value (Exp.)c |
|---|---|---|---|---|
| albA | 3.5 | 5.6 × 10−5 | 3.6 | 1.2 × 10−5 |
| albB | 4.5 | 1.2 × 10−6 | 3.6 | 9.1 × 10−6 |
| albC | 4.2 | 1.1 × 10−6 | 3.7 | 5.5 × 10−6 |
| albD | 4.6 | 1.8 × 10−6 | 4.3 | 8.0 × 10−7 |
| albE | 3.8 | 4.4 × 10−5 | 4.1 | 6.6 × 10−6 |
| albF | 4.3 | 4.4 × 10−6 | 4.4 | 4.8 × 10−6 |
| albG | 2.8 | 6.2 × 10−5 | 2.6 | 3.3 × 10−4 |
| argG | 2.1 | 7.7 × 10−4 | 1.2 | 0.2 |
| bdbA | 2.0 | 7.1 × 10−3 | 4.8 | 3.4 × 10−6 |
| bdbB | 2.1 | 1.6 × 10−3 | 1.8 | 5.0 × 10−3 |
| bhlA | 6.9 | 1.1 × 10−7 | 5.2 | 8.4 × 10−7 |
| fur | 1.9 | 9.7 × 10−3 | 1.2 | 0.1 |
| hom | 2.4 | 4.8 × 10−3 | 1.2 | 0.2 |
| rapF | 2.0 | 2.9 × 10−3 | 1.2 | 0.2 |
| rpsL | 2.3 | 4.6 × 10−3 | 1.3 | 0.3 |
| sboA | 13.3 | 1.4 × 10−9 | 5.2 | 3.3 × 10−5 |
| sboX | 12.2 | 3.3 × 10−8 | 7.2 | 2.8 × 10−6 |
| sdpB | 1.9 | 1.8 × 10−3 | 1.2 | 0.3 |
| sdpC | 1.8 | 3.9 × 10−3 | 1.1 | 0.8 |
| serA | 2.2 | 6.5 × 10−3 | 1.3 | 0.1 |
| sunA | 9.7 | 2.1 × 10−8 | 5.8 | 1.5 × 10−6 |
| sunT | 3.1 | 1.0 × 10−4 | 8.5 | 1.4 × 10−8 |
| thrC | 2.4 | 6.7 × 10−3 | 1.2 | 0.2 |
| yjcN | 2.3 | 1.4 × 10−4 | 1.4 | 1.14 × 10−2 |
| ykuJ | 2.0 | 2.9 × 10−3 | 1.3 | 0.1 |
| ykzF | 2.0 | 4.1 × 10−3 | 1.3 | 3.8 × 10−2 |
| ykzG | 2.0 | 0.3 × 10−3 | 1.3 | 0.2 |
| ylbN | 1.9 | 8.1 × 10−3 | 1.1 | 0.7 |
| yolJ | 2.7 | 4.6 × 10−4 | 5.3 | 1.5 × 10−6 |
| ypiF | 2.0 | 7.0 × 10−3 | 1.9 | 3.9 × 10−3 |
| yxaJ | 1.7 | 9.3 × 10−3 | 3.3 | 2.5 × 10−6 |
| yxaL | 1.4 | 8.9 × 10−2 | 3.5 | 1.9 × 10−6 |
| yxbC | 1.7 | 4.0 × 10−2 | 2.0 | 3.4 × 10−3 |
| yybK | 2.3 | 1.9 × 10−4 | 2.3 | 6.4 × 10−4 |
| yybL | 2.1 | 5.5 × 10−3 | 1.9 | 2.8 × 10−3 |
| yybM | 2.9 | 1.4 × 10−4 | 2.5 | 2.6 × 10−4 |
| yydH | 1.7 | 3.4 × 10−2 | 1.9 | 2.6 × 10−3 |
| yydl | 2.0 | 1.8 × 10−3 | 1.4 | 1.7 × 10−2 |
| yydJ | 3.1 | 1.9 × 10−5 | 2.6 | 2.8 × 10−5 |
| sdpAd | 1.4 | 2.4 × 10−2 | 1.0 | 0.7 |
| ykuKd | 1.4 | 0.1 | 1.2 | 0.2 |
RNA was isolated from a comK strain and from an isogenic comK rok strain growing in competence medium at the indicated times. cDNA was prepared and hybridized to microarrays prepared from oligonucleotides designed from the genome sequence of B. subtilis.
The ratios are normalized average values from the rok RNA divided by values from the rok+ RNA. T2 refers to a time 2 h after the departure from exponential growth. Exp. refers to a time during exponential growth. All of the T2 values were derived from 8 successful measurements and all of the exponential sample values from 10, except that there were 9 measurements in the case of ykuJ.
The P values are the Bayesian P values described in reference 24.
This gene was not initially called by the criteria described in the text, but it is included because it is adjacent to genes that were called and because its values fall just below these criteria.
FIG. 1.
Genetic maps of rok-regulated gene clusters. Genes are indicated with arrows, indicating the direction of transcription. Black arrows indicate genes that meet the criteria for rok regulation, as described in the text; grey arrows indicate genes that nearly met these criteria but are classified as rok regulated, based on their proximity to regulated genes; white arrows indicate genes that are judged as not rok regulated, based on the transcription profiling experiments. yybN is in brackets, since the transcription profiling for this gene was inconclusive (see the text). Dotted arrows indicate putative rok-regulated transcription units. Hairpins indicate terminators as derived from the Subtilist database (http://genolist.pasteur.fr/SubtiList/). The sizes in the figure are not proportional to the lengths of the open reading frames or intergenic regions.
Many of the candidate rok-regulated genes may comprise operons, since they are adjacent to one another. These are illustrated in Fig. 1. In addition to comK and rok itself, 41 genes in 20 putative transcription units met our criteria for potential direct or indirect regulation by Rok.
Validation of transcriptional profiling results.
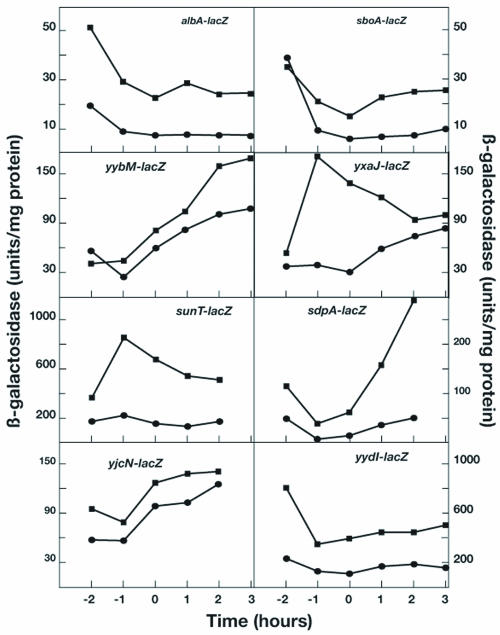
In order to validate the results of the transcriptional profiling experiments, lacZ fusions of selected candidate rok-regulated genes were tested for β-galactosidase expression in wild-type and rok backgrounds (Fig. 2) grown in competence medium (1). In all cases, the fusion data served to validate the transcriptional profiling results. The expression patterns observed fell into three categories.
FIG. 2.
Validation of transcriptional profiling results using lacZ fusions. Strains carrying fusions of lacZ to candidate rok-regulated genes were grown in competence medium, and samples were withdrawn at the indicated times for measurement of β-galactosidase specific activities. T0 refers to the time of departure from exponential growth. Each fusion was placed in both rok (▪) and rok+ (•) backgrounds for determination of β-galactosidase.
Among genes in the first category (yydI, albA, and sboA), the β-galactosidase specific activities remained constant or decreased slightly during growth, reaching a constant level after T0, the end of exponential growth. At all points tested, higher expression was observed in the rok background, and a similar differential was observed between the wild-type and rok strains throughout growth. This is consistent with the transcriptional profiling data for all nine genes in the sbo-alb operon and with the genes in the yydHIJ operon, since approximately similar ratios were observed for them during exponential growth and at T2 (Table 3). The relatively constant difference between the rok and rok+ strains indicates that Rok is active as a repressor throughout growth, consistent with the aforementioned constant expression of rok-lacZ and the similar levels of Rok detected by Western blotting during growth (18).
The second group of genes (yybM, yjcN, and sdpA) exhibited a different pattern, with a gradual increase in β-galactosidase specific activity during growth, and again the rok strain expressed higher levels, but with a similar growth dependency. These β-galactosidase expression patterns suggested that the genes in question are regulated by an additional pathway, conferring growth-dependent expression, as well as being Rok repressed.
The patterns obtained with the third group of genes (sunT and yxaJ) were again distinct, in that differing patterns of growth dependence were observed in the rok and wild-type strains. In the cases of sunT, yxaJ, and yxaL, transcriptional profiling revealed a greater differential between the rok and wild-type strains during exponential growth, consistent with the data in Fig. 1.
In all cases, the data presented in Fig. 1 confirm the transcriptional profiling results for the genes tested, demonstrating repression by Rok and suggesting that many of the genes listed in Table 3, which were not studied with lacZ fusions, are probably also regulated by rok. The lacZ fusion constructs were all studied in comK+ backgrounds, whereas the RNA samples for transcriptional profiling were isolated from isogenic rok and rok+ strains in comK backgrounds to simplify interpretation of the data. Since the two sets of results were consistent, it appears that rok repression of the genes we have studied neither requires nor is prevented by ComK. Since the fusions were made using internal gene fragments, their Campbell-like integration resulted in gene knockouts. It follows that regulation by rok in these cases does not require the cooperation of the individual gene products, although it is possible that the magnitude of the regulatory effects was affected.
Rok binds directly to the promoter regions of comK, rok, sunA, sboA, sdpA, yjcN, yybN, yxaJ, and yydH.
Although Rok apparently represses these genes, it may do so either directly, by binding to the various promoter regions, or indirectly. To distinguish between these possibilities, we performed gel electrophoresis mobility shift assays using His-tagged Rok protein (Rok-His6) (Fig. 3) and 32P-end-labeled, PCR-amplified promoter-probe fragments. For comK and sboA, the primers were designed based on published identification of the start sites of the transcription units (38, 40). For the remaining genes, we amplified fragments including 200 to 230 bp upstream from the apparent start codons of the first genes in each transcription unit.
FIG. 3.
Gel retardations. DNA fragments from the indicated genes were prepared by PCR and end labeled with 32P. In each panel the lane marked with an x corresponds to probe alone. The remaining lanes correspond to probe samples incubated with Rok-His6 at concentrations ranging from 7 to 1,792 nM. Each successive lane, from left to right, corresponds to a doubling in the concentration of Rok-His6. The arrows indicate the positions of the unshifted probe fragments. In each case, the unshifted band that migrated slower than the probe probably corresponds to single-stranded DNA, due to slightly unequal concentrations of PCR primers. This material is not shifted by Rok-His6.
In all cases, a shift was noted, suggesting that Rok was capable of direct binding to the probe fragments. The shifts observed with PcomK (Fig. 3A) and Prok (Fig. 3B) fragments, included as positive controls, yielded apparent Kd values of 26 to 56 nM, corresponding well with the previously reported estimate of 50 nM for both promoters (18).
We regard the binding observed with the newly identified Rok target fragments as specific, with the probable exceptions of PykuJ (Fig. 3K) and PbhlA (Fig. 3N). Several arguments serve to justify this conclusion. First, the gel shifts were carried out in the presence of a molar excess of poly(dI-dC). Second, several negative controls were used. No specific binding was observed to the promoter region of the ComK-regulated promoter comG (Fig. 3D), to a fragment amplified from within the coding region of rok (Fig. 3C), or to the promoter region of the arbitrarily selected gene skfA (Fig. 3E), except for some smearing of the probe at the highest concentration of Rok. Finally, the concentrations of Rok required to give half-maximal shifts were in the range from 30 to 100 nM for all but PbhlA (Fig. 3N) and PykuJ (Fig. 3K). This high-affinity binding, as well as the results obtained with the control fragments, is most consistent with the conclusion that Rok acts directly on the transcription units in question, repressing transcription.
The case of the yybN-yybM-yybL-yybK cluster (Fig. 1) deserves special mention. A 200-bp fragment derived from the sequence upstream from the start of yybM did not shift in the presence of Rok-His6 (data not shown), although this gene is clearly repressed by Rok. A probe fragment was generated containing sequences preceding yybN, which lies immediately upstream from yybM, and a half-maximal shift was observed at a Rok-His6 concentration below 100 nM (Fig. 3H). Evidence for the regulation of yybN by rok could not be obtained from the transcriptional profiling data, since few reliable data points were obtained for this gene (5 out of 10 attempts with the exponential sample and 2 out of 8 for the T2 sample). In spite of this failure, which was most likely due to a problem with the yybN oligonucleotide, we tentatively conclude that yybN is most likely the first gene in a Rok-repressed operon that also contains yybM, yybL, and yybK (Fig. 1).
Another feature worthy of mention, and evident in Fig. 3, is that Rok binding in all cases yields more than one shifted band, as noted previously for PcomK and Prok (18). This is most likely due to the association of more than one protein molecule with each regulatory sequence.
DISCUSSION
Nature of the rok-regulated genes.
Transcription profiling, gel shift experiments, and the use of fusions to lacZ have established a set of genes that are repressed by Rok, most likely by direct binding. In addition to comK, the master regulator of competence development, and rok itself, we have confirmed nine Rok-repressed transcription units, seven of which appear to be operons with two or more open reading frames. These transcription units apparently include 30 new Rok-repressed genes. It is interesting that aside from rok and comK, none of the confirmed Rok-repressed genes appear to encode regulatory proteins. This is consistent with our conclusion, based on gel shift experiments, that nearly all of the genes tested appear to be directly regulated by Rok. In addition to the confirmed genes, Table 3 lists several that remain candidates for regulation by Rok, although they were not further validated. Several additional genes may well be Rok regulated, since their transcriptional profiling data fell just below our criteria for inclusion in Table 3 (see http://www.phri.org/research/res_pidubnau.asp).
Several of the newly identified genes encode proteins needed for the production of confirmed or probable extracellular molecules, including several bacteriocin-like antibiotics. Antibiotic-like exported molecules may play important roles in development and cell-cell signaling, as demonstrated by a recent report concerning the Streptomyces coelicolor product SapB (20), by the role of surfactin in fruiting body formation in B. subtilis (7), by the activity of secreted pheromones in the development of genetic competence (16, 25), and by the effects of subinhibitory concentrations of antibiotics on transcription (10). It is also possible that antibiotics induce the lysis of neighboring bacteria and the release of DNA, suggesting a possible link between competence and antibiotic synthesis.
The sbo-alb operon encodes the genes required for the synthesis and export of the peptide antibiotic subtilosin. The precursor of subtilosin is encoded by sboA, while the albA-G genes are needed to modify and transport this molecule, as well as to confer immunity to subtilosin (39, 40). An additional bacteriocin-like precursor peptide is apparently encoded by sboX, which partially overlaps sboA, but is not required for the production of subtilosin (39). All of the genes in this operon are repressed by the rok gene product, and the flanking genes, ywiB and ywhL, are not.
The Rok-repressed gene cluster, consisting of sunA, sunT, bdbA, yolJ, and bdbB, is involved in the production and secretion of sublancin, a lantibiotic peptide antibiotic (8, 28), and is part of the SPβ prophage. sunA encodes presublancin, and sunT encodes an apparent ABC transporter, surmised to export the antibiotic (28). sunT is a dual function protein, since it also includes a novel domain which is most likely responsible for the proteolytic processing of presublancin (28). bdbA and bdbB are thiol-disulfide oxido-reductases (6), and at least bdbB is needed for the production of active sublancin, which contains two disulfide bonds (8). YolJ is similar to a large family of glycosyl transferases, including one, PlnO (accession number NC_004567.1), which is annotated as a biosynthetic protein for the production of the Lactobacillus plantarum lantibiotic plantaricin.
BhlA, also encoded by the lysogenic phage SPβ, is a holin-like protein (29) and is similar to proteins annotated as involved in the production and release of bacteriocin-like molecules. If this is the role of BhlA, its substrates have not been identified.
The sdp operon has been shown to encode an extracellular factor, derived from SdpC, that results in a delay in sporulation (11). It appears likely that this signaling molecule stimulates increased energy production, thereby delaying the onset of sporulation. Although the SdpC-derived product is apparently not an antibiotic, it may be regarded as an extracellular signaling molecule.
The putative operon consisting of yydJ, yydI, and yydH encodes two polytopic membrane proteins (YydJ and YydH), predicted by TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) to contain six and five membrane-spanning sequences, respectively. YydI is not predicted to be a membrane protein but is similar to the family of ABC transporter, ATP binding proteins. It appears likely that this operon is involved in transport, probably in export, since it does not encode an obvious substrate binding protein.
The yybN, yybM, yybL, yybK cluster encodes one predicted protein with a likely N-terminal signal sequence (YybN) and three permease-like predicted polytopic membrane proteins, each with six likely membrane-spanning sequences. Again, it is plausible to suggest that this operon has a transport function.
The putative operon containing yxaJ and yxaL encodes two proteins, the first of which, YxaJ, is predicted to contain four transmembrane sequences and the second of which contains motifs that are typically found in bacterial dehydrogenases. YxaL has been shown to be a secreted protein, and the YxaL preprotein contains a signal sequence (34). The open reading frames in the ykuJ-ykuK-ykzF putative operon do not encode proteins with suggestive similarities, and all are predicted to be cytosolic gene products. Finally, yjcN encodes a protein with a single predicted transmembrane segment.
It appears, then, that nearly all of the newly identified genes that we have confirmed to be repressed by Rok encode proteins that are involved in secretion or transport, or are themselves secreted products. The competence proteins, which are indirectly regulated by Rok, fall into this category, since they are located in the membrane or associated with the cell wall and mediate the transport of DNA. In particular, several of the Rok-regulated proteins are involved in the production of secreted antibiotics or signaling molecules.
Although we have reported that the overproduction of Rok is lethal (18), none of the genes listed in Table 3 was shown to be essential for viability (19). We may consider several possibilities for this discrepancy. Most obviously, an essential gene may simply have failed to meet our criteria for inclusion in Table 3. Alternatively, an essential gene may be repressed by Rok, which binds to its promoter with low affinity. If so, we would not have identified this gene in our transcriptional profiling experiments, but overproduction of Rok would prevent adequate expression. It is also possible that a Rok-regulated gene may be essential under our growth conditions (competence medium) (18), but not in the rich medium used for the systematic search for essential genes (19). Finally, it is possible that the lethality we detected when Rok was overexpressed was due to some combination of Rok-repressed genes.
Regulation of Rok-repressed genes.
The sbo-alb operon is known to be under complex control (26, 40). This operon is repressed by AbrB and also is induced by oxygen limitation via a mechanism that requires the two-component regulators ResD and ResE. Consistent with this is our observation that the expression of this operon is low in the rok+ background, under aerobic conditions (Fig. 2). The albA-lacZ construct was tested under anaerobic conditions in complex medium (2xYT), and a dramatic induction was observed (data not shown), as expected. Under anaerobic conditions, an even greater induction was observed in the rok background, suggesting that Rok is not required for the anaerobiosis signaling pathway and that repression by Rok is still manifest under anaerobic conditions. Although the expression of albA-lacZ in competence medium was low and apparently not growth regulated, the expression of this construct in complex medium, whether anaerobic or aerobic, was clearly growth regulated, increasing markedly at the end of exponential growth (data not shown).
Several of the Rok-repressed genes listed in Table 1 (sdpABC [15], comK [12], rok [18], hom [15], yxbC [15], and the sboA-alb operon [40]) are also repressed by AbrB. There appears to be a significant overlap between the rok and abrB regulons.
Possible binding site for Rok.
We have used a bioinformatics approach to search for a consensus Rok binding sequence. In particular, we have used BioProspector together with BioOptimizer (22, 23), as well as MEME (3). BioProspector finds candidate motifs with an improved Gibbs sampler search algorithm that allows for the introduction of gaps in the search parameters. BioOptimizer is an algorithm designed to take files containing candidate motifs identified by BioProspector as input. BioOptimizer then optimizes the candidate motif, if possible, providing a measure of confidence that the motif found by BioProspector is significant.
We have applied these programs, systematically varying the input data sets used (identities of the genes included as well as the lengths of the upstream sequences) and adjusting these data sets using information obtained from DNase protection footprinting experiments (data not shown). In each case we failed to obtain an optimized sequence, perhaps due to the subtle nature of the Rok binding motif and the inadequate size of our data set.
In spite of this failure to obtain an optimizable sequence, the motif 5′-GATAG-3′ produced a relatively high score with both BioProspector and MEME and was found in repeated attempts with varied approaches. This motif is located in the DNase-protected regions for comK, sboA, and sunA, as well as in the upstream regions of the other six genes in our core data set. The significance of this motif awaits further experimental work.
The effect of rok on sporulation is not mediated solely by repression of sdp.
Mutational inactivation of rok results in an oligosporogenic phenotype (18). Since Rok appears to be a direct repressor of the sdp operon, the possibility was considered that the oligosporogenic phenotype is explained by overexpression of sdp, which would be expected to delay spore formation (11). To test this, we determined whether an sdp null mutation would suppress the oligosporogenic phenotype of a rok knockout (data not shown). In fact, the sporulation frequency, measured by colony formation and by microscopic examination, was no higher in an sdp rok double mutant than in a rok single mutant. It appears that the oligosporogenic phenotype of the rok knockout mutation cannot be explained only by overexpression of the sdp operon.
Evolution of Rok.
As noted above, Rok has an ortholog among sequenced genomes in B. subtilis, B. licheniformis, and B. amyloliquefaciens, but not in other bacilli and spore-forming gram-positive bacteria, such as B. halodurans, B. anthracis, B. cereus, or Oceanobacillus iyensis. In fact yknT, the convergently transcribed gene immediately downstream from rok, is also absent from these organisms, whereas flanking genes (ykuV and mobA) are present. It is interesting that yknT (also called cse15) is dependent on sigma factor E for its expression in the spore mother cell, but no sporulation phenotype was observed in a gene knockout (17). It has been suggested that the yknT gene product plays a subtle role in spore coat formation (17), consistent with its absence from related spore-forming bacilli. rok and yknT may have been introduced into the B. subtilis-B. licheniformis lineage by horizontal transmission, or they may have been lost together from some common ancestor of B. halodurans, B.cereus, and B. anthracis. The combined average base composition of the rok and yknT coding sequences is 40.2%, which is less than the average for the entire B. subtilis genome (43.5%) (21). However, rok and yknT are located in a large region of lower-than-average percent GC (21), and the base composition differences therefore neither support nor refute the hypothesis of horizontal transmission.
Attention to the phylogenetic tree (Fig. 4) is more revealing. The rok determinant is present in the closely related B. subtilis-B. licheniformis-B. amyloliquefaciens group, but absent from the B. cereus-B. anthracis group. This permits two possibilities, if we assume a single relevant event. rok may have been introduced just before the branch point that led to the B. subtilis group and would therefore not be present in other more distantly related gram-positive spore formers. Alternatively, it may have been lost specifically from the B. cereus branch, in which case it would be present in the more distantly related spore-forming gram positives. Since rok is also missing from the outliers B. halodurans and O. iyensis, as well as from the gram-positive nonsporulating Listeria monocytogenes, the most parsimonious hypothesis is that rok was introduced by horizontal transfer into an ancestor of the branch that contains B. subtilis, B. licheniformis, and B. amyloliquefaciens.
FIG. 4.
Relationships among group 1 bacilli (adapted from reference 2). Species in which a rok ortholog have been identified are boxed, and those in which it is known to be absent are underlined.
Acknowledgments
We thank Saleena Ghanny, Pat Soteropoulos, and Tongsheng Wang from the Center for Applied Genomics at Public Health Research Institute for spotting the microarrays, for letting us use their scanner, and for general advice. We thank Peter Zuber and Michiko Nakano for providing ORB3147 and ORB3162 and members of our various labs, and especially Leendert Hamoen, for discussion and advice. We also thank Randy Berka of Novozymes for providing access to the B. licheniformis genome sequence before publication and Rainer Borriss of Humboldt University for access to the B. amyloliquefaciens genome sequence. Finally, we thank Shane T. Jensen for the use of a new version of BioProspector and for advice in its implementation.
This work was supported by National Institutes of Health grant GM57720 awarded to D.D., by grant 811.35.002 from The Netherlands Organization of Scientific Research (NWO-ALW), which supported the work of W.K.S., and by grant number 3211-01-831/467 from the Slovenian Ministry for Education, Science and Sport, which supported the work of B.K.
REFERENCES
- 1.Albano, M., J. Hahn, and D. Dubnau. 1987. Expression of competence genes in Bacillus subtilis. J. Bacteriol. 169:3110-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ash, C., J. A. E. Farrow, S. Walbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 3.Bailey, T. L., and W. S. Noble. 2003. Searching for statistically significant regulatory modules. Bioinformatics 19(Suppl. 2):II16-II25. [DOI] [PubMed] [Google Scholar]
- 4.Baldi, P., and G. W. Hatfield. 2002. DNA microarrays and gene expression. Cambridge University Press, Cambridge, United Kingdom.
- 5.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 6.Bolhuis, A., G. Venema, W. J. Quax, S. Bron, and J. M. van Dijl. 1999. Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J. Biol. Chem. 274:24531-24538. [DOI] [PubMed] [Google Scholar]
- 7.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorenbos, R., T. Stein, J. Kabel, C. Bruand, A. Bolhuis, S. Bron, W. J. Quax, and J. M. Van Dijl. 2002. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 277:16682-16688. [DOI] [PubMed] [Google Scholar]
- 9.Dubnau, D., and C. M. Lovett, Jr. 2001. Transformation and recombination, p. 453-471. In J. A. Hoch, R. Losick, and A. L. Sonenshein (ed.), Bacillus subtilis and its relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 10.Goh, E. B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 12.Hamoen, L. W., D. Kausche, M. A. Marahiel, D. van Sinderen, G. Venema, and P. Serror. 2003. The Bacillus subtilis transition state regulator AbrB binds to the−35 promoter region of comK. FEMS Microbiol. Lett. 218:299-304. [DOI] [PubMed] [Google Scholar]
- 13.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamoen, L. W., A. F. Van Werkhoven, J. J. E. Bijlsma, D. Dubnau, and G. Venema. 1998. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev. 12:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52:847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriques, A. O., E. M. Bryan, B. W. Beall, and C. P. Moran, Jr. 1997. cse15, cse60, and csk22 are new members of mother-cell-specific sporulation regulons in Bacillus subtilis. J. Bacteriol. 179:389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoa, T. T., P. Tortosa, M. Albano, and D. Dubnau. 2002. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43:15-26. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kodani, S., M. E. Hudson, M. C. Durrant, M. J. Buttner, J. R. Nodwell, and J. M. Willey. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 101:11448-11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., D. L. Brutlag, and J. S. Liu. 2001. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001:127-138. [PubMed] [Google Scholar]
- 23.Liu, Y., L. Wei, S. Batzoglou, D. L. Brutlag, J. S. Liu, and X. S. Liu. 2004. A suite of web-based programs to search for transcriptional regulatory motifs. Nucleic Acids Res. 32:W204-W207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 25.Magnuson, R., J. Solomon, and A. D. Grossman. 1994. Biochemical and genetic characterization of a competence pheromone. Cell 77:207-216. [DOI] [PubMed] [Google Scholar]
- 26.Nakano, M. M., G. Zheng, and P. Zuber. 2000. Dual control of sbo-alb operon expression by the Spo0 and ResDE systems of signal transduction under anaerobic conditions in Bacillus subtilis. J. Bacteriol. 182:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogura, M., H. Yamaguchi, K. Kobayashi, N. Ogasawara, Y. Fujita, and T. Tanaka. 2002. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paik, S. H., A. Chakicherla, and J. N. Hansen. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 273:23134-23142. [DOI] [PubMed] [Google Scholar]
- 29.Regamey, A., and D. Karamata. 1998. The N-acetylmuramoyl-l-alanine amidase encoded by the Bacillus subtilis 168 prophage SP beta. Microbiology 144(Pt. 4):885-893. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Smith, I. 1993. Regulatory proteins that control late-growth development, p. 785-800. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 32.Strauch, M. 1993. AbrB, a transition state regulator, p. 757-764. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 33.Susanna, K. A., A. F. van der Werff, C. D. den Hengst, B. Calles, M. Salas, G. Venema, L. W. Hamoen, and O. P. Kuipers. 2004. Mechanism of transcription activation at the comG promoter by the competence transcription factor ComK of Bacillus subtilis. J. Bacteriol. 186:1120-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tortosa, P., L. Logsdon, B. Kraigher, Y. Itoh, I. Mandic-Mulec, and D. Dubnau. 2001. Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J. Bacteriol. 183:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 37.van Sinderen, D., A. Luttinger, L. Kong, D. Dubnau, G. Venema, and L. Hamoen. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 38.van Sinderen, D., A. ten Berge, B. J. Hayema, L. Hamoen, and G. Venema. 1994. Molecular cloning and sequence of comK, a gene required for genetic competence in Bacillus subtilis. Mol. Microbiol. 11:695-703. [DOI] [PubMed] [Google Scholar]
- 39.Zheng, G., R. Hehn, and P. Zuber. 2000. Mutational analysis of the sbo-alb locus of Bacillus subtilis: identification of genes required for subtilosin production and immunity. J. Bacteriol. 182:3266-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng, G., L. Z. Yan, J. C. Vederas, and P. Zuber. 1999. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol. 181:7346-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]