Abstract
We have investigated recombination mechanisms promoting the completion of replication in the face of unrepaired DNA damage by transforming an isogenic set of uvrA6 excision-defective Escherichia coli strains with pUC-based plasmids in which each strand carried, at staggered positions, a single thymine-thymine pyrimidine (6-4) pyrimidinone lesion. The distance between the lesions was 28 or 8 bp in one orientation relative to the unidirectional ColE1 origin of replication or, in the other orientation, 30 or 10 bp. C-C mismatches placed opposite each of the T-T photoproducts permit unambiguous detection of the three events that can lead to the completion of replication: sister-strand recombination, translesion replication (TR) on the leading strand, and TR on the lagging strand. We find that E. coli possesses a largely constitutive, recA-independent sister-strand recombination mechanism that allows 9% or more of these severely compromised plasmids to be fully replicated. In one orientation, such recombination depends partly on recG and priA but not on ruvA, ruvB, ruvC, or mutS and is largely independent of recF. In the other orientation, recombination is dependent on none of the genes. The strains used did not contain the cryptic phage encoding recET, which encodes enzymes that promote interplasmid recombination. The nature of the recA-independent recombination mechanism is not known but could perhaps result from a template-strand-switching, or copy choice, process.
In addition to DNA repair processes, which accurately remove damage arising spontaneously or from overt mutagen exposure, Escherichia coli possesses tolerance mechanisms that promote the completion of genome replication in the presence of unrepaired DNA damage that might otherwise block this process (reviewed in reference 14). Apart from blocking replication, unrepaired damage may also result in the collapse of the replication fork and the production of double-strand breaks, a particularly damaging outcome (13). Two general categories of mechanism exist to overcome the potential lethality arising from incomplete replication of the genome, one employing recombination and the other using translesion replication (TR). In the latter case, DNA polymerase V is used to replicate past the lesion, or for some lesions and circumstances, DNA polymerase IV or DNA polymerase II is used (23). Recombination-based mechanisms serve to rectify two different conditions associated with incomplete replication, the occurrence of single-stranded gaps that remain after reinitiation of replication beyond the site of the lesion and the occurrence of double-strand breaks. Although the first of these requires the activities of the recF pathway and the second depends on the recBC pathway, they both depend on the RecA protein, which is also required for TR (reviewed in reference 14).
A third recombination-based mechanism, involving transient template strand switching or copy choice, has been proposed (6) but has proved difficult to investigate experimentally in vivo. Unlike the other mechanisms, in which segments of DNA are physically exchanged between strands, transient template strand switching entails informational, but not physical, transfer. Because such transfer occurs between genetically identical sister duplexes, exchange cannot be detected by means of genetically marked strains. As a consequence, there is little in vivo evidence to support the occurrence of such a mechanism or to indicate its genetic requirements, such as dependence on RecA, though in vitro results with E. coli and phage T4 proteins strongly support such a model (5, 7, 8, 9, 16, 17, 19-21).
We have investigated the possible occurrence of such an alternative recombination mechanism for the completion of replication in the face of unrepaired DNA damage by transforming various E. coli strains with plasmids in which each strand carried, at specific staggered positions, a single thymine-thymine pyrimidine (6-4) pyrimidinone [T-T (6-4)] lesion. The distance between the lesions was either 28 or 8 bp apart in orientation 1 or, in orientation 2, 30 or 10 bp apart, with the orientation depending on the positions of the lesions relative to the unidirectional ColE1 origin of replication (Fig. 1). Plasmids that carry defined lesions at specific locations have the useful property that they can focus analysis on one or a small number of damage tolerance mechanisms and can provide precise sequence data that characterize individual events. For example, previous work with single-stranded vectors that carried a UV photoproduct or a basic site examined translesion replication events exclusively, both from the point of view of the types of the nucleotide insertions that occurred and of the contrasting properties of different DNA polymerases that carried out the lesion bypass (1, 15, 28). Double-stranded plasmids that carry a lesion in both strands extend such analysis and offer the possibility of studying TR and recombination mechanisms simultaneously. We find that E. coli possesses a significant capacity to complete the replication of these seriously compromised plasmids by a mechanism that is largely constitutive and, in contrast to previously described mechanisms, RecA independent.
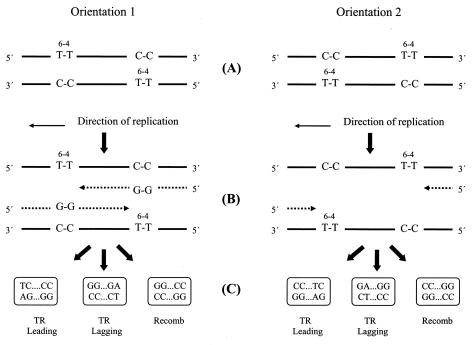
FIG. 1.
(A) Positions of T-T (6-4) photoadducts and opposing C-C mismatches in leading and lagging strands for orientations 1 and 2. (B) Proposed replication intermediates following stalling of forks at T-T (6-4) photoadducts. (C) Sequence motifs resulting from translesion replication on leading (TR leading) or lagging (TR lagging) strand templates or from recombination (recomb).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Table 1 lists the bacterial strains used in this study and those used to construct these strains. Where noted, strains were constructed by generalized P1 transduction with P1vir. Because RecA protein is required for recombination during P1 transduction, the recA mutations were introduced into the double-mutant strains at the last stage of construction. All mutant strains were checked by PCR to verify that they contained the correct genomic alteration, and their phenotype examined to ensure it was consistent with expectation. In a few cases where this was necessary, verification of the alteration was carried out by sequence analysis. The plasmids pES1 and pES2 used for this work were derived from pUC19 by digestion with EcoRI and PstI followed by ligation of a synthetic adaptor between these sites. For pES1, the adaptor was formed by annealing two complementary oligonucleotides, 5′-AATTGTGGAATTCCTGCAGGCATGCA-3′ and 5′-TGCCTGCAGGAATTCCAC-3′. For pES2, 5′-AATTGCAGAATTCCTGCAGTCATGCA-3′ and 5′-TGACTGCAGGAATTCTGC-3′ were used. Ligation of either of these adaptor sequences into the doubly digested plasmid destroys the original EcoRI and PstI sites but generates these sites within the adaptor sequence itself. pES1 and pES2 differ, however, with regard to the two base pairs immediately flanking the restriction sites within the adaptor sequence, which are T-A-G-C, and G-C-C-G in pES1 but C-G-A-T and G-C-A-T in pES2. These sequence markers were used to detect interplasmid recombination when strains were simultaneously transformed with both pES1 and pES1 constructs.
TABLE 1.
Bacterial strains used in this work
| E. coli strain | Relevant genotype | Source |
|---|---|---|
| TK603a | uvrA6 | R. Woodgate |
| G48382 | recA938::cat | S. Sandler |
| JC15359 | ΔrecF349 | S. Sandler |
| GM5555 | mutS205::Tn10 | R. Woodgate |
| BT12 | recF400::Tn5 | R. Woodgate |
| N3793 | recG263::Kan | R. G. Lloyd |
| JC19018 | priA2::Kan | R. Woodgate |
| N2057 | ruvA60 (polar) | M. Berlynb |
| N4453 | ruvC67 | M. Berlynb |
| DV08 | Δ(araD-polB) | R. Woodgate |
| RW564 | ΔrecA306 | R. Woodgate |
| ES200 | TK603 with recF349 | This study |
| ES201 | TK603 with recA938 | This study |
| RW540 | TK603 with lexA51(Def) | R. Woodgate |
| AO1 | TK603 with mutS215 | This study |
| AO6 | TK603 with lexA51(Def) recA306 | This study |
| AO7 | TK603 with recG263 | This study |
| AO10 | TK603 with ruvC67 | This study |
| AO11 | TK603 with ruvA60 (polar) | This study |
| AO12 | RW540 with lexA51(Def) recF400 | This study |
| AO13 | TK603 with priA2 | This study |
| AO14 | TK603 with Δ(araD-polB) | This study |
uvrA6 thi-1 thr-1 araD-14 leuB6 lacY1 ilv323(Ts) Δ(gpt-proA)62 mtl-1 xyl-5 rpsL31 tsx-33 supE44 galK2(Oc) hisG4(Oc) rfbD1 kdgK51.
E. coli Genetic Stock Center, Yale University.
Production and purification of 11-mer containing a specifically located T-T pyrimidine (6-4) pyrimidinone photoadduct.
A T-T (6-4) photoadduct was induced at the unique T-T site in the sequence 5′-GCAAGTTGGAG-3′ as described previously (15). Briefly, 25-ml samples of an aqueous solution containing 100 μg of this 11-mer per ml in a glass petri dish were exposed to ∼300 kJ of 254-nm UV/m2. The solution was stirred continuously and covered by polyethylene film to prevent evaporation. After UV exposure, samples were concentrated by evaporation, and the different species were separated by high-performance liquid chromatography on a 25-mm by 10-cm radial compression Prep Nova-Pak HR C18 column (Waters Corp., Milford, Mass.), using linear gradients from 94% A (100 mM triethylammonium acetate, pH 7.00), 6% B (acetonitrile) to 92% A, 8% B over the first 30 min to 90% A, 10% B over the next 10 min and to 30% A, and 70% B over the last 5 min, all at 6 ml per min. Fractions of the T-T (6-4)-containing species, identified by its absorbance at 325 nm, and photoproduct-free 11-mer were collected and evaporated to dryness and repurified on a Machery-Nagel Nucleogen 60-7 DEAE 10- by 125-mm column (The Nest Group, Southboro, Mass.), using linear gradients from 75% A (20 mM NaAc buffer, pH 6.5, containing 20% acetonitrile), 25% B (1 M LiCl in A) to 65% A, 35% B over 20 min to 60% A, 40% B over the next 5 min, and to 100% B over a final 5 min, all at 1 ml per min. The fractions collected were desalted with PD-10 columns (Amersham Pharmacia Biotech AB, Uppsala, Sweden), and further purified on an 8- by 100-mm radial compression Nova-Pak C18 column (Waters Corp.), using linear gradients from 94% A (100 mM triethyl ammonium acetate, pH 7.00), 6% B (acetonitrile) to 91% A, 9% B over 20 min, and to 30% A, 70% B over a further 10 min. Fractions containing either the T-T (6-4) species or the photoproduct-free oligomer were evaporated to dryness, and the 11-mer was phosphorylated with T4 polynucleotide kinase followed by two further rounds of purification with the Nova-Pak C18 column. The T-T (6-4)-containing 11-mer was >99% pure as estimated by a digestion assay (15). Samples of photoproduct-free 11-mer, derived from the same irradiated material, were purified identically to act as controls.
Assembly of plasmid insert sequences.
The strands that constitute the 72- and 80-bp (interlesion spacing, 28 or 30 bp) or 57- and 65-bp (interlesion spacing, 8 or 10 bp) complementary sequences to be inserted into either pES1 or pES2 were assembled individually by ligating together oligonucleotides annealed to complementary scaffold oligomers and then purifying the required single-stranded species by electrophoresis through a 12.5% denaturing polyacrylamide gel. The strands were designed to place the photoproducts in two orientations and, in each orientation, at two distances from each other. In orientation 1, the T-T (6-4) lesion was placed closer to the ColE1 origin of replication in the lagging strand template than in the leading strand template, a disposition that was reversed in orientation 2 (Fig. 1). Photoproduct spacings were 28 or 8 bp in orientation 1 and 30 or 10 bp in orientation 2. For the wider photoproduct spacing, the strands were assembled from the following oligonucleotides in the order given and numbered as in Table 2: complementary strands for orientation 1; oligonucleotides 2 (19-mer), 1 (11-mer), and 3 (50-mer); oligonucleotides 4 (15-mer), 1 (11-mer), and 5 (46-mer); complementary strands for orientation 2; oligonucleotides 10 (50-mer), 1 (11-mer), and 11 (19-mer); and oligonucleotides 12 (46-mer), 1 (11-mer), and 13 (15-mer). These oligomers were annealed to scaffolds of easily differentiated size at molar ratios, relative to the scaffold, of 1.2:1, except for the T-T (6-4) lesion-containing 11-mer, where the ratio was 2.5:1. Ligation was performed at 15°C for 48 h. For the narrower photoproduct spacing, the complementary strands were assembled as follows: orientation 1; oligonucleotides 6 (19-mer), 1 (11-mer), and 7 (35-mer); oligonucleotides 8 (20-mer), 1 (11-mer), and 9 (26-mer); orientation 2; oligonucleotides 14 (30-mer), 1 (11-mer), and 15 (24-mer); and oligonucleotides 16 (31-mer), 1 (11-mer), and 17 (15-mer). Assembly of strands for the narrower photoproduct spacing required ligation to be carried out in two steps because of the formation of snap-back hairpin structures between the 11-mer and its complementary sequence. In the first step, the 11-mer was ligated to the flanking oligomer that did not contain a sequence complementary to the 11-mer. The product from this reaction was then purified and ligated to the other flanking oligonucleotide at ∼21°C, which is above the melting temperature of the 11-mer sequence. In addition to using an 11-mer carrying the site-specific T-T (6-4) photoproduct, a complete set of strands was assembled using lesion-free 11-mer sequences, for both the wider and the narrower spacing, to act as controls. In all cases, the duplex insert molecules resulting from the annealing of full-length complementary strands possessed NsiI and MfeI ends, compatible with PstI and EcoRI cohesive ends. All oligonucleotides used to assemble full-length strands were treated with polynucleotide kinase, except for those forming the MfeI terminus. These molecules also possessed a double C-C mismatch opposite either the T-T (6-4) lesion or the lesion-free T-T doublet that acted as sequence markers.
TABLE 2.
Oligonucleotides used to assemble plasmid inserts
| No. | Purpose | Sequence |
|---|---|---|
| 1 | All strands | 5′-GCAAGTTGGAG-3′a |
| 2 | Orientation 1, 80-mer, 28-bp spacing | 5′-AATTGGTCTTGCTGGACTC-3′ |
| 3 | Orientation 1, 80-mer, 28-bp spacing | 5′-GGTACGAGCTACTGGTAACCCTCCCCCTTGCCTAAGGTACCGTGGATGCA-3′ |
| 4 | Orientation 1, 72-mer, 28-bp spacing | 5′-TCCACGGTACCTTAG-3′ |
| 5 | Orientation 1, 72-mer, 28-bp spacing | 5′-GGTTACCAGTAGCTCGTACCCTCCCCCTTGCGAGTCCAGCAAGACC-3′ |
| 6 | Orientation 1, 65-mer, 8-bp spacing | 5′-AATTGGTAGCTGCTGGATC-3′ |
| 7 | Orientation 1, 65-mer, 8-bp spacing | 5′-CTCCCCCTTGCCTAAGGTACGCGTCATCGGATGCA-3′ |
| 8 | Orientation 1, 57-mer, 8-bp spacing | 5′-TCCGATGACGCGTACCTTAG-3′ |
| 9 | Orientation 1, 57-mer, 8-bp spacing | 5′-CTCCCCCTTGCGATCCAGCAGCTACC-3′ |
| 10 | Orientation 2, 80-mer, 30-bp spacing | 5′-AATTGAGCTACTGGTAACCCTCCCCCTTGCCTAAGGTACCTGCTGGACTC-3′ |
| 11 | Orientation 2, 80-mer, 30-bp spacing | 5′-GGTACGAGCTACTGATGCA-3′ |
| 12 | Orientation 2, 72-mer, 30-bp spacing | 5′-TCAGTAGCTCGTACCCTCCCCCTTGCGAGTCCAGCAGGTACCTTAG-3′ |
| 13 | Orientation 2, 72-mer, 30-bp spacing | 5′-GGTTACCAGTAGCTC-3′ |
| 14 | Orientation 2, 65-mer, 10-bp spacing | 5′-AATTGCTGCGCATGTAACCCTCCCCCTTGC-3′ |
| 15 | Orientation 2, 65-mer, 10-bp spacing | 5′-GGTACGAGCTACTGATCGGATGCA-3′ |
| 16 | Orientation 2, 57-mer, 10-bp spacing | 5′-TCCGATCAGTAGCTCGTACCCTCCCCCTTGC-3′ |
| 17 | Orientation 2, 57-mer, 10-bp spacing | 5′-GGTTACATGCGCAGC-3′ |
The bold underlined sequence is the site of the T-T (6-4) adduct.
Construction of lesion-containing plasmids.
The inserts with NsiI and MfeI cohesive ends, assembled as described above, were ligated into the PstI and EcoRI sites of either pES1 or pES2 using a two-step procedure designed to maximize ligation efficiency and production of the desired construct. In the first step, a high DNA concentration, an excess of insert material, and the presence of restriction enzymes were used to very efficiently ligate insert material to each end of PstI-linearized plasmid. In the second step, EcoRI digestion released the insert from one arm of the plasmid, and recircularization was promoted by decreasing the DNA concentration by more than 100-fold. For the first step, a fivefold molar excess of the insert was ligated to PstI-linearized plasmid DNA at a DNA concentration of ∼600 ng/μl, using 33 U of high-concentration T4 ligase/μl, and 1 U each of PstI and NsiI/μl. The presence of the PstI restriction enzyme and the competing molar excess of PstI-compatible (NsiI) cohesive ends of the insert DNA promote ligation of the insert to the plasmid ends and prevent recircularization of the PstI-linearized plasmid DNA. Additionally, ligation efficiency is promoted by the presence of the NsiI restriction enzyme, which inhibits insert dimerization and thus maintains the pool of insert monomer. Inserts cannot dimerize by ligation of their MfeI cohesive ends because these are not phosphorylated. Under these conditions, insert was ligated to both ends of the linearized plasmid with an efficiency that was close to 100%, as shown by the detection of PvuII restriction fragments that were 209 and 273 bp in length but not those that were 76 bp shorter. Following the first-stage ligation, the linear plasmid product was digested with EcoRI to generate a plasmid end compatible with the MfeI terminus of the insert and to release a fragment containing one of the inserts. Recircularization of the plasmid was achieved by phosphorylation of the MfeI end of the construct, and ligation at a DNA concentration of <5 ng/μl using 2 U of T4 ligase/μl in the presence of 0.08 U of MfeI and EcoRI. After the free construct ends were removed and the reaction mix was concentrated using Centricon YM-30 spin columns (Millipore Corporation, Bedford, Mass.), the covalently closed circular species was purified by electrophoresis through a 0.7% agarose gel in Tris-acetate-EDTA buffer containing ethidium bromide, run in the dark to prevent DNA damage. To excise the desired covalently closed circular species, the gel was shielded with polyester film, and all but the outermost lanes were entirely protected from the brief exposure to 300-nm UV used to visualize the bands, to minimize UV damage in the plasmid. Such damage can appreciably reduce transformation frequencies, particularly in uvrA6 ΔrecA strains. Covalently closed circular DNA was isolated from the gel slices with a QIAQuick gel extraction kit (QIAGEN, Valencia, Calif.) following the manufacturer's protocol but with increased digestion time, additional column washes, and additional elution from the column. The purified plasmid vector was quantitated with a PicoGreen double-stranded DNA quantitation kit (Molecular Probes, Eugene, Oreg.) and a Turner Quantech digital filter fluorometer (Barnstead/Thermolyne, Dubuque, Iowa) according to the kit protocols.
Transformation with duplex vectors.
Twenty-five milliliters of each experimental strain at a concentration of ∼2 × 108/ml, either SOS induced by exposure to 4 J/m2 of 254-nm UV or uninduced, was made competent by suspension in 60 mM HEPES-buffered CaCl2 (pH 7.0) and concentrated 10-fold. Fifty microliters from each strain and condition was then transformed with 3 ng of control or lesion-containing construct and plated on ampicillin-containing plates, and the Ampr colonies were counted the next day. The fraction of replicated plasmids was estimated by normalization of the number of colonies from the lesion-containing constructs to the number of colonies from the controls. Analyses of the types of DNA damage tolerance mechanism used to complete replication, made possible by tandem double cytosine mismatches opposite the T-T (6-4) photoadducts (Fig. 1), were carried out by DNA sequence determinations of the insert region, using fluorescence-tagged cycle sequencing reactions.
Confirmation that TK603 does not contain recET.
To detect the possible presence of recET by PCR, genomic DNA was prepared from four different E. coli strains, including the AB1157 derivative TK603 used in these experiments and from which all subsequent mutant strains were derived by P1 transduction, and the three control strains SMH10 (another AB1157 derivative), JM101, and DH5α. The absence of recET was demonstrated by using the forward and reverse primers 5′ TTCAACAAGCCATTGCCC 3′ and 5′ CGTCCGTAAAAGAAGCACC 3′, with an expected product length of 1,193 bp for the presence of recET. The forward and reverse primers for the dbpA gene, used as an internal control, were 5′ CCCAACTCACGAACCTTAATGAG 3′ and 5′ CACGGGCGCTACCGTTAG 3′, respectively, with an expected product of 843 bp. In addition, we also used PCR to look for a signal that indicated the absence of the cryptic Rac prophage, using forward and reverse primers 5′ GCGAGAACACAGTGAGCAAG 3′, and 5′ ACTTATCTGCTCGGTTCCAAC 3′, respectively. In a Rac− strain, a product of between 628 and 630 bp is expected, but in a Rac prophage-containing strain the product would be 23.3 kb, a size unattainable with standard PCR. The genomic DNA was extracted and purified from these strains using a Wizard genomic DNA purification kit (Promega Corporation, Madison, Wisc.) according to instructions for gram-negative bacteria. Reactions for each primer pair were performed for 40 cycles, using annealing and melting temperatures appropriate for the primer pairs, and the products were analyzed by electrophoresis through 1% agarose gels.
RESULTS
Experimental design.
The goal of this work was to investigate DNA damage tolerance mechanisms in E. coli that enable the completion of replication by processes that utilize recombination between sister strands. To this end, we transformed various excision-repair-deficient (uvrA6) strains with pUC-based plasmids that carry specifically located T-T (6-4) photoadducts at staggered positions in each strand, under conditions in which almost all transformants contained plasmids originating from a single transforming molecule. Photoproducts were placed in two orientations relative to the unidirectional ColE1 origin of replication in the plasmid (Fig. 1). In orientation 1, the photoproduct in the template for lagging strand synthesis was placed nearer to the origin than the photoproduct in the template for leading strand synthesis, whereas in orientation 2 this disposition was reversed. The interphotoproduct region can be replicated on both strands in orientation 1 but, at first sight, on neither in orientation 2. To examine the influence of photoproduct spacing on completion of plasmid replication, the T-T (6-4) lesions were placed closer or further apart, by 28 and 8 bp in orientation 1 and 30 and 10 bp in orientation 2. Estimates of the fraction of fully replicated plasmids were obtained by normalizing the number of Ampr colonies resulting from transformation with the photoproduct-containing construct to the number obtained from transformation with an equal amount of lesion-free construct; transformant colonies can be established only if plasmid replication is completed. Such estimates were obtained both from unirradiated cells and, where possible, from those in which the SOS regulon had been induced by exposure to 4 J per m2 of 254-nm UV. However, strains carrying deletions of a number of genes studied were too UV sensitive for this procedure. The types of events that were used to overcome the block to fork progression and allow replication to be completed were determined by sequence analysis. C-C mismatches placed opposite the T-T photoadduct enabled unambiguous identification of the DNA strand, or part of a strand, from which replicated plasmids originated, and they differentiated between recombination events within the interlesion region and translesion replication events on either the leading or lagging strands (Fig. 1). Leading and lagging strands can be identified because the plasmid constructs carry a unidirectional ColE1 origin of replication. We chose T-T (6-4) lesions because, in the absence of tolerance processes, they essentially block replication fork progression completely; without SOS induction, almost no molecules are replicated following transfection of E. coli with single-stranded vectors carrying this photoproduct (15). As a consequence, duplex plasmids carrying such a lesion in each strand are particularly suitable tools for detecting tolerance mechanisms using sister-strand recombination. We also chose the T-T (6-4) photoadduct because translesion replication past this lesion is highly mutagenic, resulting in a 3′ T →C substitution in close to 100% of bypass events (15; unpublished data); the occurrence of this mutation therefore acts as an additional marker for this event (Fig. 1).
When replication is blocked on both strands, it can be restarted by a recombination process that does not employ RecA or RecF.
Although T-T (6-4) photoproducts in both strands of the plasmid at staggered positions only 28 or 30 bp apart might seem to present a severe block to replication, we found that replication could nevertheless be completed in an appreciable fraction of plasmids. With the photoproducts in orientation 1, 8.3% of the plasmid constructs were fully replicated, even in the absence of SOS induction, and following SOS induction this percentage rose to 32.5% (Table 3, TK603). In uninduced cells, this was achieved solely by recombination; as expected, no translesion replication was observed. Both mechanisms were found in SOS-induced cells, however, with recombination being employed in 14.8% of the transformants and translesion replication on the leading and lagging strand templates being used in 7.3 and 10.4% of the transformants, respectively. Surprisingly, and in contrast to other recombination mechanisms for tolerating unrepaired DNA damage (14), the frequency of recombination in the uvrA6 recA938 strain ES201 was 8.8%, indicating that the recombination mechanism did not employ RecA. It was also largely independent of RecF, since 6.1% recombination occurred in the uvrA6 recF349 strain, ES200. Although the data from TK603 suggested that there might be an SOS-inducible component to this RecA- and RecF-independent recombination, other results indicate that the amount of induction is at best very small. We investigated this issue in LexA-deficient strains carrying the lexA51 mutation, because the extreme sensitivity of the uvrA6 recA938 and uvrA6 recF349 strains precluded the use of DNA damaging agents such as UV to induce the SOS regulon. Little or no evidence of induction, either LexA dependent or damage inducible but LexA independent, can be seen in results from the uvrA6 lexA51 strain RW540, an isogenic derivative of TK603. In this strain, recombination frequencies were 8.3% in uninduced cells and 11.9% after exposure of the cells to 4 J per m2. Results from AO6 and AO12, a recA306 and a recF400 derivative of RW540, respectively, also confirmed that the recombination process was not LexA repressible. In these strains, recombination frequencies were 9.4 and 7.0%, respectively (Table 3). Lastly, the events detected truly result from recombination and are not artifacts arising from mismatch repair. Even though the C-C double mismatches opposite the T-T (6-4) photoproducts may well constitute targets for the binding of mismatch repair proteins, the activities of these proteins do not generate the recombinant sequences we observe; recombination frequencies are no smaller in the mutS205 strain, AO1, than in mismatch repair-proficient strains (Table 3).
TABLE 3.
Total fraction of plasmids replicated and percent replicated following a recombination or TR event in isogenic excision-deficient (uvrA6) strains of E. coli carrying recA, recF, lexA, or mutS mutations
| Orientation and strain | Relevant genotype | Amt of UV to cells (J/m2) | Plasmids replicated (%)
|
Total no. of transformants sequencedb | |||
|---|---|---|---|---|---|---|---|
| Totala | By recombination | By leading strand TR | By lagging strand TR | ||||
| 1 (photoproducts 28 bp apart) | |||||||
| TK603 | uvrA6 | 0 | 8.3 ± 0.7 | 8.3 | 0 | 0 | 155 |
| 4 | 32.5 ± 4.2 | 14.8 | 7.3 | 10.4 | 287 | ||
| ES201 | uvrA6 recA938 | 0 | 8.8 ± 1.9 | 8.8 | 0 | 0 | 72 |
| ES200 | uvrA6 recF349 | 0 | 6.1 ± 1.2 | 6.1 | 0 | 0 | 50 |
| RW540 | uvrA6 lexA51 | 0 | 11.2 ± 1.4 | 8.3 | 1.7 | 1.2 | 132 |
| 4 | 32.7 ± 4.7 | 11.9 | 10.8 | 10.0 | 171 | ||
| AO6 | uvrA6 lexA51 recA306 | 0 | 9.4 ± 1.4 | 9.4 | 0 | 0 | 56 |
| AO12 | uvrA6 lexA51 recF400 | 0 | 7.5 ± 0.8 | 7.0 | 0.2 | 0.3 | 70 |
| AO1 | uvrA6 mutS205 | 0 | 13.6 ± 4.5 | 13.6 | 0 | 0 | 53 |
| 4 | 30.4 ± 4.7 | 9.8 | 10.3 | 10.3 | 106 | ||
| 2 (photoproducts 30 bp apart) | |||||||
| TK603 | uvrA6 | 0 | 5.9 ± 0.9 | 5.9 | 0 | 0 | 46 |
| 4 | 19.8 ± 3.6 | 5.8 | 5.2 | 8.8 | 99 | ||
| ES201 | uvrA6 recA938 | 0 | 7.5 ± 1.6 | 7.5 | 0 | 0 | 53 |
| ES200 | uvrA6 recF349 | 0 | 6.2 ± 1.1 | 6.2 | 0 | 0 | 48 |
| RW540 | uvrA6 lexA51 | 0 | 8.1 ± 1.9 | 5.3 | 1.3 | 1.5 | 122 |
| 4 | 23.8 ± 6.6 | 5.8 | 7.5 | 10.5 | 55 | ||
| AO6 | uvrA6 lexA51 recA306 | 0 | 9.7 ± 2.7 | 9.7 | 0 | 0 | 34 |
| AO12 | uvrA6 lexA51 recF400 | 0 | 8.2 ± 2.3 | 8.2 | 0 | 0 | 50 |
The numbers of independent experiments per strain for orientation 1 were as follows: TK603, 24 and 20; ES201, 12; ES200, 10; RW540, 24 and 22; AO6, 11; AO12, 8; AO1, 16 and 14. The numbers of independent experiments per strain for orientation 2 were as follows: TK603, 15 and 13; ES201, 12; ES200, 13; RW540, 10 and 10; AO6, 4; AO12, 4. The number of colonies counted in each experiment (control plasmid) ranged from 615 to 30,167.
Number of transformants from which plasmids were extracted and sequenced to determine the type of event (recombination or TR on a leading or lagging strand) that allowed completion of replication.
Completion of plasmid replication by a recombination process seems to be much less probable when the photoproducts are placed in orientation 2 because, unlike in orientation 1, it appears that the interphotoproduct region cannot be replicated (Fig. 1), which would preclude recombination. Surprisingly, recombination not only occurs, but on average its frequency is about 73% of that in orientation 1 (Table 3); for orientation 1, the average of recombination frequencies in all strains except AO1 is 9.3%, whereas the comparable frequency for orientation 2 is 6.8%. In uninduced cells of TK603, a recombination frequency of 5.9% was observed, compared to 8.3% with photoproducts in orientation 1. Moreover, as with orientation 1, the recombination mechanism used does not employ RecA or RecF, as shown by the results with strains ES201 and ES200 (7.5 and 6.2% recombination, respectively, compared to 8.8 and 6.1% with orientation 1). Again, as observed with photoproducts in orientation 1, this recombination was constitutive; recombination frequencies in RW540 (uvrA6 lexA51), AO6 (uvrA6 lexA51 recA306), and AO12 (uvrA6 lexA51 recF400) were 5.3, 9.7, and 8.2%, respectively. With the photoproducts in orientation 1, the corresponding frequencies were 8.3, 9.4, and 7.0%. We therefore conclude that E. coli possesses a largely constitutive recombination mechanism that does not employ RecA or RecF and which facilitates the completion of replication when this is blocked on both DNA strands.
In photoproduct orientation 2, the RecA- and RecF-independent recombination mechanism depends partly on PriA and RecG but not on RuvABC or polymerase II (Pol II); in orientation 1, however, none of these genes appears to have a significant influence.
We investigated the possible role of other gene products in the RecA- and RecF-independent recombination mechanism by transforming isogenic derivatives of TK603 that were deficient in PriA, RecG, RuvAB, RuvC, or PolB with plasmids carrying photoproducts in each orientation (Table 4). With respect to orientation 1, none of these strains exhibited recombination frequencies that were significantly different from the frequency in uninduced cells of TK603 (8.3%), though the frequency in the uvrA6 priA2 strain AO13 appeared somewhat low. Nevertheless, even if a better estimate of the recombination frequency is obtained by averaging all of the data for this orientation in Table 3, the uvrA6 priA2 strain still retains >50% of this value (5.5/9.3 = 59%). A very different result was found with orientation 2. In this case, recombination frequencies in both the uvrA6 priA2 strain AO13 and the uvrA6 recG263 strain AO7 were significantly lower than in TK603 (2.4 and 1.9 versus 5.9%; P ≪ 0.01; P ≪ 0.01), indicating that 59 and 68% of the recombination depended on the activities of PriA and RecG, respectively. If a better estimate of the wild-type recombination frequency is obtained by averaging all of the data for orientation 2 in Table 3, these values increase to 65 and 72% (6.8−2.4/6.8 = 65%; 6.8−1.9/6.8 = 72%). In both orientations, isogenic derivatives of TK603 deficient in RuvAB, RuvC, or PolB give results very similar to those obtained with TK603 (Table 4), suggesting that these gene products do not play a role in recombinational bypass of replication blocks observed in this experimental system. Thus, we conclude that the recombination mechanism used in orientation 2 partly depends on PriA and RecG activities though not on those of RuvABC or Pol II, whereas for recombination in orientation 1 none of these activities are significantly involved.
TABLE 4.
Total fraction of plasmids replicated and percent replicated following a recombination or TR event in isogenic excision-deficient (uvrA6) strains of E. coli carrying recG, ruvC, ruvA, priA, and polB mutations.
| Orientation and strain | Relevant genotype | Amt of UV to cells (J/m2) | Plasmids replicated (%)
|
Total no. of transformants sequencedb | |||
|---|---|---|---|---|---|---|---|
| Totala | By recombination | By leading strand TR | By lagging strand TR | ||||
| 1 (photoproducts 28 bp apart) | |||||||
| TK603 | uvrA6 | 0 | 8.3 ± 0.7 | 8.3 | 0 | 0 | 155 |
| 4 | 32.5 ± 4.2 | 14.8 | 7.3 | 10.4 | 287 | ||
| AO-7 | uvrA6 recG263 | 0 | 7.3 ± 1.1 | 7.3 | 0 | 0 | 62 |
| AO-10 | uvrA6 ruvC67 | 0 | 9.7 ± 1.8 | 9.7 | 0 | 0 | 36 |
| AO-11 | uvrA6 ruvA60 (polar) | 0 | 11.6 ± 1.4 | 11.6 | 0 | 0 | 30 |
| AO-13 | uvrA6 priA2 | 0 | 6.0 ± 1.0 | 5.5 | 0.4 | 0.1 | 129 |
| AO-14 | uvrA6 ΔpolB | 0 | 11.3 ± 1.6 | 11.3 | 0 | 0 | 24 |
| 4 | 31.3 ± 4.6 | 8.7 | 10.2 | 12.4 | 41 | ||
| 2 (photoproducts 30 bp apart) | |||||||
| TK603 | uvrA6 | 0 | 5.9 ± 0.9 | 5.9 | 0 | 0 | 46 |
| 4 | 19.8 ± 3.6 | 5.8 | 5.2 | 8.8 | 99 | ||
| AO-7 | uvrA6 recG263 | 0 | 1.9 ± 0.4 | 1.9 | 0 | 0 | 101 |
| AO-10 | uvrA6 ruvC67 | 0 | 5.6 ± 1.7 | 5.6 | 0 | 0 | 72 |
| AO-11 | uvrA6 ruvA60 (polar) | 0 | 5.1 ± 2.2 | 5.1 | 0 | 0 | 33 |
| AO-13 | uvrA6 priA2 | 0 | 2.4 ± 0.5 | 2.4 | 0 | 0 | 73 |
| AO-14 | uvrA6 ΔpolB | 0 | 5.9 ± 1.7 | 5.9 | 0 | 0 | 14 |
| 4 | 14.8 ± 4.9 | 4.8 | 5.2 | 4.8 | 56 | ||
The numbers of independent experiments per strain for orientation 1 were as follows: TK603, 24 and 20; AO7, 7; AO10, 4; AO11, 3; AO13, 11; AO14, 3 and 3. The numbers of independent experiments per strain for orientation 2 were as follows: TK603, 15 and 13; AO7, 10; AO10, 5; AO11, 3; AO13, 12; AO14, 2 and 2.
Number of transformants from which plasmids were extracted and sequenced to determine the type of event (recombination or TR on a leading or lagging strand) that allowed completion of replication.
Decreasing the interphotoproduct distance reduces the frequency of recombination about equally for each orientation.
The occurrence of recombination in orientation 2, and moreover at a frequency only marginally lower than that in orientation 1, was unexpected because orientation 2 appeared to provide no substrate that would promote this event. A possible solution to this puzzle might be that replication is reinitiated within the 30-bp interphotoproduct region on at least the lagging strand, at a site sufficiently close to the photoproducts to generate an overlapping DNA sequence that can engage in recombination. Since RNA primers are thought to be ∼9 to 14 ribonucleotides long (18, 32), recombination might be expected to be absent in orientation 2 if the interphotoproduct distance was reduced to 10 bp, because primers must presumably be greater than some minimum size to be effective. With orientation 1, however, reduction of the interlesion distance to 8 bp should allow some recombination, even if at a lower frequency. The interphotoproduct distance cannot be made identical in the two orientations because the T-T (6-4) photoadduct is placed 5 nucleotides from the 5′ end of the 11-mer used to construct the plasmid inserts and 4 nucleotides from the 3′ end. We found, however, that although recombination frequencies were appreciably reduced in both orientations, recombination was still observed in orientation 2 and indeed was slightly but consistently greater than in orientation 1, the reverse of the prediction (Table 5). Since recombination as detected by our method appears to depend absolutely on replication within the interphotoproduct region on at least one of the templates, such a result presumably implies that at least a minor subset of primers can be only a few ribonucleotides in length. As for the greater interphotoproduct spacing, the recombination observed is largely RecA and RecF independent.
TABLE 5.
Total fraction of plasmids replicated and percent replicated following a recombination or TR event in isogenic excision-deficient (uvrA6) strains of E. coli carrying recA or recF mutations
| Orientation and strain | Relevant genotype | Amt of UV to cells (J/m2) | Plasmids replicated (%)
|
Total no. of transformants sequencedb | |||
|---|---|---|---|---|---|---|---|
| Totala | By recombination | By leading strand TR | By lagging strand TR | ||||
| 1 (photoproducts 8 bp apart) | |||||||
| TK603 | uvrA6 | 0 | 0.7 ± 0.2 | 0.66 | 0.04 | 0.00 | 40 |
| 4 | 6.2 ± 1.4 | 0.56 | 2.76 | 2.88 | 111 | ||
| ES201 | uvrA6 recA938 | 0 | 0.5 ± 0.04 | 0.45 | 0.03 | 0.02 | 33 |
| ES200 | uvrA6 recF349 | 0 | 0.8 ± 0.1 | 0.75 | 0.05 | 0.00 | 30 |
| 2 (photoproducts 10 bp) apart) | |||||||
| TK603 | uvrA6 | 0 | 1.5 ± 0.6 | 1.41 | 0.05 | 0.05 | 33 |
| 4 | 6.5 ± 3.0 | 0.84 | 3.13 | 2.53 | 185 | ||
| ES201 | uvrA6 recA938 | 0 | 1.0 ± 0.4 | 0.93 | 0.03 | 0.03 | 30 |
| ES200 | uvrA6 recF349 | 0 | 0.9 ± 0.4 | 0.88 | 0.00 | 0.02 | 45 |
Three independent experiments per strain were conducted for both orientations and all strains. Total numbers of colonies counted for each strain (control plasmid) ranged from 6,661 to 34,293 with orientation 1 and from 4,315 to 18,254 with orientation 2.
Number of transformants from which plasmids were extracted and sequenced to determine the type of event (recombination or TR on a leading or lagging strand) that allowed completion of replication.
Almost all replicated plasmids are derived from a single transforming molecule and result in homogeneous sequences.
The sequence of the recombinants and the low ratio of plasmid molecules to competent cells used in the experiments strongly suggest that the recombination event took place between sister strands following partial replication of a single plasmid, as must be the case if transformation resulted from the uptake of one plasmid molecule. Evidence indicating that transformants rarely originated from more than one plasmid, and that interplasmid recombination could therefore at best be only a relatively rare event, was obtained by transforming TK603 with an equimolar mixture of photoproduct containing plasmid constructed with pES1 and pES2. These two vectors are identical except for the sequence of a base pair doublet flanking each side of the 80-mer/72-mer insertion site (see Materials and Methods), which constitutes markers that can be used to detect interplasmid recombination. Sequence analysis of a total of 112 recombinant plasmids, comprising 34 and 55 recovered from unirradiated and UV-irradiated TK603 cells, respectively, and 13 and 10 recovered from unirradiated and UV-irradiated RW540 cells, respectively, all contained the marker combinations found in either pES1 or pES2; no recombination between pES1 and pES2 markers was observed. Similarly, no recombination between these markers was observed in a total of 74 replicated plasmids deriving from translesion replication (UV-irradiated TK603, 23 leading strands, 33 lagging strands; UV-irradiated RW540, 1 leading strand, 6 lagging strands; unirradiated RW540, 6 leading strands, 5 lagging strands). Sequence analysis of 250 colonies obtained by transforming TK603 with an equimolar mixture of photoproduct-free pES1 and pES2 vectors also failed to detect any heterogeneity. Further, only a single type of plasmid appears to exist within most transformants, as shown by sequence analysis of plasmid DNA isolated from subclones of transformant colonies. Only a single sequence was found in an average of 24 subclones (range, 18 to 27) from each of six transformants, identified as arising from a recombination event in the interlesion region. Analysis of up to 12 subclones from a further five transformants of the same type also showed complete genetic homogeneity. In addition, genetic homogeneity was observed in the plasmids derived from six of seven transformants resulting from a translesion replication event, from which 12 to 17 subclones were analyzed. Presumably, plasmid replication following the generation of one lesion-free strand, either by recombination or translesion replication, is sufficiently rapid that products from the other template, or from other events, fail to contribute more than a very small fraction to the replicated population. The single exceptional, heterogeneous, transformant contained a mixed population of plasmids resulting from leading and lagging strand translesion replication events. This transformant came from UV-irradiated cells of the LexA-deficient strain RW540, in which the level of DNA polymerase V, the enzyme that is probably responsible for lesion bypass, is expected to be high.
TK603 does not contain the Rac cryptic prophage or the recET genes.
The products of the recET genes, carried on the Rac cryptic prophage, permit recombination between plasmids in recA-deficient cells (12). As a direct clonal derivative of AB1157 (11), a strain known not to carry the Rac prophage (10), TK603 is not expected to carry this defective lambdoid genome. We nevertheless used PCR to confirm that the recET genes, and the cryptic prophage itself, were in fact absent from this strain. No PCR product of appropriate size to indicate the existence of recET could be found using genomic DNA from TK603, even though this was found in JM101 and DHα5 and product from the dbpA internal control was found in all cases. Additionally, the generation of PCR product of predicted size from TK603 DNA, accomplished using primers designed to span the Rac prophage insertion site, indicates the absence of the prophage; if present, the size of the prophage would have precluded creation of a PCR product. Such evidence indicates that the RecA-independent recombination is not the result of RecET activities.
DISCUSSION
The goal of this investigation was to use plasmid constructs carrying a T-T (6-4) photoproduct at staggered positions in each strand of the DNA duplex to explore the possibility that E. coli possessed novel mechanisms for circumventing such serious impediments to replication. More specifically, we wished to investigate whether there was evidence for E. coli possessing a transient template-switching mechanism of the kind first proposed by Higgins, Kato and Strauss in 1976 (6) for mammalian cells and later proposed for E. coli (4, 5, 16, 19, 24). In addition, a mechanism of this kind has been reconstructed in vitro, using purified phage T4 proteins (8, 9). However, this model has been difficult to explore experimentally in vivo because the recombination event depends on informational, rather than physical, exchange between DNA strands and because it occurs between identical sister strands. To overcome this problem, we placed a double C-C mismatch opposite the T-T (6-4) photoproducts, which allowed the unambiguous identification of the three events observed in completely replicated plasmids: recombination within the interphotoproduct region, TR on the leading strand template, and TR on the lagging strand template. The plasmid construct employed is not intended to simulate the result of UV irradiation, since the close proximity of T-T (6-4) photoproducts is unlikely following such a treatment, but rather any circumstance in which both strands are damaged. Two-strand DNA damage has been inferred to occur in almost all E. coli cells in every division cycle (2, 22) and is produced by some agents, such as ionizing radiation (31). The presence of T-T (6-4) photoproducts in each strand of the DNA duplex in the plasmids used in our experiments has the potential to seriously compromise replication, since they almost completely block fork progression; almost no bypass of this lesion occurs in single-stranded phages unless the SOS regulon is induced (15).
Using this approach, we find that excision-repair-defective (uvrA6) E. coli can complete the replication of plasmids carrying closely opposed T-T (6-4) photoproducts in each strand by a recombination process which does not employ RecA protein and, in addition, is also largely independent of RecF. This process is largely constitutive and is sufficiently efficient to permit replication to be completed in about 8 to 10% of the plasmids, even though the T-T photoproducts are only 28 or 30 bp apart and there is no access to a separate, undamaged, duplex plasmid with which to recombine. These properties suggest that in this circumstance the mechanism responsible for the completion of replication is different from the well-investigated recombination-dependent DNA damage tolerance process first described by Rupp et al. (26), particularly with regard to the lack of a requirement for RecA protein. Two processes are now recognized, daughter-strand gap filling and double-strand end repair (14), and while the former depends on RecF and the latter depends on RecBC, they both employ RecA. Apart from its various regulatory roles, RecA-mediated strand invasion is an essential function in each of these processes, leading to the formation of Holliday junctions, branch migration, and the incorporation of template DNA into newly synthesized strands after resolution of the Holliday junctions by a junction-specific endonuclease, the resolvase. The absence of a requirement for RecA in the recombination-dependent DNA damage tolerance phenomenon that we observe suggests that it does not involve these processes.
To investigate this issue further, and in general to examine the possible involvement of other gene products in the RecA-independent process, we transformed isogenic mutant strains deficient in RecG, RuvAB, RuvC, PriA, or PolII (Table 1) with constructs carrying lesions in orientation 1 or 2 (Fig. 1). Each of these proteins has been implicated in restoration of replication at blocked forks. RecG is a helicase that is thought to facilitate fork regression and the formation of a chicken foot structure, allowing the elongation of the previously blocked strand (19). However, recovery of DNA synthesis after UV irradiation has been found to occur normally in RecG-deficient cells, perhaps indicating that fork regression is not required, that the process requiring RecG is only a minor mechanism in the recovery, or that another helicase can also perform this function (3). The RuvAB complex interacts directly with the Holliday junction and drives branch migration with its helicase activity (30), whereas RuvC is the resolvase that cuts Holliday junctions (30, 33). PriA has come to be understood as an important link between replication and recombination in that the restart of stalled replication forks requires PriA to load the DnaB replicative helicase at branched DNA structures (27). Further, it is proposed that RecG and PriA helicase activities, together with the primosome assembly function of PriA, may rescue stalled replication forks independently of RecBCD and RuvABC (7). PolB encodes the catalytic subunit of Pol II, which is believed to be involved in replication restart in cells exposed to UV irradiation (25).
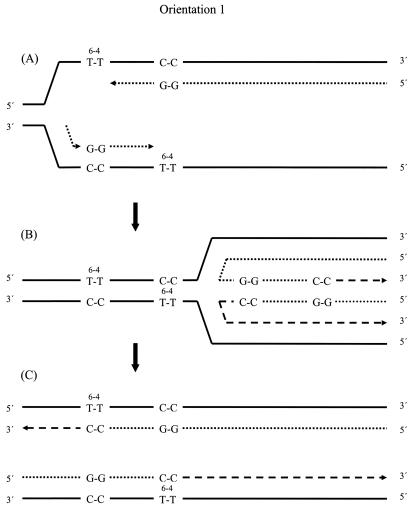
Since none of the mutant strains has a recombination frequency that is significantly lower than the wild-type frequency, none of these gene products appears to play an appreciable role in generating recombinants when the photoproducts are in orientation 1. This suggests that, with photoproducts in this orientation, the recombination mechanism does not employ strand invasion, the formation of Holliday junctions, or the incorporation of template DNA in the nascent strands. The data are therefore consistent with a transient template strand-switching model, though the specific process employed is not yet known. However, following the stalling of the replication fork (Fig. 2A), a possible mechanism might involve fork regression, the annealing together of nascent strands, and the extension of each of the 3′OH ends by at least a few nucleotides (Fig. 2B). Following the reannealing of the nascent strands back to their original templates, both nascent strands possess the sequence motif, indicating the occurrence of recombination, and replication can be completed (Fig. 2C). Such recombination depends on informational, rather than physical, exchange between DNA segments. A subsequent round of replication will generate lesion-free plasmids. The lesion-containing templates presumably contribute very little to further plasmid replication. There is no obvious role for RecA protein in this process, though it might employ as-yet-unidentified proteins that promote the various melting and annealing steps that the model proposes. Although the model depicted in Fig. 2 entails fork regression, it can also be drawn without this feature, in a fashion analogous to the model depicted in Fig. 3. In view of the uncertainties surrounding the in vivo role of RecG, it is unclear which of these depictions is a better reflection of cellular events, and the observation (Table 4) that recombination in orientation 1 occurs independently of RecG is not decisive in the choice of models.
FIG. 2.
Model for production of recombinants in orientation 1. (A) Proposed replication intermediate at stalled forks. (B) Fork regression, annealing of nascent strands, and formation of a chicken foot structure, which permits continued synthesis beyond the region of the blocking lesions. (C) Reannealing of nascent strands to their original templates. In the following round of plasmid replication, each nascent strand will give rise to a lesion-free duplex plasmid. Continuous lines, original templates; dotted lines, nascent strand sequences synthesized before stalling of the replication fork; dashed lines, nascent strand sequences synthesized after resumption of replication.
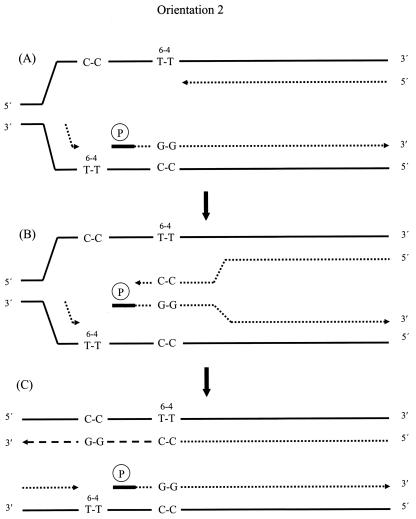
FIG. 3.
Model for production of recombinants in orientation 2. (A) Proposed replication intermediate at stalled forks, accompanied by synthesis of a primer within the interlesion region on the lagging strand, and its extension. (B) Annealing of nascent strands in the absence of replication fork regression, which permits limited extension of the leading strand. (C) Reannealing of nascent strands to their original templates. In the following round of plasmid replication, the nascent leading strand will give rise to a lesion-free duplex plasmid. The nascent lagging strand is likely to remain incompletely replicated and incapable of generating a lesion-free plasmid. However, this might be achieved by an additional recombination event between the downstream Okazaki fragment and the nascent leading strand. Continuous lines, original templates; dotted lines, nascent strand sequences synthesized before stalling of the replication fork; dashed lines, nascent strand sequences synthesized after resumption of replication. The thick bar and circled P indicate the RNA primer.
At first sight, neither of the above models appears to be applicable when the photoproducts are in orientation 2, because in this circumstance replication within the interphotoproduct region appears to be impossible on either strand (Fig. 1), and thus no structures that might promote recombination appear to exist. Nevertheless, recombination occurs with this photoproduct orientation at a frequency that is only about 27% lower than that for orientation 1. Recombination-promoting structures might, however, be generated by the initiation of priming on the lagging strand template close to the T-T (6-4) photoproduct (Fig. 3A). Extension from this primer could then provide a nascent strand with which its complementary nascent strand might pair without fork regression. Fork regression appears unlikely in lesion orientation 2 because it would require the displacement of the downstream Okazaki fragment. Annealing of the two nascent strands permits elongation of the leading strand which, although limited, is nonetheless sufficient to extend beyond the lesion site and to form the sequence marking the recombination event (Fig. 3B). Once reannealed to its original template, the leading strand template can then be fully replicated (Fig. 3C), and a subsequent round of replication will generate a lesion-free plasmid. Replication on the lagging strand template might remain incomplete, or the downstream Okazaki fragment might engage in a second recombination event with the nascent leading strand which, since it would occur outside of the genetically marked region, would not be detectable in our system. The substantial dependence of recombination on PriA in this lesion orientation is perhaps because this protein facilitates the priming event on the lagging strand. RecG, on which recombination also depends substantially, may be required for the displacement of the RNA primed end of the lagging strand Okazaki fragment that is initiated in the interphotoproduct region, since this enzyme is effective on RNA/DNA hybrids (29). An attempt to gain evidence for the existence of priming within the interphotoproduct region by reducing its length to 10 nucleotides failed to provide a decisive result. Based on the assumptions that replication in this region is an absolute requirement for recombination to occur, that primers are 9 to 14 ribonucleotides in length (18, 32), and that the primer is unlikely to abut the lagging strand template photoproduct, we predicted that no recombination would occur in photoproduct orientation 2, although a low frequency would be found in orientation 1. We found, on the contrary, that recombination frequencies in orientation 2 were slightly, but consistently, greater than in the other orientation. Such a result most probably implies that a minor fraction of primers are appreciably shorter than 9 ribonucleotides, allowing the continuing DNA to overlap the C-C sequence that corresponds to the location of the leading strand template T-T (6-4) photoadduct. Following the reannealing of the nascent leading strand back to its original template, the addition of only two guanine residues would probably be sufficient for further extension. Because the T-T (6-4) photoadduct substantially distorts DNA structure, such extension would presumably initially require the activity of DNA polymerase V rather than DNA polymerase III, but this would not be detectable by the present method. If the DNA extended a few additional nucleotides beyond the added G-G sequence, DNA polymerase III should alone be capable of elongation.
The recombination observed in either photoproduct orientation appears to be intramolecular and taking place between sister DNA duplexes generated by partial replication of a single plasmid rather than intermolecular and occurring between two plasmid molecules that might be introduced simultaneously into a transformed cell. No interplasmid recombinants were detected following transformation with an equimolar mixture of two plasmids that differed by sequence markers flanking the photoproduct sites but which were otherwise identical to one another. Similarly, analysis of subclones from individual transformants showed that they were homogeneous and carried plasmids of identical sequence, providing no evidence for the uptake and propagation of more than one plasmid in a given transformant. Although the mismatches may well be a substrate for mismatch repair enzymes, the exchanges observed clearly result from recombination and not from mismatch repair, because they occurred as frequently in a mismatch-deficient strain (Table 3). In summary, we conclude that the completion of plasmid replication by the RecA-, RecF-, RuvABC-, and DNA polymerase II-independent recombination process that we observe is likely to entail a copy choice mechanism in which the nascent strands, whose elongation is blocked by the photoproducts, transiently anneal with one another. It remains to be seen whether such a mechanism can be employed on the E. coli genome, where replication is bidirectional rather than unidirectional, but at first sight there appear to be no reasons why it should not be.
Acknowledgments
This work was supported by grant GM32885 to C.W.L. from the National Institutes of Health.
We thank Roger Woodgate for strains and advice on building strains used in this study; Robert Lloyd, Steven Sandler, and Mary Berlyn (E. coli Genetic Stock Center) for strains; and Grace Poh, Candace Brayfield, Michelle Coleman, Jessica Barzideh, Vanessa Moore, Erin Zahradnik, Erin Bressler, Michelle Villasmil, and Kimberly Colern for their help in the preparation and sequencing of replicated plasmids.
REFERENCES
- 1.Banerjee, S. K., R. B. Christensen, C. W. Lawrence, and J. E. LeClerc. 1998. Frequency and spectrum of mutations produced by a single cis-syn thymine-thymine dimer in a single-stranded vector. Proc. Natl. Acad. Sci. USA 85:8141-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherrat, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 3.Donaldson, J. R., C. T. Courcelle, and J. Courcelle. 2004. RuvAB and RecG are not essential for recovery of DNA synthesis following UV-induced DNA damage in Escherichia coli. Genetics 166:1631-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Echols, H. 1982. Mutation rate: some biological and biochemical considerations. Biochimie 64:571-575. [DOI] [PubMed] [Google Scholar]
- 5.Gregg, A. V., P. McGlynn, R. P. Jaktaji, and R. G. Lloyd. 2002. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell 9:241-251. [DOI] [PubMed] [Google Scholar]
- 6.Higgins, N. P., K. Kato, and B. Strauss. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417-425. [DOI] [PubMed] [Google Scholar]
- 7.Jaktaji, R. P., and R. G. Lloyd. 2003. PriA supports two distinct pathways for replication restart in UV-irradiated Escherichia coli cells. Mol. Microbiol. 47:1091-1100. [DOI] [PubMed] [Google Scholar]
- 8.Kadyrov, F. A., and J. W. Drake. 2003. Properties of bacteriophage T4 proteins deficient in replication repair. J. Biol. Chem. 278:25247-25255. [DOI] [PubMed] [Google Scholar]
- 9.Kadyrov, F. A., and J. W. Drake. 2004. UvsX recombinase and Dda helicase rescue stalled bacteriophage T4 replication forks in vitro. J. Biol. Chem. 279:35735-35740. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser, K., and N. E. Murray. 1979. Physical characterization of the “Rac prophage” in E. coli K12. Mol. Gen. Genet. 175:159-174. [DOI] [PubMed] [Google Scholar]
- 11.Kato, T., and Y. Shinoura. 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet. 156:121-131. [DOI] [PubMed] [Google Scholar]
- 12.Kolodner, R., S. D. Hall, and C. Luisi-DeLuca. 1994. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol. Microbiol. 11:23-30. [DOI] [PubMed] [Google Scholar]
- 13.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16:373-384. [DOI] [PubMed] [Google Scholar]
- 14.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeClerc, J. E., A. Borden, and C. W. Lawrence. 1991. The thymine-thymine pyrimidine-pyrimidinone (6-4) ultraviolet light photoproduct is highly mutagenic and specifically induces 3′ thymine-to-cytosine transitions in Escherichia coli. Proc. Natl. Acad. Sci. USA 88:9685-9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louarn, J. M., J. Louarn, V. François, and J. Patte. 1991. Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J. Bacteriol. 173:5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovett, S. T., R. L. Hurley, V. A. Sutera, R. H. Aubuchon, and M. A. Lebedeva. 2002. Crossing over between regions of limited homology in Escherichia coli: RecA-dependent and RecA-independent pathways. Genetics 160:851-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marians, K. J. 1992. Prokaryotic DNA replication. Annu. Rev. Biochem. 61:673-719. [DOI] [PubMed] [Google Scholar]
- 19.McGlynn, P., and R. G. Lloyd. 1999. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 27:3049-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101:35-45. [DOI] [PubMed] [Google Scholar]
- 21.McGlynn, P., and R. G. Lloyd. 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 98:8227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel, B., M.-J. Flores, E. Viguera, G. Crompone, M. Seigneur, and V. Bidnenko. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napolitano, R., R. Janel-Bintz, J. Wagner, and R. P. P. Fuchs. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV, and Pol V) are involved in induced mutagenesis. EMBO J. 19:6259-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postow, L., C. Ullsperger, R. W. Keller, C. Bustamante, A. V. Vologodskii, and N. R. Cozzarelli. 2001. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 276:2790-2796. [DOI] [PubMed] [Google Scholar]
- 25.Rangarajan, S., R. Woodgate, and M. F. Goodman. 1999. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc. Natl. Acad. Sci. USA 96:9224-9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rupp, W. D., C. E. Wilde III, D. L. Reno, and P. Howard-Flanders. 1971. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J. Mol. Biol. 61:25-44. [DOI] [PubMed] [Google Scholar]
- 27.Sandler, S. J., and K. J. Marians. 2000. Role for PriA in replication fork reactivation in Escherichia coli. J. Bacteriol. 182:9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szekeres, E. S., Jr., R. Woodgate, and C. W. Lawrence. 1996. Substitution of mucAB or rumAB for umuDC alters the relative frequencies of the two classes of mutations induced by a site-specific T-T cyclobutane dimer and the efficiency of translesion DNA synthesis. J. Bacteriol. 178:2559-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent, S. D., A. A. Mahdi, and R. G. Lloyd. 1996. The RecG branch migration protein of Escherichia coli dissociates R-loops. J. Mol. Biol. 264:713-721. [DOI] [PubMed] [Google Scholar]
- 30.West, S. C. 1996. The RuvABC proteins and Holliday junction processing in Escherichia coli. J. Bacteriol. 178:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokoya, A., S. M. T. Cunniffe, and P. O'Neill. 2002. Effect of hydration on the induction of strand breaks and base lesions in plasmid DNA films by γ-radiation. J. Am. Chem. Soc. 124:8859-8866. [DOI] [PubMed] [Google Scholar]
- 32.Zechner, E. L., C. A. Wu, and K. J. Marians. 1992. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J. Biol. Chem. 267:4045-4053. [PubMed] [Google Scholar]
- 33.Zerbib, D., C. Mézard, H. George, and S. C. West. 1998. Coordinated actions of RuvABC in Holliday junction processing. J. Mol. Biol. 281:621-630. [DOI] [PubMed] [Google Scholar]