Abstract
Duplication of transporter genes is apparent in the genome sequence of the hyperthermophilic bacterium Thermotoga maritima. The physiological impacts of these duplications are not well understood, so we used the bacterium's two putative maltose transporters to begin a study of the evolutionary relationship between a transporter's function and the control of expression of its genes. We show that the substrate binding proteins encoded by these operons, MalE1 and MalE2, have different substrate specificities and affinities and that they are expressed under different growth conditions. MalE1 binds maltose (dissociation constant [KD], 24 ± 1 μM), maltotriose (KD, 8 ± 0.5 nM), and β-(1→4)-mannotetraose (KD, 38 ± 1 μM). In contrast, MalE2 binds maltose (KD, 8.4 ± 1 μM), maltotriose (KD, 11.5 ± 1.5 μM), and trehalose (KD, 9.5 ± 1.0 μM) confirming the findings of Wassenberg et al. (J. Mol. Biol. 295:279-288, 2000). Neither protein binds lactose. We examined the expression of these operons at both the transcriptional and translational levels and found that MalE1 is expressed in cells grown on lactose or guar gum and that MalE2 is highly expressed in starch- and trehalose-grown cells. Evidence is provided that malE1, malF1, and perhaps malG1 are cotranscribed and so constitute an operon. An open reading frame encoding a putative transcriptional regulatory protein adjacent to this operon (TM1200) is also up-regulated in response to growth on lactose. These evolutionarily related transporter operons have diverged both in function and expression to assume apparently different physiological roles.
Annotation of the complete genome sequence of the hyperthermophilic, heterotrophic bacterium Thermotoga maritima indicates that 24% of the bacterium's open reading frames (ORFs) are most closely related to archaeal sequences (16). Subsequent studies have provided evidence that these genes were acquired by horizontal gene transfer (HGT). The closest known relative of T. maritima, Thermotoga species RQ2, acquired sugar ABC transporter and polysaccharide hydrolase genes independently of T. maritima, and this acquisition presumably provides selective nutritional advantages to this organism in its natural habitats (18). Intradomain gene acquisition or even gene duplication may confer novel selective advantages as well. This may be evident in the two putative maltose ABC transporters encoded in distant locations in the T. maritima genome (16). The amino acid sequences of the individual components of both transporters are very similar, suggesting a common evolutionary history (16). The transporter gene clusters (putative operons) may have arisen through operon duplication or horizontal acquisition of a second, orthologous operon by the ancestor of T. maritima. Evidence of a similar intradomain HGT of ABC transporter genes was found in two closely related archaea, Thermococcus litoralis and Pyrococcus furiosus (4). Regardless of their mechanism of duplication, the functions of these two T. maritima ABC transporters must confer distinct selective advantages since both have been retained.
An organism will acquire a new catabolic trait after acquisition of new genes only if the genes are expressed and regulated in concert with other catabolism genes. Consequently, the evolution of the regions upstream of new genes and the binding of transcription factors to these regions are important processes that dictate the addition of novel traits. The processes by which new regulatory connections are made following gene acquisition are not well understood. An examination of the role of gene duplication in the evolution of transcriptional regulators and their regulated operons in Escherichia coli suggests that regulatory networks often evolve through duplications and that about half of those examined evolved new regulatory connections not present in the ancestral regulatory element (30). Those studies showed that new connections can arise either from new DNA binding activities of duplicated transcription factor proteins or from changes in upstream regulatory sites that allow other proteins to bind there. For operons like the T. maritima mal operons that originated either through operon duplication or horizontal acquisition of an orthologous operon, the new operon could fall under the control of a new promoter downstream of which it recombined. This phenomenon has been observed in Lactococcus lactis, where lac genes acquired from a gram-negative organism fell under the control of an apparently preexisting gal promoter (31). Acquisition of a new ABC transporter may cause selection for an altered periplasmic binding protein that responds to the same ligand as the transcription factor that controls its expressions. It has been postulated that periplasmic binding proteins and their cognate transcriptional repressors acquire their specificities for the same ligands through independent mutations rather than through domain swapping (7).
We sought to learn about the divergence of these mal operons in terms of their transport functions and their transcriptional regulatory controls. To do so, we have determined the substrate affinities of their corresponding binding proteins and compared their transcriptional and translational expression patterns. Maltose transport has been the subject of previous investigations into the processes of sugar transport and its regulation in T. maritima. Maltose binding activities in periplasmic extracts of T. maritima cells grown on different sugars have been measured (15). MalE2, one of two putative periplasmic maltose binding proteins identified in the T. maritima genome sequence, has been shown to bind maltose, maltotriose, and trehalose (32). In this report, we describe how we have determined the substrate affinities of the other maltose binding protein, MalE1. We show that, although similar in sequences, MalE1 and MalE2 have different substrate binding specificities. We also present transcriptional and proteomic analyses showing that each binding protein is expressed under different conditions. Based on our findings, we conclude that these transporters have assumed different cellular functions while still retaining some features of their common origin.
MATERIALS AND METHODS
Chemicals.
Unless otherwise noted, all sugars were a minimum of 99% pure. Glucose, galactose, mannose, maltose, lactose, raffinose, and melibiose were of high-performance liquid chromatography grade and were obtained from Sigma Chemical Co. Maltotriose, trehalose, and α-(1→3)-arabinogalactoside were supplied by ICN Biomedicals. 61-Galactosyl-mannobiose, 63-64-digalactosyl mannopentose, β-(1→4)-mannobiose, β-(1→4)-mannotriose, and β-(1→4)-mannotetraose purified from carob galactomannan hydrolysates were from Megazyme and were 95% pure.
Organism and growth conditions.
T. maritima MSB8T (DSM 3109T) was obtained from the Deutsche Sammlung von Mikroorganismen and Zellkulturen, Braunschweig, Germany. Cells were grown on defined basal media as described previously (15). Sugars used as carbon and energy sources were filter sterilized and added separately to media at a final concentration of 5 g/liter. Typically, cells were grown to an optical density at 600 nm (OD600) of 0.2 at 77°C in 300 ml of anoxic defined medium. The cells were harvested by centrifugation at 5,500 × g for 20 min and then washed in buffer (30 mM KCl, 2 mM MgSO4, 40 mM potassium phosphate [pH 7.0]) before periplasmic protein and mRNA extractions.
Cloning, expression, and purification of MalE1 and MalE2.
The periplasmic binding protein-encoding gene malE1 was amplified from T. maritima genomic DNA by using synthetic oligonucleotide primers 5′ CATATGCAACCGAAACTCACC 3′ and 5′ CTCGAGTTACTGAATCTGAGC 3′ for PCR using Pfu DNA polymerase (Invitrogen). The NdeI and XhoI sites in the primers are underlined. The PCR primers were designed so that the nucleotide sequence encoding the first 19 amino acids, including the putative signal peptide, was not incorporated into the PCR product (22). Initially, the PCR product was cloned into the pGEMeasy vector (Promega) and then digested with NdeI and XhoI. The 1,200-bp product was subcloned into the pET15b expression vector previously digested with the same enzymes. The malE2 gene, with its signal peptide-encoding sequence removed, was previously cloned into an expression vector. The resulting plasmid, pET21c-MalE2, was kindly provided by W. Liebl (32). The malE2 gene was removed from this plasmid by digestion with restriction endonucleases NdeI and BamHI. The resulting 1,200-bp product was cloned into pET15b previously digested with these enzymes. A single colony of the expression host E. coli strain BL21(DE3) harboring either pET15b-malE1 or pET15b-malE2 was inoculated into 10 ml of Luria-Bertani medium with ampicillin (50 μg/ml) and incubated at 37°C to an OD600 of 0.3. The cells were harvested by centrifugation, and the cell pellets were inoculated into fresh medium (100 ml of Luria-Bertani, 50 μg of ampicillin/ml) and incubated with shaking at 37°C. When the culture reached an OD600 of 0.5, expression was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside; to 1 mM) and cells were further incubated for 4 h at 37°C. The cells were then harvested by centrifugation (Beckman GA10 instrument; 5,500 × g) for 20 min. The soluble recombinant proteins were purified by nickel chelation chromatography using Ni-nitrilotriacetic acid resin as per the instructions of the manufacturer (Novagen). The pure proteins were dialyzed against 15 liters of 50 mM sodium phosphate, pH 7.5, at 4°C to remove imidazole. The dialyzed samples were stored at 4°C until needed.
Determination of protein concentrations.
The molar extinction coefficients of MalE1 and MalE2 were determined from their predicted amino acid sequences (8). Protein concentrations were determined by measuring the absorbance of solutions at 280 nm and applying the calculated molar extinction coefficients (44,520 M−1 cm−1 for MalE1 and 45,800 M−1 cm−1 for MalE2).
Fluorescence spectroscopy.
All fluorescence measurements were performed using an SLM Aminco-Bowman 2 spectrofluorimeter. During each experiment, the temperature of the cuvette was held constant at 20°C. Fluorescence emission spectra were measured at an excitation wavelength of 280 nm. Emission intensities were measured over the wavelength range of 300 to 360 nm in the presence (final concentration, 1 mM) and absence of the carbohydrate. The excitation and emission slit widths were 1 and 8 nm, respectively. The dissociation constants were measured by adding increasing amounts of selected carbohydrates into a stirred cuvette at 20°C. In a typical experiment, 3 μl of different stock solutions of the carbohydrate was added to 1.5 ml of a 0.3 μM protein solution (50 mM sodium phosphate, 300 mM sodium chloride [pH 7.5]). After the addition, the sample was stirred for 3 min to reach equilibrium and then the fluorescence intensity at the predetermined emission wavelength was recorded for 2 min. The mean intensity signal for 2 min after each addition was corrected for dilution (5). The program KaleidaGraph was used to plot relative fluorescence (F/Fo) against total carbohydrate concentration (Lo). The dissociation constant (KD) was determined by nonlinear fitting of the data to the following equation: F = Fo + ΔF/2Po{(KD + Po + Lo) − [(KD + Po + Lo)2 − 4LoPo]1/2}, where F is the measured fluorescence of the protein in the presence of the ligand, Fo is the fluorescence of the ligand-free protein, ΔF is the change in protein fluorescence at saturation with the ligand, Po and Lo are the total concentrations of protein and ligand, respectively, and KD is the dissociation constant. The data were fitted using the Marquardt-Levenberg algorithm.
Northern hybridization.
RNA was extracted from approximately 109 cells by using the QIAGEN RNeasy mini kit as per the manufacturer's instructions. Total RNA (5 μg/lane) was resolved on FMC Reliant precast gels. RNA was transferred by capillary transfer onto Hybond (Amersham) nylon membranes in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membranes were hybridized with digoxigenin-labeled probes. Labeling, hybridization, and detection were done as per the Boehringer Mannheim Genius system instructions. The hybridization signals were detected by exposing the blots to X-ray films. The signal intensities on scanned images were measured using Bio-Rad Molecular Analyst software. The probes for malE1 and malE2 were derived by PCR amplification of the genes and gave products of 350 and 347 bp. The primers for malE1 were 5′ CCTTTCATTTTCGGATACGG 3′ and 5′ AGTTTGTTTGGGGATTTTGC 3′, and the primers for malE2 were 5′ TGACATCCTTCAGAAACTCG 3′ and 5′ CACCTTTGACTGCTCCTTCG 3′. These hybridization probes do not cross-hybridize between the malE genes as shown by hybridization to restriction digests of chromosomal DNA (data not shown). Equal amounts of RNA were applied to each lane of the gels. This was confirmed by stripping each blot with boiling 0.1% sodium dodecyl sulfate (SDS) and rehybridizing with labeled 16S rRNA gene probe. The16S rRNA gene probe was prepared by PCR amplification of the gene with the primers 5′ CACAAGGGCACTGAGACACG 3′ and 5′ CCCTACACCAGCAGTTCCGT 3′.
Analysis of expression of MalE1 and MalE2 by two-dimensional (2-D) gel electrophoresis.
A freeze-thaw procedure was used to selectively release periplasmic proteins from T. maritima cells grown on different sugars (glucose, maltose, lactose, trehalose, starch, and guar gum) as described previously (15). Periplasmic extract resulting from 0.5 g (wet weight) of cell pellet was dialyzed against 3 liters of 10 mM Tris · HCl (pH 7) to remove magnesium chloride. The dialyzed periplasmic extract was concentrated by ultrafiltration (10-kDa cutoff), and the protein concentration in the dialysate was estimated by the Bradford method (2). The 50 μg of resulting protein was solubilized in 125 μl of solubilization buffer {8 M urea, 65 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 40 mM dithiothreitol, 0.2% (vol/vol) Bio-Lyte 3/10 ampholyte, 2 mg of bromophenol blue/liter}. The sample was used to rehydrate overnight 7-cm ReadyStrip immobilized pH gradient strips, pI 4 to 7 (Bio-Rad). Isoelectric focusing with a Protean isoelectric focusing cell (Bio-Rad) was performed in three steps as follows: step 1, 0 to 250 V for 15 min in rapid ramp mode; step 2, 250 V to 8 kV for 1 h in slow ramp mode; and step 3, 8 kV for 10 kV · h in rapid ramp mode. After focusing, the strips were suspended in equilibration buffer 1 (6 M urea, 0.375 M Tris · HCl, 2% SDS, 20% glycerol, 2% dithiothreitol) and equilibration buffer 2 (6 M urea, 0.375 M Tris · HCl, 2% SDS, 20% glycerol, 2.5% iodoacetamide) for 10 min in each buffer. The second dimension was run on a 10% SDS-polyacrylamide gel as described by Laemmli (12). The gels were stained with Coomassie blue R-250 for 30 min and destained with destaining solution (40% methanol, 10% acetic acid) for 8 h with intermittent changes of destaining solution. The gel images were analyzed using Bio-Rad PDQuest software. The reference condition was growth on starch. For each condition, the average spot quantities were obtained from three Gaussian gel images from independently grown cultures. The data were normalized by total intensity of all the pixels in the image. The spots corresponding to 41 kDa and pI 4.8 were cut from gels and identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric analysis at the University of Connecticut Microchemistry Laboratory, Storrs, and the Proteomics Research Facility of the Integrated Biotechnology Laboratories, University of Georgia, Athens. The peptide mass fingerprints were compared to those of T. maritima in the T. maritima genome database (at The Institute for Genomic Research) by using ProteinLynx1.1 (Micromass).
Sequence alignments and motif searches.
AlignAce 3.0 was used to search for common regulatory motifs in the regions 500 nucleotides upstream of genes reported to be up-regulated in response to the indicated sugars. Up-regulated genes used for the lactose group were TM0060, TM0061, TM0070, TM0071, TM0110, TM0309, TM1199, TM1204, and TM1839. The starch group included TM0364, TM1069, TM1835, TM1839, TM1840, and TM1845. The genes in the guar gum group were TM1192, TM1204, TM1227, and TM1624. In addition, a gene not differentially expressed in response to the given carbon source was included in each analysis to serve as a negative control. These genes were TM1835 (lactose group), TM1204 (starch group), and TM1839 (guar gum group). Consensus sequences were derived from alignments of the putative regulatory motifs.
RESULTS
Sequence comparisons and operon organization of the putative maltose transporters.
The amino acid sequences of the transporter components encoded by the two mal operons are very similar to each other, suggesting a common evolutionary history (16). Both correspond to genes encoding a periplasmic substrate binding protein (MalE) and the MalG inner membrane protein. The other inner membrane protein, MalF, is encoded by mal1, but the mal-2 homolog contains a malF pseudogene with a frameshift mutation. The ATP binding protein-encoding genes typical of mal operons in gram-negative organisms are missing from each T. maritima mal operon. This feature, however, is not uncommon, particularly among gram-positive bacteria (9, 25). Indeed, many of the putative ABC transporter operons in T. maritima lack an ATP binding protein-encoding gene. The periplasmic binding protein homologs MalE1 (TM1204) and MalE2 (TM1839) are 80% identical (91% similar), the inner membrane protein homologs MalF1 (TM1203) and MalF2 (TM1837) are 59% identical (74% similar), and MalG1 (TM1202) and MalG2 (TM1836) are 41% identical (62% similar). For this comparison, an authentic frameshift mutation in the malF2 gene was corrected and the resulting translated amino acid sequence was aligned with that of MalF1.
The high sequence identity between the maltose binding protein homologs is unusual since, of the three components of ABC transporters, the substrate binding proteins are usually the least conserved (29). MalE1 and MalE2 differ at only 79 amino acid positions, and MalE2 contains two additional amino acids. Many of the amino acid sequence differences between MalE1 and MalE2 are in the 20 N-terminal amino acids, which correspond to their signal peptides, a region that can perhaps tolerate more changes and still retain its function. The sequences of MalE1 and MalE2 are 36% identical to that of the maltose binding protein from E. coli, 32% identical to that of the maltose-trehalose binding protein of Thermococcus litoralis, and 33% identical to that of the maltose binding protein from Alicyclobacillus acidocaldarius (9). The sequence alignment of T. maritima MalE1 and MalE2 with E. coli MalE shows that most of the amino acids involved in hydrogen bonding with hydroxyl residues of maltose are conserved in MalE1 and MalE2. The evolutionary reason for the anomalously high sequence similarity between the T. maritima MalE paralogs is unknown.
Experimental determination of the substrate specificities of MalE1 and MalE2.
Since both maltose transporter operons were retained following either operon duplication or intralineage HGT, they likely serve different physiological purposes. To determine what these may be, we measured the binding properties of MalE1 and MalE2 by using fluorescence spectroscopy (1, 26). The dissociation constants that we derived for each substrate by measuring changes in fluorescence in response to sugar binding may not reflect the in vivo ligand affinities since, for technical reasons, our measurements were made at a temperature almost 60°C lower than the optimal growth temperature for T. maritima. However, the dissociation constants that we obtained provide measures of comparative ligand specificities and likely provide upper limits for their physiologically relevant affinities.
Wassenberg et al. reported that recombinant MalE2 binds to maltose (KD, 7.2 ± 0.2 μM), maltotriose (KD, 12.6 ± 3.4 μM), and trehalose (KD, 15.3 ± 1.5 μM) (32). We obtained almost identical values in our analyses using their recombinant protein (Table 1). We found that recombinant MalE1 also bound maltose and maltotriose with dissociation constants of 24 ± 1 μM and 8 ± 0.5 nM, respectively (Table 1). In contrast to MalE2, MalE1 does not bind trehalose. The fact that MalE2 binds trehalose is interesting in light of the fact that a nearby gene (aglA; TM1834) encodes an α-glucosidase that can hydrolyze trehalose (27). The fact that MalE2, but not MalE1, binds both maltose and trehalose is consistent with our findings of such activity in periplasmic extracts of maltose- and glucose-grown cells (15). This lends support to the physiological relevance of our binding data derived from these recombinant proteins.
TABLE 1.
Apparent binding affinities of MalE1 and MalE2 for different sugars as measured by changes in intrinsic fluorescence upon ligand binding
| Ligand |
KDa (μM at 20°C) of:
|
|
|---|---|---|
| MalE1 | MalE2 | |
| Maltose | 24 ± 1 | 8.4 ± 1 |
| Maltotriose | 0.008 ± 0.0005 | 11 ± 1.5 |
| Trehalose | − | 9.5 ± 1 |
| β-(1→4)-Mannotetraose | 38 ± 1 | − |
| 63-64-Digalactosyl mannopentose | + | − |
−, no change in fluorescence was detected. +, a change in fluorescence was detected, but data could not be fitted to a binding model. Other sugars that elicited no change in fluorescence for either protein were glucose, galactose, mannose, fructose, xylose, ribose, sorbitol, mannitol, arabinose, sucrose, cellobiose, lactose, raffinose, melibiose, α-(1→3)-arabinogalactoside, and β-(1→4)-mannobiose.
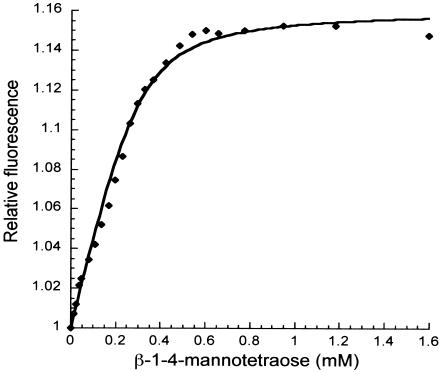
The mal1 operon is adjacent to genes encoding enzymes that hydrolyze galactosides, and two downstream genes, encoding an α-arabinogalactosidase (TM1201) and a LacI homolog (TM1200), are highly expressed in cells grown on carob galactomannan {[Man β-(1→4) Man]n Gal-[α-(1→6)]} (3). We measured affinities of MalE1 and MalE2 for oligosaccharides that may result from enzymatic hydrolysis of galactomannan, including 61-galactosyl-mannobiose, 63-64-digalactosyl mannopentose, β-(1→4)-mannobiose, β-(1→4)-mannotriose, and β-(1→4)-mannotetraose. We also measured the affinities for other galactosides [galactose, lactose, raffinose, melibiose, and α-(1→3)-arabinogalactoside] and glucose. MalE2 did not bind any of these sugars (Table 1). Intrinsic fluorescence increases were observed for MalE1 only in the presence of 61-galactosyl-mannobiose, β-(1→4)-mannotriose, β-(1→4)-mannotetraose, and 63-64-digalactosyl mannopentose (by 7, 9, 16, and 17%, respectively). The addition of 63-64-digalactosyl mannopentose and β-(1→4)-mannotetraose also resulted in blue shifts from 344 to 337 nm. The dissociation constant of MalE1 for β-(1→4)-mannotetraose was 38 ± 1 μM (Fig. 1 and Table 1). We were unable to determine dissociation constants for MalE1 binding to 61-galactosyl-mannobiose and β-(1→4)-mannotriose since high final concentrations of these ligands (1.5 and 1.0 mM, respectively) were necessary to observe changes in MalE1 fluorescence. We were able to detect the signal from 63-64-digalactosyl mannopentose binding with as little as 11 μM, but attempts to describe the binding by using the model as described in Materials and Methods were unsuccessful and fits to other models that assume two noninteracting binding sites or two identical independent binding sites were similarly unsuccessful (data not shown) (5). Since 63-64-digalactosyl mannopentose, 61-galactosyl-mannobiose, and β-(1→4)-mannotriose preparations were 95% pure (the best commercially available grade), we cannot rule out that minor contaminating oligosaccharides in these carob galactomannan hydrolysate products may have been responsible for the changes in fluorescence. However, these contaminants are only β-(1→4)-linked mannooligosaccharides (analyses reported by the manufacturer Megazyme), so the general conclusion that MalE1 binds mannooligosaccharides is valid.
FIG. 1.
Fluorescence emission of MalE1 in response to binding β-(1→4)-mannotetraose. A titration of 0.3 μM MalE1 with β-(1→4)-mannotetraose at 20°C is shown (excitation λ = 280 nm; emission λ = 337 nm).
Maltose operon expression patterns support divergent functions.
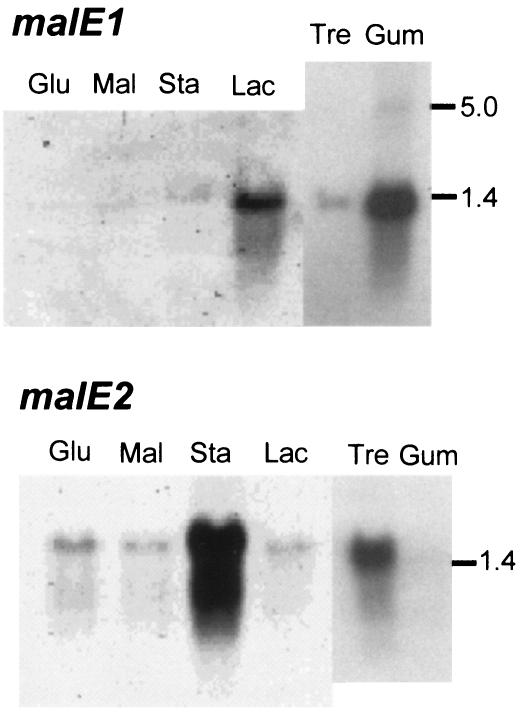
The different binding affinities of MalE1 and MalE2 demonstrate that the two operons have diverged in function. We sought further evidence for this diversion in the patterns of expression of the mal operons in response to growth on different sugars by examining malE transcripts and MalE proteins harvested from periplasmic extracts. We analyzed the expression of malE1 and malE2 by Northern blot analyses of RNA extracted from T. maritima cells grown on starch, maltose, glucose, lactose, trehalose, and guar gum. malE1 expression was detectable in lactose- and guar gum-grown cells, and malE2 was expressed most abundantly in starch- and trehalose-grown cells (Fig. 2). In trehalose-grown cells, a very faint signal corresponding to the malE1 transcript was also observed but at a level considerably lower than that in lactose- and guar gum-grown cells. Under all conditions, the most abundant transcripts were monocistronic since hybridized band sizes were similar to the sizes of the malE1 and malE2 genes (Fig. 2). It is common for a transcript encoding just the binding protein to be expressed to a greater extent than a transcript encoding the other ABC components (19, 28). These results are consistent with those obtained using DNA microarrays, Northern blots, dot blots, and real-time PCR for cells grown on glucose, maltose, and lactose in chemostats (21).
FIG. 2.
Northern blot analyses of RNA extracted from T. maritima cells grown on glucose (Glu), maltose (Mal), starch (Sta), lactose (Lac), trehalose (Tre), and guar gum (Gum). Blotted RNAs were visualized with labeled hybridization probes for malE1 and malE2 as indicated. Equal amounts of RNA were applied to each lane as confirmed by a final hybridization with a labeled probe for the 16S rRNA transcript. The migration of RNA markers (sizes in kilobases) is indicated.
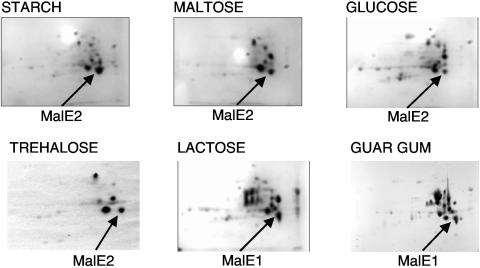
The patterns of expression observed in Northern analyses were confirmed by analyzing gene expression at the protein level by using periplasmic extracts resolved by 2-D gel electrophoresis. Expression was examined in cells grown on the same sugars. A spot corresponding to a protein of 41 kDa with a pI of 4.8 was apparent on each gel, and the protein(s) in each spot was identified by MALDI-TOF (Fig. 3). Although both MalE1 and MalE2 are of this mass and isoelectric point, MALDI-TOF analysis can distinguish between them without ambiguity since their peptide mass fingerprints are different. Our 2-D protein analyses confirmed our Northern analysis data showing that MalE2 is detectable in starch-, maltose-, glucose-, and trehalose-grown cells. MalE2 is the most abundant protein in periplasmic extracts of starch-grown cells (Fig. 3), consistent with the saturating signal for malE2 observed in Northern analysis (Fig. 2). Based upon quantitation of the 2-D gel electrophoresis images, MalE2 is over twice as abundant in starch-grown cells as in maltose- and trehalose-grown cells, and the level of MalE2 in starch-grown cells is almost fivefold higher than that in glucose-grown cells. Although MalE2 levels are the same in maltose- and trehalose-grown cells, the malE2 transcript is not as abundant in maltose-grown cells. This may be due to differences in mRNA stability under these two conditions. malE1 is expressed primarily in lactose- and guar gum-grown cells (Fig. 3).
FIG. 3.
Results of 2-D gel electrophoresis of periplasmic extracts derived from cells grown on lactose, guar gum, starch, maltose, trehalose, and glucose. The spots corresponding to either MalE1 or MalE2 are indicated. The spot from trehalose-grown cells showed mass peaks corresponding to both proteins. Proteins were separated based upon molecular mass (vertically) and isoelectric point (horizontally).
The area corresponding to the MalE proteins in extracts from cells grown on guar gum appeared to show two spots with slightly different apparent molecular masses. MALDI-TOF analysis of both spots showed that they were isoforms of MalE1. Based upon quantitation of 2-D gel electrophoresis data, MalE1 is present at the same concentrations in guar gum-grown cells and lactose-grown cells. This expression pattern is consistent with the substrate specificities of the proteins.
Putative regulatory sequences upstream of the mal operons.
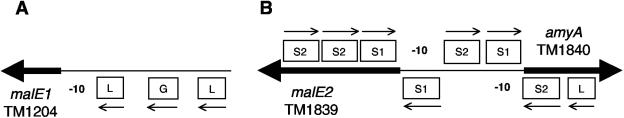
The regions surrounding the start codons of the malE genes were compared to search for possible transcriptional regulatory motifs. Other regions upstream of genes coordinately expressed with the mal operons were also included in our alignments to strengthen the identification of these motifs (3). A summary of the motifs identified in those alignments is shown in Table 2, and the locations of the motifs relative to the malE genes is shown schematically in Fig. 4. malE1 is preceded by motifs common to genes up-regulated in response to lactose and mannooligosaccharides, and malE2 is preceded by the same lactose-responsive motif and two starch-responsive motifs. The divergently transcribed amylase gene (amy) is preceded by these same starch motifs.
TABLE 2.
Consensus sequences of putative regulatory motifs found in the upstream regions of genes up-regulated in lactose-, starch-, and guar gum-grown cells relative to those in glucose-grown cells
| Consensusa in genes up-regulated by the indicated sugar | Locus | Function and/or identity of corresponding peptideb | Distance(s) from −10 box or start codon (nt) |
|---|---|---|---|
| Lactose | |||
| TTTCGGGAAA | TM0060 | Oligopeptide ABC transporter; PBP | 377 and 187 |
| CT | |||
| A | |||
| TM0070 | Endo-1,4-β-xylanase, XynB | 172, 128, and 103 | |
| TM0309 | Oligopeptide ABC transporter; PBP | 261 and 43 | |
| TM1204 | Maltose ABC transporter; PBP; MalE1 | 398 and 110 | |
| TM1839 | Maltose ABC transporter; PBP; MalE2 | 419 | |
| Starch 1 | |||
| TGTGATAAAAT | TM1069 | Transcriptional regulator; DeoR family | 26c |
| GTC | |||
| T | TM1839 | Maltose ABC transporter; PBP; MalE2 | 63c |
| TM1840 | α-Amylase, AmyA | 238 and 54c | |
| Starch 2 | |||
| TCCACCTGGA | TM1069 | Transcriptional regulator; DeoR family | 101 |
| ATGG | |||
| TA | |||
| TM1839 | Maltose ABC transporter; PBP; MalE2 | 285 | |
| TM1840 | α-Amylase, AmyA | 332, 293, and 92 | |
| TM1845 | Pullulanase, PulA | 199, 187, and 49 | |
| Guar gum | |||
| CCTTTGAGGAG | TM1204 | Maltose ABC transporter; PBP; MalE1 | 358 |
| GGGATC | |||
| AAC T | |||
| TM1227 | Endo-1,4-β-mannosidase | 296, 263, 211, and 142 | |
| TM1624 | β-Mannosidase | 125 and 6 |
Nucleotides in the consensus sequences are present in at least 70% of the aligned sequences. Nucleotides below the line are alternative nucleotides at the indicated positions in the consensus. Underlining indicates potential palindromes.
PBP, periplasmic substrate binding protein.
Distance is relative to the start codon of the corresponding gene since the −10 box is part of those motifs.
FIG. 4.
Schematic representation of TM1839, TM1840, and TM1204 and the regulatory motifs described in Table 2. The boxes depict the relative locations of the motifs given in the table: L, lactose-associated motif; S1, first starch-associated motif; S2, second starch-associated motif; and G, guar gum-associated motif. The thin arrow over or under each box indicates the 5′ to 3′ (arrowhead) orientation of each motif. The 5′ ends of ORFs are depicted as heavy arrows in the direction of their transcription. The diagram is not drawn to scale.
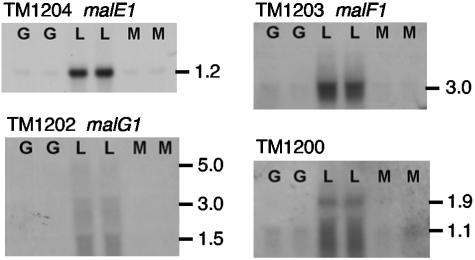
We examined the transcription of the genes adjacent to malE1 to investigate their expression in more detail. The data in Fig. 5 show that probes directed against malE1, malF1, and malG1 (1.2, 1.7, and 2.5 kbp, respectively) hybridized to multiple transcripts. Probes for each gene showed the same patterns of expression in response to growth on the three sugars. It is typical for the periplasmic substrate binding protein to be encoded by a monocistronic, high-copy-number transcript (19, 28), so the malE1 1.2-kb transcript is expected. From the size of the malF1-labeled transcript, malE1 and malF1 appear to be cotranscribed (2.9 kb). The malG1-labeled transcripts were only a smear, so there is no definitive evidence that malG1 is transcribed with the other two genes. However, the mal operon of E. coli shows similar transcript instability, where mal transcripts are preferentially degraded from their 3′ ends (20). The facts that the smear begins near the 5-kb position and that a similar band was observed when transcripts from guar gum-grown cells were probed with malE1 (Fig. 2) may indicate that all the mal1 genes are cotranscribed (5.4 kb). We have no evidence that the mal2 genes are cotranscribed. We detected monocistronic transcripts only by using malE2-derived hybridization probes against mRNA extracts (Fig. 2). In a previous study, transcripts containing only malE2 and not malF2 or malG2 were detected (21). Since malF2 contains an authentic frameshift mutation, transcription after malE2 may terminate in a Rho-dependent manner (23) or a longer transcript including all three genes is more readily processed to contain only malE2 since the other genes may not be translated.
FIG. 5.
Northern hybridization of total RNA to digoxygenin-labeled PCR products of genes of the mal1 operon and a putative LacI-type transcriptional repressor, TM1200. Total RNAs were loaded in duplicates. G, glucose; L, lactose; M, maltose. The migration of RNA markers (sizes in kilobases) is indicated.
Genes adjacent to the mal-1 operon do not appear to be included in its transcripts, but they are expressed in response to the same conditions (21). TM1200 (1 kbp) encoding a putative LacI-like transcriptional regulator was also up-regulated only in lactose-grown cells (Fig. 5). This signal was indistinct, perhaps indicating transcript degradation, but hybridization was detected only in lactose-grown cells. If TM1200 is a repressor, then it may be responsible for the repression of expression of malE2 and amy, perhaps binding to one or more of the starch-responsive elements shown in Fig. 4. An intracellular α-glucoside may act as an inducer, relieving the repression in starch-grown cells.
DISCUSSION
Several lines of evidence suggest that the evolution of T. maritima has been marked by significant HGT events, particularly interdomain exchanges with archaea (16-18, 33). Many of these events involved genes encoding putative ABC transporters (18), though a sizable fraction of T. maritima's ABC transporter operons appear to have a bacterial origin. These bacterial genes may have undergone horizontal transfer as well, but their evolutionary histories are not as easily discerned as are those of the genes shared with archaea. Two bacterial transporter operons, those currently assigned as encoding maltose transporters, appear to have arisen either as the result of operon duplication or intradomain HGT. Since both operons have been retained, they presumably provide distinct selective advantages to T. maritima. We studied these operons to better define their cellular roles and to determine whether their diverging functions are reflected in their transcriptional regulatory patterns.
The ligand specificities and expression patterns of the periplasmic substrate binding proteins MalE1 and MalE2 suggest that their cognate operons serve different functions in T. maritima. Both proteins bind maltose and maltotriose with high affinities, but MalE2 uniquely binds trehalose. MalE1, but not MalE2, binds β-(1→4)-mannotetraose and shows a change in fluorescence in the presence of mannooligosaccharides. These differences are significant since the mal1 operon is adjacent to genes encoding enzymes involved in the hydrolysis of galactomannans and the mal2 operon is in the vicinity of genes encoding alpha glucoside catabolism, including that encoding AglA that hydrolyzes trehalose (27). Consequently, it appears that the ancestral mal operon may have encoded their common function, maltoside transport, and following a duplication event, mal1 diverged to encode a mannooligosaccharide transporter. mal2 may have expanded its repertoire to include trehalose, or it may have retained this ability from the ancestral mal operon while the mal1 transporter subsequently lost this capability.
One may expect that a substrate binding protein would be maximally expressed in cells grown on the corresponding substrate, as is the case for many sugar ABC transporters (14). Indeed, MalE2 shows high affinity for the α-glucosides maltose, maltotriose, and trehalose and is expressed to a greater extent in starch-, trehalose-, and maltose-grown cells than MalE1. Consequently, MalE2 is likely involved in maltose and trehalose transport. A previous study reported lower maltose binding activity in proteins extracted from the periplasm of T. maritima cells grown on maltose than in those from glucose-grown cells (15). However, the maltose binding protein extracted from maltose-grown cells may have been saturated with maltose already and this may have diminished its measurable binding capacity. That study also demonstrated that maltose binding activity isolated from periplasmic extracts of maltose- and glucose-grown cells was inhibited by trehalose but not significantly by lactose, galactose, glucose, or cellobiose (15). This finding provides proof that the native protein expressed in T. maritima in response to growth on maltose is MalE2 since MalE2, but not MalE1, would competitively bind trehalose.
Although MalE1 binds to maltose and maltotriose with affinities similar to or higher than those of MalE2, it is not expressed under conditions in which maltosaccharide transport is necessary. The high level of expression of MalE1 in guar gum-grown cells and the ability of MalE1 to bind mannooligosaccharides suggest that the role of MalE1 is to transport oligosaccharides generated from galactomannan hydrolysis. Both endo-1-4-β-mannanase and β-galactosidase are extracellular in T. maritima, so β-1-4-linked mannooligosaccharides would be available for transport into cells (24). MalE1 binds only β-(1→4)-mannotetraose and not β-(1→4)-mannobiose and β-(1→4)-mannotriose, showing that it has affinity only for larger mannooligosaccharides. Such a transport system would be more efficient energetically since more monomers would be transported for every ATP unit utilized to energize transport. It is likely that T. maritima requires an additional transporter to transport mannobiose and mannotriose generated by hydrolysis of galactomannans. A study of gene expression in T. maritima in response to growth on galactomannans using DNA microarrays with a subset of all identified ORFs postulated that the ABC transporter encoded by TM1746 to TM1750 transports galactomannans (3). Those microarrays did not include malE1 and malE2, so the patterns we observed were not found. The transporter encoded by TM1746 to TM1750 may transport mannobiose and mannotriose.
The archaeal hyperthermophile Thermococcus litoralis has a maltose-trehalose ABC transporter that has also been found in Pyrococcus furiosus (4). Pyrococcus furiosus has an additional ABC transporter for maltodextrins (10, 11). Expression of the Thermococcus litoralis transporter operon is controlled by a repressor (TrmB) and is derepressed by maltose and trehalose (13). These maltose-trehalose substrate binding proteins are similar to that encoded by the E. coli mal operon according to results of BLAST similarity searches (4). The T. maritima MalE proteins also show sequence similarity to these archaeal binding proteins in BLAST searches, but we have found through phylogenetic analyses that the archaeal transporters are only very distantly related to the E. coli maltose transporter proteins and that the T. maritima Mal proteins are more closely related to those from E. coli and related organisms (D. M. Nanavati and K. M. Noll, unpublished data). Consequently, the T. maritima and archaeal maltose transporters are related by functional characteristics but not by direct ancestry.
As noted above, the relatively high amounts of MalE2 in maltose- and starch-grown cells suggest that this binding protein may be responsible for transport of maltooligosaccharides resulting from starch hydrolysis. However, it is not clear what transmembrane proteins MalE2 would interact with during maltose transport. One of the adjacent membrane-spanning protein-encoding genes, malF2, encodes a natural frameshift mutation, which would produce a truncated and presumably inactive protein. Since it has high sequence similarity to MalF2, MalF1 may potentially provide the necessary component of the transmembrane transporter. Periplasmic substrate binding proteins can share membrane permeases, as evidenced by the histidine and lysine-arginine-ornithine binding proteins in E. coli (6). However, it was shown previously that neither malF2 and malG2 nor the mal1 genes are up-regulated along with malE2 in maltose-grown cells compared with those in glucose-grown cells (21). Thus, it is not clear which membrane-spanning components MalE2 would use when cells grow on maltose.
Clearly, the regulation of expression of these paralogous operons differs. Possible upstream regulatory motifs reflect their differential expression patterns. Transcriptional control proteins that respond to catabolites likely evolve separately of their cognate periplasmic substrate binding proteins (7), but both must respond to similar ligands. The proteins that regulate transcription of the mal operons are unknown, but it is of interest to identify them to determine whether their evolution parallels that of these transporters.
Acknowledgments
We thank Carol Teschke for her helpful assistance and advice with the binding studies, Wolfgang Liebl for his generous provision of the MalE2 clone, and Dennis W. Hill and Albert J. Kind of the University of Connecticut Microchemistry Laboratory facility for their assistance with the MALDI-TOF analyses.
This work was supported by funds from the NASA Exobiology Program (NAG5-12367).
REFERENCES
- 1.Boos, W., A. S. Gordon, R. E. Hall, and H. D. Price. 1972. Transport properties of the galactose-binding protein of Escherichia coli. Substrate-induced conformational change. J. Biol. Chem. 247:917-924. [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Chhabra, S. R., K. R. Shockley, S. B. Conners, K. L. Scott, R. D. Wolfinger, and R. M. Kelly. 2003. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J. Biol. Chem. 278:7540-7552. [DOI] [PubMed] [Google Scholar]
- 4.DiRuggiero, J., D. Dunn, D. L. Maeder, R. Holley-Shanks, J. Chatard, R. Horlacher, F. T. Robb, W. Boos, and R. B. Weiss. 2000. Evidence of recent lateral gene transfer among hyperthermophilic Archaea. Mol. Microbiol. 38:684-693. [DOI] [PubMed] [Google Scholar]
- 5.Eftink, M. R. 1997. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 278:221-257. [DOI] [PubMed] [Google Scholar]
- 6.Ehrmann, M., R. Ehrle, E. Hofmann, W. Boos, and A. Schlosser. 1998. The ABC maltose transporter. Mol. Microbiol. 29:685-694. [DOI] [PubMed] [Google Scholar]
- 7.Fukami-Kobayashi, K., Y. Tateno, and K. Nishikawa. 2003. Parallel evolution of ligand specificity between LacI/GalR family repressors and periplasmic sugar-binding proteins. Mol. Biol. Evol. 20:267-277. [DOI] [PubMed] [Google Scholar]
- 8.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 9.Hulsmann, A., R. Lurz, F. Scheffel, and E. Schneider. 2000. Maltose and maltodextrin transport in the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius is mediated by a high-affinity transport system that includes a maltose binding protein tolerant to low pH. J. Bacteriol. 182:6292-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imamura, H., B. S. Jeon, and T. Wakagi. 2004. Molecular evolution of the ATPase subunit of three archaeal sugar ABC transporters. Biochem. Biophys. Res. Commun. 319:230-234. [DOI] [PubMed] [Google Scholar]
- 11.Koning, S. M., W. N. Konings, and A. J. M. Driessen. 2001. Biochemical evidence for the presence of two α-glucoside ABC-transport systems in the hyperthermophilic archaeon Pyrococcus furiosus. Archaea 1:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. J., A. Engelmann, R. Horlacher, Q. H. Qu, G. Vierke, C. Hebbeln, M. Thomm, and W. Boos. 2003. TrmB, a sugar-specific transcriptional regulator of the trehalose/maltose ABC transporter from the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 278:983-990. [DOI] [PubMed] [Google Scholar]
- 14.Lucht, J. M., and W. Boos. 1996. Periplasmic binding protein-dependent ABC transporters, p. 1175-1209. In F. C. N. R. Curtiss (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 15.Nanavati, D., K. M. Noll, and A. H. Romano. 2002. Periplasmic maltose- and glucose-binding protein activities in cell-free extracts of Thermotoga maritima. Microbiology 148:3531-3537. [DOI] [PubMed] [Google Scholar]
- 16.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 17.Nelson, K. E., J. A. Eisen, and C. M. Fraser. 2001. Genome of Thermotoga maritima MSB8. Methods Enzymol. 330:169-180. [DOI] [PubMed] [Google Scholar]
- 18.Nesbo, C. L., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newbury, S. F., N. H. Smith, and C. F. Higgins. 1987. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51:1131-1143. [DOI] [PubMed] [Google Scholar]
- 20.Newbury, S. F., N. H. Smith, E. C. Robinson, I. D. Hiles, and C. F. Higgins. 1987. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell 48:297-310. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen, T. N., A. D. Ejaz, M. A. Brancieri, A. M. Mikula, K. E. Nelson, S. R. Gill, and K. M. Noll. 2004. Whole-genome expression profiling of Thermotoga maritima in response to growth on sugars in a chemostat. J. Bacteriol. 186:4824-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Opperman, T., and J. P. Richardson. 1994. Phylogenetic analysis of sequences from diverse bacteria with homology to the Escherichia coli rho gene. J. Bacteriol. 176:5033-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker, K. N., S. R. Chhabra, D. Lam, W. Callen, G. D. Duffaud, M. A. Snead, J. M. Short, E. J. Mathur, and R. M. Kelly. 2001. Galactomannanases man2 and man5 from Thermotoga species: growth physiology on galactomannans, gene sequence analysis, and biochemical properties of recombinant enzymes. Biotechnol. Bioeng. 75:322-333. [DOI] [PubMed] [Google Scholar]
- 25.Puyet, A., and M. Espinosa. 1993. Structure of the maltodextrin-uptake locus of Streptococcus pneumoniae. Correlation to the Escherichia coli maltose regulon. J. Mol. Biol. 230:800-811. [DOI] [PubMed] [Google Scholar]
- 26.Quiocho, F. A., and P. S. Ledvina. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 20:17-25. [DOI] [PubMed] [Google Scholar]
- 27.Raasch, C., W. Streit, J. Schanzer, M. Bibel, U. Gosslar, and W. Liebl. 2000. Thermotoga maritima AglA, an extremely thermostable NAD+-, Mn2+-, and thiol-dependent alpha-glucosidase. Extremophiles 4:189-200. [DOI] [PubMed] [Google Scholar]
- 28.Stern, M. J., E. Prossnitz, and G. F. Ames. 1988. Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol. Microbiol. 2:141-152. [DOI] [PubMed] [Google Scholar]
- 29.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teichmann, S. A., and M. M. Babu. 2004. Gene regulatory network growth by duplication. Nat. Genet. 36:492-496. [DOI] [PubMed] [Google Scholar]
- 31.Vaughan, E. E., R. D. Pridmore, and B. Mollet. 1998. Transcriptional regulation and evolution of lactose genes in the galactose-lactose operon of Lactococcus lactis NCDO2054. J. Bacteriol. 180:4893-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wassenberg, D., W. Liebl, and R. Jaenicke. 2000. Maltose-binding protein from the hyperthermophilic bacterium Thermotoga maritima: stability and binding properties. J. Mol. Biol. 295:279-288. [DOI] [PubMed] [Google Scholar]
- 33.Worning, P., L. J. Jensen, K. E. Nelson, S. Brunak, and D. W. Ussery. 2000. Structural analysis of DNA sequence: evidence for lateral gene transfer in Thermotoga maritima. Nucleic Acids Res. 28:706-709. [DOI] [PMC free article] [PubMed] [Google Scholar]