Abstract
The anti-TRAP protein (AT), encoded by the rtpA gene of Bacillus subtilis, can bind to and inhibit the tryptophan-activated trp RNA-binding attenuation protein (TRAP). AT binding can prevent TRAP from promoting transcription termination in the leader region of the trp operon, thereby increasing trp operon expression. We show here that AT levels continue to increase as tryptophan starvation becomes more severe, whereas the TRAP level remains relatively constant and independent of tryptophan starvation. Assuming that the functional form of AT is a trimer, we estimate that the ratios of AT trimers per TRAP molecule are 0.39 when the cells are grown under mild tryptophan starvation conditions, 0.83 under more severe starvation conditions, and approximately 2.0 when AT is expressed maximally. As the AT level is increased, a corresponding increase is observed in the anthranilate synthase level. When AT is expressed maximally, the anthranilate synthase level is about 70% of the level observed in a strain lacking TRAP. In a nutritional shift experiment where excess phenylalanine and tyrosine could potentially starve cells of tryptophan, both the AT level and anthranilate synthase activity were observed to increase. Expression of the trp operon is clearly influenced by the level of AT.
In Bacillus subtilis expression of seven genes is required for the biosynthesis of l-tryptophan from chorismic acid, the common aromatic amino acid precursor (2, 12). Six of these seven genes are organized as a trp operon, a suboperon within the aromatic supraoperon (16, 36). The seventh trp gene, trpG (pabA), is located in the unlinked folate operon, and its polypeptide product participates in biosynthesis of both tryptophan and folate (12, 16). Expression of all seven genes is regulated by the trp RNA-binding attenuation protein (TRAP) in response to the accumulation of l-tryptophan (2, 12). When TRAP is activated by tryptophan, it binds to a transcript antiterminator segment containing specific trinucleotide repeats, promoting terminator formation and transcription termination (2). Activated TRAP also binds to similar RNA trinucleotide repeat segments that precede the start codons for three coding regions, trpG (11, 35), trpP (yhaG) (21, 34), and ycbK (22; our unpublished data); when bound to these repeats, TRAP inhibits translation initiation (1).
The accumulation of uncharged tRNATrp in a temperature-sensitive tryptophanyl-tRNA synthetase (trpS1) mutant increases trp operon expression, despite the presence of excess tryptophan (17, 28). The at operon was identified as being responsible for this increase (6, 22, 30). Transcription of the structural genes of the at operon is regulated by the T-box antitermination mechanism in response to changes in the charging of tRNATrp (22). However, expression of the at operon is subject to translational as well as transcriptional sensing of uncharged tRNATrp (6). Thus, immediately preceding the start codon of rtpA, the coding region for the anti-TRAP protein (AT), there is a 10-codon leader peptide-coding region, rtpLP, containing three tandem Trp codons. Attempted translation of these three Trp codons is believed to provide the cell with the translational opportunity to sense and respond to the availability of charged tRNATrp (6, 7). When the tRNATrp in the cell is largely charged, the ribosome translating rtpLP completes translation of rtpLP and appears to inhibit rtpA translation (22). However, when the cell is deficient in charged tRNATrp, the translating ribosome is believed to stall over one of the rtpLP Trp codons, exposing the rtpA Shine-Dalgarno region for efficient translation initiation (6, 7, 22). The increased AT protein produced then binds to tryptophan-activated TRAP, blocking TRAP's RNA binding ability, thereby inhibiting TRAP's role in regulating trp gene expression (30, 31).
Many events and several proteins are already known to contribute to regulation of tryptophan biosynthesis in B. subtilis. These events are as follows: (i) availability of the tryptophan precursors chorismate, glutamine, phosphoribosylpyrophosphate, and serine (12); (ii) the rate of transcription initiation at the respective aroF and trp operon promoters (4); (iii) synthesis of the TRAP protein (13); (iv) activation of TRAP by tryptophan (3, 20); (v) TRAP binding to leader RNA, promoting transcription termination (19); (vi) inactivation of TRAP, allowing antiterminator formation, thereby preventing transcription termination (2); (vii) active TRAP binding to leader RNA inducing formation of a hairpin structure that sequesters the trpE Shine-Dalgarno sequence, inhibiting trpE translation (10, 19); (viii) active TRAP binding at the trpG translation start site, inhibiting trpG translation (11); (ix) uncharged tRNATrp accumulation causing antitermination in the at operon leader region, allowing transcription of the at operon to proceed (22); (x) uncharged tRNATrp accumulation leading to stalling of the ribosome translating rtpLP, activating translation of rtpA, which increases the cellular level of the AT protein (6); (xi) AT binding to tryptophan-activated TRAP, preventing TRAP from promoting transcription termination in the trp operon leader region (30) and inhibiting translation of trpG; (xii) uncharged tRNATrp accumulation activating transcription antitermination in the trpS leader region, presumably resulting in increased synthesis of tryptophanyl-tRNA synthetase and production of increased levels of charged tRNATrp (14, 15, 22); (xiii) tryptophan feedback inhibition of anthranilate synthase, reducing tryptophan biosynthesis (12). Other metabolic features also contribute to regulation of tryptophan biosynthesis: for example, the trp leader transcript forms a 5′ hairpin structure that has been shown to facilitate TRAP binding to leader RNA (26). In addition, TRAP bound to leader RNA can be released by RNase action in vivo, rendering TRAP free and available for RNA binding (9).
In this article we describe measurements of the cellular levels of AT and TRAP proteins and the tryptophan biosynthetic enzyme anthranilate synthase in a variety of strains of B. subtilis grown under different conditions. Some of the growth conditions examined cause mild or severe tryptophan starvation. We observed that the AT level increases upon tryptophan starvation and that this increase is associated with increased anthranilate synthase production. Deletion of the AT structural gene reduces this increase in trp operon expression.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The B. subtilis strains used in this study are listed in Table 1. CYBS400 is a prototroph. CYBS318 (Δat Spr) is a strain lacking the ability to synthesize the AT protein (7, 22). CYBS223 (mtrBΩTcr) is a strain lacking the ability to synthesize the TRAP protein (19). CYBS542 is a Δat strain in which the natural at operon has been deleted and in which a modified at operon with a deletion removing the leader and rtpLP region [P-ΔL-Δ(SD and AUG for rtpLP)-ΔrtpLP (SD is the Shine-Dalgarno region)] has been integrated into the chromosomal amyE locus (7). 1A353 (trpS1) is a temperature-sensitive mutant strain which produces a heat-labile tryptophanyl-tRNA synthetase (28, 29). Strains with gene fusions were prepared by transformation and selection using relevant antibiotics. Disruption of amyE, the site of gene insertion, was confirmed by testing for amylase production using iodine straining (24). Cultures were generally grown to mid-log phase with shaking in minimal medium (33) supplemented with 0.5% glucose and appropriate trace elements plus various additional supplements and antibiotics at the appropriate temperatures. Where indicated, the following supplements were added: 50 μg of l-tryptophan per ml, 50 μg of l-phenylalanine per ml, 50 μg of l-tyrosine per ml, 0.2% acid-hydrolyzed casein, or 20 or 40 μg of indole acrylic acid (IAA) per ml. In the nutritional shift experiments, strains CYBS400, CYBS318, and CYBS223 were grown in minimal medium with 20 μg of l-tryptophan per ml to a optical density at 525 nm (OD525) of 0.3, and the cells were harvested and washed with an aqueous solution containing 0.9% NaCl. The washed cells were then shifted into minimal medium containing 100 μg of l-phenylalanine per ml and 50 μg of l-tyrosine per ml or minimal medium containing 0.2% acid-hydrolyzed casein. Cells in these two media were harvested at an OD525 of 0.4 and 0.8, respectively.
TABLE 1.
Strains of B. subtilis used in this study
| Strain | Genotype or phenotypea | Reference(s) |
|---|---|---|
| CYBS400 | Prototroph | 21 |
| CYBS318 | Δat Spr | 7, 21 |
| CYBS223 | mtrBΩTcr | 19 |
| CYBS542 | amyE::[P-ΔL-Δ(SD and AUG for rtpLP)-ΔrtpLP]Cmr | 7 |
| 1A353 | trpS1 | 28 |
Δat, at operon deletion. Spr, spectinomycin resistance; Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; SD, Shine-Dalgarno region. See Materials and Methods for additional description of the various strains examined.
Cell growth and preparation of protein extracts.
Cultures were grown under various conditions at 30, 37, or 42°C with shaking overnight and then subcultured in a modified medium and grown to mid-log phase (OD525 of 0.8). The resulting cultures were chilled on ice, and 20-ml portions were harvested by centrifugation at 6,000 × g for 10 min. Each cell pellet was resuspended in 5 ml of 0.9% NaCl and centrifuged again, and the pellet was frozen and stored at −20°C.
Frozen cells were resuspended in 0.3 ml of sonication buffer (0.1 M Tris [pH 7.8] containing 1 mM dithiothreitol) and were disrupted by sonication. Cell debris was removed by centrifugation at 12,000 × g for 15 min at 4°C, and the supernatants collected were used for various assays. The protein concentration of each extract was determined using the Advance protein assay (Cytoskeleton, Inc.). The levels of AT and TRAP proteins in each sample were measured by Western blot analysis. Known concentrations of pure AT or TRAP in the sonication buffer were used as reference bands and as controls.
Determination of cell numbers and AT and TRAP protein standards.
To determine the number of cells per milliliter of culture, cultures were diluted 1:105, and 20 μl of each solution was plated on nutrient broth agar plates in triplicate and incubated overnight at 30 or 37°C. The colonies were counted, and these values were used to calculate the number of cells per milliliter of culture. Microscopic counting was performed on several samples, and the values obtained agreed with colony counts. Pure AT protein was prepared as previously described (30), and pure TRAP protein was generously provided by Paul Gollnick (State University of New York at Buffalo). The exact concentrations of the pure AT and TRAP proteins used to generate standard curves were determined by hydrolysis and analysis of pure samples by the Amino Acid Analysis Service Laboratory (Boring, Oreg.). Standard curves for known concentrations of AT or TRAP protein were established by serial dilution and detection using specific antisera by Western blotting. The standard curve for each blot was run on an identical gel and transferred to the same membrane for each assay.
Western blot analysis of AT and TRAP levels.
The supernatant of each sonicated sample was mixed with an equal volume of 2× Tricine sample buffer (0.1 M Tris-HCl [pH 6.8], 24% glycerol, 8% sodium dodecyl sulfate [SDS], 0.2 M dithiothreitol, 0.02% Coomassie blue G-250) and heated in a boiling water bath for 10 min. After centrifugation, samples were electrophoresed on SDS-15% polyacrylamide gels in Tris-Tricine buffer systems (23) and then electrophoretically transferred to a nitrocellulose membrane (pore size, 0.2 μm; Bio-Rad, Inc.). Immunoblotting was performed as described previously (5) with rabbit polyclonal antisera directed against the AT protein (antisera prepared by Covance, Inc.) or the TRAP protein (provided by Paul Gollnick, State University of New York at Buffalo). Bound antibody was visualized by the use of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Rockland) and SuperSignal West Pico chemiluminescence detection reagents (Pierce). The Western blot bands were quantified with the Molecular Analyst 2.1 software package (Bio-Rad). Other cross-reacting bands on the Western blots were used as internal controls for quantitation.
Anthranilate synthase assays.
Cultures were grown under the same conditions described above in “Western blot analysis of AT and TRAP levels.” The specific activity of anthranilate synthase was assayed fluorometrically as described previously (8). This assay procedure requires independent fluorescence calibration for each experiment. Therefore, the values obtained in different experiments often varied, although the relative values within an experiment were highly reproducible. Two tubes containing the control sample and the standard reaction mixture were used to set a range of 0 to 100 fluorometer units (FU). These tubes were incubated with the assayed tubes and contained the standard reaction mixture. After incubation and reaction termination, the fluorescence in the tube with the standard level of anthranilate was set at 100 FU, and the control sample was set at 0 FU. The fluorescence in each of the sample tubes was then determined. Since the 100 and 0% values are relative and the fluorescence of the blank sample often varies slightly, there is generally some variation in the values obtained in identical experiments performed on different days. Nevertheless, when one assays duplicate samples or compares the values within an experiment, the variation is low. Because of these peculiarities, the standard deviations shown for anthranilate synthase levels measured in repeat experiments performed on different days are greater than the true variation observed using this assay with the same reference standards. Nevertheless, it is essential to perform the assay on several days and use samples obtained in repeat experiments to be certain of the relative values. Each assay contained duplicates of each sample or two levels of each sample. Each assay was repeated on several occasions. The data are reported as FU per milligram of protein.
RESULTS
Detection and quantification of AT and TRAP proteins.
It is not yet known whether the AT protein functions as a trimer, hexamer, or dodecamer (31). The trimeric species was the most prominent in our previous studies (31). Snyder et al. have shown that AT exists in an equilibrium state, part trimer and part dodecamer, with formation of the dodecamer being concentration dependent (27). Preliminary crystallographic data have been obtained for an AT complex in which four trimers are associated in a dodecamer (25). We shall assume in the present study that AT functions in vivo as a trimer.
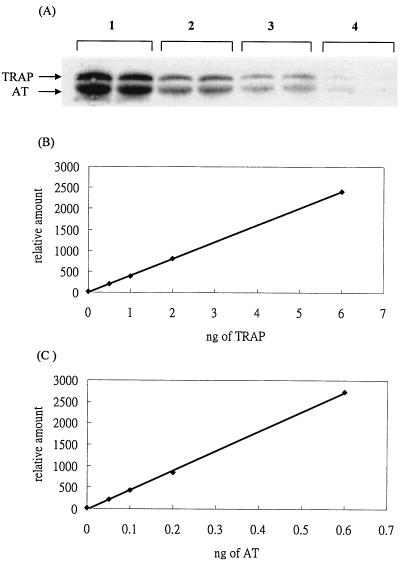
To obtain precise quantification of the pure AT and TRAP proteins used in establishing standard curves, the exact concentration of each protein was determined by hydrolysis and analysis by the Amino Acid Analysis Service Laboratory. Standard curves for known concentrations of TRAP (Fig. 1B) or AT (Fig. 1C) were established by serial dilution and detection using specific antisera by Western blotting (Fig. 1A). The Western blot bands were quantified with the Molecular Analyst 2.1 software package (Bio-Rad). The detection limit for AT protein was 0.05 ng, and the detection limit for TRAP protein was 0.5 ng. The recovery of AT or TRAP proteins extracted from cells was estimated by adding known concentrations of pure protein in sonication buffer to cells of B. subtilis strain CYBS318 (at deletion) or CYBS223 (mtrB deletion). The mixtures were then sonicated, and the AT or TRAP protein concentrations were measured and quantified by Western blot analysis. Our results show that recovery of either protein by this extraction method is over 90% (data not shown).
FIG. 1.
Standard curves using Western blotting and specific antisera to detect denatured, pure TRAP and AT proteins. Dilutions of purified standard proteins (TRAP and AT) were used for Western blotting and immunodetection with TRAP- and AT-specific antibodies. Each sample was assayed in duplicate, and the background value was used to calculate the value with no protein added (A). Quantification of TRAP and AT was performed as described in Materials and Methods. Standard curves were generated by using dilutions of purified standard proteins TRAP (B) and AT (C).
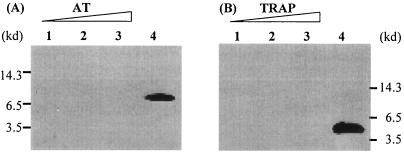
The anti-AT and anti-TRAP antisera were mixed and used to detect AT and TRAP proteins in the same sample to reduce the quantitative variation that might result from separate Western blot assays. However, the prerequisite of this assay is that each separate antiserum should not show cross-reaction with TRAP or AT. Figure 2A presents the results obtained with anti-TRAP antiserum, when used to detect a band with increasing amounts of AT protein (lanes 1 to 3) or 6 ng of TRAP protein (lane 4). Figure 2B presents results using the anti-AT serum with increasing amounts of TRAP protein (lanes 1 to 3) or 0.6 ng of AT protein (lane 4). The results indicate that the separate antisera do not cross-react with the other protein. Therefore, it is feasible to use an antibody mixture to quantify AT and TRAP proteins that are present in the same sample.
FIG. 2.
Lack of cross-reaction of TRAP and AT antisera. Western bolt analysis using anti-TRAP antiserum (A) and anti-AT antiserum (B), respectively. (A) Lanes 1 to 3: 4, 6, and 8 ng of AT protein, respectively; lane 4: 6 ng of TRAP protein. (B) Lanes 1 to 3: 0.4, 0.6, and 0.8 ng of TRAP protein, respectively; lane 4: 0.6 ng of AT protein. The positions of molecular mass markers (in kilodaltons [kd]) are shown to the sides of the blots.
Effects of adding IAA on the cellular AT and TRAP levels.
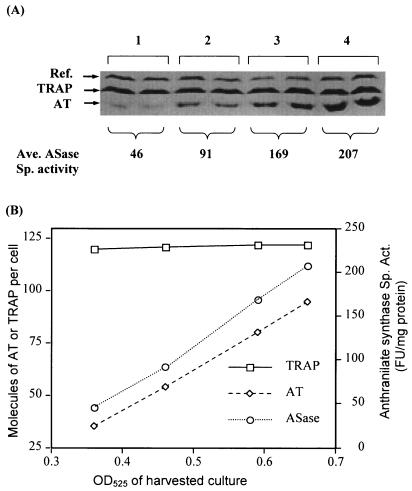
Previous results demonstrated that the rtpA gene was expressed significantly when wild-type cultures were grown in the presence of 30 μg of IAA per ml, an inhibitor of tRNATrp charging (4). To obtain a better understanding of the kinetics of expression of the AT and TRAP proteins in the presence of IAA and the consequences on trp operon expression, B. subtilis strain CYBS400 (wild type) was grown in minimal medium supplemented with 40 μg of IAA per ml and 0.5% glucose. Cultures were harvested at different time points during growth and assayed for AT and TRAP by Western blot analysis and for anthranilate synthase activity by measuring enzyme activity. Figure 3A presents the results of the Western blot analysis of the AT and TRAP levels at different cell densities; the average anthranilate synthase levels are also shown. Figure 3B is a diagrammatic representation of these values. The results obtained indicate that AT expression and anthranilate synthase activity both increase with increasing cell density upon growth with IAA. However, there is no significant change in the TRAP level. The molecules of AT (trimer) detected increased from about 35 to 90 per cell under the conditions examined.
FIG. 3.
Effects of adding IAA on the cellular AT and TRAP levels. B. subtilis strain CYBS400 (wild type) was grown in minimal medium with 40 μg of IAA per ml and 0.5% glucose added. Cultures were harvested, and the cell concentration was determined at different optical densities. Cells were then washed, frozen, and subsequently disrupted by sonic oscillation, and the supernatants were assayed using Western blot analysis and anthranilate synthase measurements. Anthranilate synthase specific activity is presented in fluorometer units (FU) per milligram of protein. (A) Western blot analysis of TRAP and AT levels at different time points using anti-AT and anti-TRAP sera. Each sample was assayed in duplicate. The reference (Ref.) band shown was produced by an unidentified protein in the extracts that cross-reacts with the antiserum to the TRAP protein. The average anthranilate synthase specific activity (Ave. ASase Sp. activity) is shown below the braces. (B) The estimated number of molecules of AT (trimer) or TRAP (11-mer) per cell and the average anthranilate synthase (ASase) specific activity (Sp. Act.) calculated from panel A are shown.
Contribution of AT synthesis to levels of anthranilate synthase production.
The results in Fig. 3 show that increases in AT expression are associated with increases in the specific activity of anthranilate synthase. To understand the contribution of AT synthesis to anthranilate synthase production, cultures of B. subtilis CYBS400 (wild-type strain) and CYBS318 (AT deletion strain) were grown in minimal medium with or without l-tryptophan (50 μg/ml) or IAA (40 μg/ml) with 0.5% glucose. The data in Table 2 show that the anthranilate synthase activities of wild-type cultures were higher than those of cultures of the at deletion strain under each of the three conditions examined. In both assays performed, the anthranilate synthase activity were lower in cultures of the at deletion strain than in the wild-type cultures. These results indicate that the at operon does influence the level of anthranilate synthase that is produced in wild-type cultures and that AT production can up-regulate trp operon expression.
TABLE 2.
Effects of deleting the at operon on anthranilate synthase productiona
| Strain | Growth conditionb | Anthranilate synthase sp actc |
|---|---|---|
| CYBS400 (wild type) | min − Trp | 31 ± 9 |
| min + Trp | 16 ± 2 | |
| min + IAA40 | 480 ± 130 | |
| CYBS318 (Δat) | min − Trp | 18 ± 15 |
| min + Trp | 8 ± 5 | |
| min + IAA40 | 326 ± 74 |
Cultures of B. subtilis strains CYBS400 (wild type) and CYBS318 (Δat deletion) were grown to mid-log phase (OD525 of 0.8) at 37°C in minimal medium with or without l-tryptophan or IAA with glucose. Cultures were harvested, washed, and frozen, and the cells were subsequently disrupted by sonic oscillation and assayed for anthranilate synthase activity.
Cells were grown in minimal medium (min) in the presence (+) or absence (−) of l-tryptophan (50 μg/ml) or IAA (40 μg/ml) (IAA40) with 0.5% glucose.
The specific activity is shown in fluorometer units (FU) per milligram of protein. The values presented are the averages of results from two experiments.
Cellular levels of AT and TRAP proteins in various strains grown under different conditions and their effects on trp operon expression.
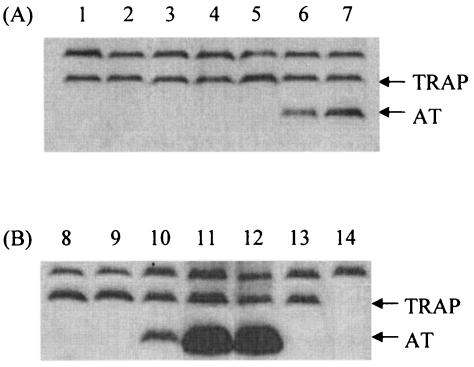
B. subtilis wild-type and mutant cultures were grown under a variety of conditions, some of which imposed mild to severe tryptophan starvation. Cells were harvested, extracts were prepared, and Western blot analyses were performed (Fig. 4) to measure the numbers of AT trimers and TRAP protein molecules per cell (Table 3). Extracts were also assayed for anthranilate synthase activity (Table 3). A typical Western blot analysis for the strains examined, under the growth conditions employed, is shown in Fig. 4. Table 3 presents a summary of the quantitation based on these data and reveals that no AT was detected per cell when wild-type cultures were grown in the absence (Fig. 4, lane 1) or presence (Fig. 4, lane 2) of tryptophan, or in the presence of phenylalanine (Fig. 4, lane 3), phenylalanine and tyrosine (Fig. 4, lane 4), or acid-hydrolyzed casein (Fig. 4, lane 5). Acid-hydrolyzed casein is a mixture of all the amino acids except tryptophan, which is acid labile. AT was detected when wild-type cultures were grown in the presence of IAA (Fig. 4, lanes 6 and 7). IAA inhibits tRNATrp charging and increases trp operon expression. Its presence also reduces the cell growth rate; cultures grown with 20 or 40 μg of IAA/ml required 8 and 10 h of growth, respectively, to reach the same cell density reached by the control culture in 6 h of growth. Cultures grown with IAA, as expected, contained elevated levels of anthranilate synthase (Table 3). Mutant 1A353 was also examined; this mutant forms a temperature-sensitive tryptophanyl-tRNA synthetase (28, 29). Mutant 1A353 did not contain detectable levels of AT when grown at 30°C (Fig. 4, lanes 8 and 9), but it did form significant levels of AT when grown at 42°C (Fig. 4, lane 10). At this elevated temperature, its anthranilate synthase level also was elevated (Table 3). Strain CYBS542, which was engineered to produce AT maximally, formed the highest levels of AT detected (Fig. 4, lanes 11 and 12); it contained approximately two AT trimers per molecule of TRAP (Table 3). Its anthranilate synthase level was approximately 70% that of a TRAP-deficient mutant, CYBS223 (Fig. 4, lane 14), a mutant that expresses the trp operon maximally (Table 3). Expression of TRAP by strain CYBS318 (Fig. 4, lane 13), a strain lacking the at operon, was similar to that of the wild-type strain. These findings demonstrate that the cellular AT level increases as a function of the severity of tryptophan starvation. As the AT level increases, the anthranilate synthase level also is elevated. The observed TRAP level was relatively independent of tryptophan starvation, indicating that TRAP formation is not appreciably affected by tryptophan starvation.
FIG. 4.
Western blot analysis of AT and TRAP levels under various growth conditions. B. subtilis cultures were grown to mid-log phase (OD525 of 0.8) in minimal medium supplemented with 0.5% glucose with the additions indicated below. Cells were harvested, washed, frozen, disrupted by sonic oscillation, and assayed by Western blot analysis. Quantification of TRAP and AT was performed as described in Materials and Methods. The top band in each lane was produced by an unknown protein that cross-reacts with the anti-TRAP antiserum; this band was used as the reference band. (A)Strain CYBS400 (wild type) was grown at 37°C with the following additions: lane 1, no addition; lane 2, l-tryptophan (50 μg/ml); lane 3, l-phenylalanine (50 μg/ml); lane 4, l-phenylalanine and l-tyrosine (50 μg/ml each); lane 5, acid-hydrolyzed casein (50 μg/ml); lane 6, IAA (20 μg/ml); lane 7, IAA (40 μg/ml). (B) Cultures were grown under the following conditions: lane 8, strain 1A353, 30°C, without addition; lane 9, strain 1A353, 30°C, with l-tryptophan (50 μg/ml); lane 10, strain 1A353, 42°C, with l-tryptophan (50 μg/ml); lane 11, strain CYBS542, 37°C, without addition; lane 12, strain CYBS542, 37°C, with l-tryptophan (50 μg/ml); lane 13, strain CYBS318, 37°C, without addition; lane 14, strain CYBS223, 37°C, with l-phenylalanine (50 μg/ml).
TABLE 3.
Measurement of the number of molecules of AT and TRAP protein per cell and the specific activity of anthranilate synthase in cultures grown under various conditions
| Strain | Conditionc | Temp (°C) | No. of moleculesa
|
AT/TRAP (trimer/11-mer) | Anthranilate synthase sp actb (FU/mg of protein) | ||
|---|---|---|---|---|---|---|---|
| AT (trimer)d | TRAP (11-mer) | ||||||
| CYBS400 | −Trp | 37 | <10e | 115 ± 6 | 33 ± 8 | ||
| CYBS400 | +Trp | 37 | <10 | 119 ± 8 | 17 ± 3 | ||
| CYBS400 | +Phe | 37 | <10 | 120 ± 11 | 76 ± 12 | ||
| CYBS400 | +Phe, Tyr | 37 | <10 | 122 ± 8 | 104 ± 23 | ||
| CYBS400 | +ACH | 37 | <10 | 131 ± 10 | 84 ± 19 | ||
| CYBS400 | +IAA20 | 37 | 47 ± 7 | 122 ± 9 | 0.39 | 230 ± 51 | |
| CYBS400 | +IAA40 | 37 | 102 ± 6 | 123 ± 10 | 0.83 | 470 ± 77 | |
| 1A353 | −Trp | 30 | <10 | 117 ± 10 | 150 ± 27 | ||
| 1A353 | +Trp | 30 | <10 | 119 ± 5 | 63 ± 8 | ||
| 1A353 | +Trp | 42 | 145 ± 5 | 120 ± 7 | 1.21 | 515 ± 88 | |
| CYBS542 | −Trp | 37 | 327 ± 12 | 171 ± 10 | 1.91 | 2,080 ± 340 | |
| CYBS542 | +Trp | 37 | 334 ± 16 | 166 ± 12 | 2.01 | 2,530 ± 660 | |
| CYBS318 | −Trp | 37 | 0 | 120 ± 11 | 0 | 7 ± 2 | |
| CYBS223 | +Phe | 37 | <10 | 0 | 3,270 ± 502 | ||
The AT and TRAP values presented are the averages for at least three samples and two determinations.
Anthranilate synthase levels were determined in six repeat experiments; each strain and condition was examined at least three times.
Cultures were grown in minimal medium in the presence (+) or absence (−) of the indicated supplements. The final concentration of each supplement was as follows: l-tryptophan, 50 μg/ml; l-phenylalanine, 50 μg/ml; l-tyrosine, 50 μg/ml; acid-hydrolyzed casein (ACH), 0.2%; IAA, 20 or 40 μg/ml.
Assuming AT is bound as a trimer.
The minimum level of AT detectable in the samples examined in our assay is 10 molecules/cell.
AT and anthranilate synthase levels in cultures subjected to a nutritional shift.
The strategy used by B. subtilis to regulate the flow of metabolites into the common aromatic amino acid pathway differs from that used by Escherichia coli. In B. subtilis, the presence of an excess of phenylalanine and tyrosine would decrease the further metabolism of prephenate by feedback inhibition, leading to prephenate accumulation. Prephenate would then inhibit 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase, the enzyme catalyzing the first reaction of the common aromatic precursor pathway, thereby reducing the flow of carbon into the common aromatic pathway (16). This scenario could potentially starve the cell of tryptophan, depending on the effectiveness of feedback inhibition.
To examine the effects of growth with excess phenylalanine and tyrosine on AT production and trp operon expression, we shifted cultures growing in a medium containing tryptophan to media containing phenylalanine and tyrosine or acid-hydrolyzed casein as described in Table 4. The data show a striking response to a nutritional shift from a medium containing tryptophan to a medium lacking tryptophan and containing phenylalanine and tyrosine or acid-hydrolyzed casein. The anthranilate synthase activity of a wild-type culture grown with tryptophan was about 15 FU/mg of protein (Tables 2 and 3). After the shift to minimal medium plus tyrosine (50 μg/ml) and phenylalanine (100 μg/ml) or minimal medium plus 0.2% acid-hydrolyzed casein, the anthranilate synthase activity increased to 650 to 820 FU/mg of protein (Table 4). AT expression was also high, reaching approximately 155 molecules of AT trimer per cell (Table 4). Most importantly, after the shift we observed no significant increase in the anthranilate synthase level with strain CYBS318, which lacks the at operon, or with strain CYBS223, which lacks the TRAP protein. These results suggest that feedback inhibition of the common aromatic pathway resulting from the presence of excess phenylalanine and tyrosine could potentially starve cells of tryptophan and charged tRNATrp, activating AT production. Apparently, when phenylalanine and tyrosine or acid-hydrolyzed casein is added and cultures are shifted from a high-tryptophan medium to a medium lacking tryptophan, insufficient chorismate is produced to satisfy the tryptophan requirement for growth. This then triggers AT production in an effort to increase the rate of tryptophan synthesis.
TABLE 4.
Anthranilate synthase specific activity and AT protein levels in shifted culturesa
| Shift and strain | OD525 when harvested | Anthranilate synthase sp act (FU/mg of protein) | No. of molecules of AT (trimer)/cell |
|---|---|---|---|
| Shift from minb + Trp to min + Tyr + Phec | |||
| CYBS400 | 0.436 | 820 ± 11 | 160 ± 10 |
| 0.831 | 710 ± 16 | 152 ± 8 | |
| CYBS318 | 0.445 | 1 ± 0.5 | NDd |
| 0.825 | 1 ± 0 | ND | |
| CYBS223 | 0.433 | 2,250 ± 104 | ND |
| 0.830 | 2,190 ± 101 | ND | |
| Shift from min + Trp to min + 0.2% acid-hydrolyzed casein | |||
| CYBS400 | 0.475 | 750 ± 21 | 155 ± 7 |
| 0.835 | 650 ± 26 | 148 ± 11 | |
| CYBS318 | 0.492 | 2 ± 0.5 | ND |
| 0.832 | 4 ± 0.5 | ND | |
| CYBS223 | 0.465 | 2,190 ± 93 | ND |
| 0.812 | 2,420 ± 28 | ND |
The cells in the inoculum (OD525 of 0.2) were washed, centrifuged, and resuspended in the medium indicated, and then the culture was grown and harvested at the OD525 indicated in the table. The values presented are the averages for two experiments.
min, minimal medium.
phenylalanine, 100 μg/ml; tyrosine, 50 μg/ml.
ND, not determined.
DISCUSSION
In the present study and a previous study (7), it has been shown that expression of the at operon in B. subtilis does influence trp operon expression. Synthesis of the AT protein is regulated both transcriptionally and translationally, transcriptionally by the T-box mechanism of transcription termination, common to many tRNA synthetase operons of B. subtilis (14), and translationally, by leader peptide control of translation initiation at the rtpA (AT) start codon (7, 22). Both regulatory mechanisms are used to increase the AT level in response to the accumulation of uncharged tRNATrp (6). In the studies described in this paper, we show that a variety of growth conditions that would be expected to increase the cellular content of uncharged versus charged tRNATrp do increase AT production and that this increased AT production is associated with—and probably at least partly responsible for—increased trp operon expression.
We estimated the number of AT and TRAP molecules per cell under different growth conditions. However, several of the growth conditions we examined may also have lowered the tryptophan concentration in the cell. This decrease would reduce the number of active TRAP molecules, thereby elevating trp operon expression. During growth with excess tryptophan, growth conditions where cellular TRAP should be fully activated, the presence of two AT trimers per molecule of TRAP relieved TRAP function appreciably, elevating trp operon expression to a level about 70 to 80% of that observed in a mutant lacking TRAP (Table 3). When IAA was added and the AT/TRAP ratio was 0.83, trp operon expression was increased 10-fold, not the 50-fold observed with two molecules of AT/molecule of TRAP. However, under these conditions the presence of IAA probably led to depletion of some of the tryptophan in the cell, and therefore, the TRAP molecules present probably were not fully activated. Consistent with this conclusion, the growth rate in the presence of 20 or 40 μg of IAA per ml was lower than the growth rate observed in the absence of IAA. We also observed that appreciable levels of AT, ca. 145 trimers/cell, were produced in the trpS temperature-sensitive strain grown at 42°C with high concentrations of tryptophan (Table 3). This AT level and trp operon expression level were comparable to the values obtained after growth in a medium containing 40 μg of IAA per ml. This finding with the trpS mutant indicates that although its TRAP was undoubtedly fully activated by tryptophan, the presence of one molecule of AT trimer/molecule of TRAP was sufficient to reduce the effectiveness of TRAP, increasing trp operon expression appreciably, 25-fold over the value observed with the wild-type control (Table 3).
The results presented in Fig. 3 and Table 3 show that increased AT levels are associated with increased specific activity of anthranilate synthase. Even in the presence of excess tryptophan and normal levels of TRAP, in the tryptophanyl tRNA synthetase mutant and in the AT-overproducing strain, the anthranilate synthase level increased appreciably (Table 3). Thus, despite the fact that every TRAP molecule is fully activated, the presence of AT must inactivate a sufficient number of these activated TRAP molecules so they can no longer bind RNA. To determine whether AT, once produced, is stable when it is no longer needed, we grew the wild-type strain in minimal medium plus glucose and IAA (40 μg/ml) to an OD525 of 0.6. We then divided the culture into three fractions and harvested and washed the cells in each fraction. The cells were then resuspended in the following three media at an OD525 of 0.2: (i) minimal medium plus glucose; (ii) minimal medium plus glucose plus 100 μg of tryptophan per ml; and (iii) minimal medium plus glucose and chloramphenicol, an inhibitor of translation. Portions of the cells in each culture were harvested after 20 min, 1 h, or 2 h of incubation, and the AT level was then determined. The results obtained (data not shown) establish that AT is stable; its concentration decreased to a level consistent with replication of the cells in each culture.
Most interesting and unexpected was the finding in strains lacking the at operon that trp operon expression was reduced relative to the level of expression observed in wild-type cultures (Table 2; Table 3). Perhaps this result implies that in wild-type cultures there is a basal level of AT produced per cell and this AT level is sufficient to inactivate a small fraction of the active TRAP that is present. This observation was not examined further. Our Western blotting procedure, as used, detected only AT levels greater than 10 molecules per cell; therefore, we could not detect lower AT levels, if they existed in wild-type cultures.
Obtaining an overall understanding of the interrelationships between the many processes and events that influence trp operon expression in B. subtilis is not possible at this time. As detailed in the introduction, multiple events influence trp operon expression in this organism, some of which are poorly understood. In addition, a quantitative analysis will be required of the role of each event under different physiological conditions in order to relate the various metabolic activities influencing tryptophan biosynthesis to one another.
One technical issue relevant to our findings which we believe we have addressed is whether our measurements of the AT and TRAP levels were accurate. Of some concern was whether the low-molecular-weight subunits of the TRAP and AT proteins would pass through the membranes during electrophoretic transfer. To address this concern, we used double nitrocellulose membranes back to back during electrophoretic transfer to determine whether any AT or TRAP subunits passed through the first nitrocellulose membrane. Using Western blot analysis, we did not detect any AT or TRAP on the second membrane. We also ran a standard curve for each blot on an identical gel and transferred the polypeptides to the same membrane in each set of measurements to reduce the variation between different assays.
McCabe and Gollnick have recently measured the cellular levels of TRAP protein in B. subtilis cultures grown in Luria-Bertani broth either overnight or to mid-log phase or grown in Vogel and Bonner minimal medium in the absence or presence of tryptophan (18). They estimate that there are approximately 200 to 400 molecules of TRAP 11-mer per cell. Our estimated value is slightly less than half of their value, which may be due to the different strains and methods that were used. It is known that there are at least four different mRNAs with TRAP-binding sites in B. subtilis (2). Furthermore, it has been shown that TRAP can be released from bound mRNA in vivo by polynucleotide phosphorylase action (9). The presence of TRAP-binding sites in mRNAs of several operons that are differentially expressed may be responsible for the necessity of producing several hundred molecules of TRAP per cell to achieve regulation by this protein. Our findings demonstrate the important roles played by uncharged tRNATrp sensing, and AT synthesis on TRAP function in maintaining a desired level of trp operon expression.
Acknowledgments
We are grateful to P. Gollnick for kindly providing antisera. We thank P. Gollnick, G. N. Chen, L. R. Cruz-Vera, and M. Gong for advice and helpful discussions, and we thank P. Gollnick, P. Babitzke, and A. Antson for valuable comments on the manuscript.
These studies were supported in part by funds from the National Science Foundation, MCB-0093023.
REFERENCES
- 1.Babitzke, P. 2004. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr. Opin. Microbiol. 7:132-139. [DOI] [PubMed] [Google Scholar]
- 2.Babitzke, P., and P. Gollnick. 2001. Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure. J. Bacteriol. 183:5795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babitzke, P., and C. Yanofsky. 1993. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc. Natl. Acad. Sci. USA 90:133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berka, R. M., X. Cui, and C. Yanofsky. 2003. Genomewide transcriptional changes associated with genetic alterations and nutritional supplementation affecting tryptophan metabolism in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 100:5682-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnette, W. N. 1981. “Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 6.Chen, G., and C. Yanofsky. 2003. Tandem transcription and translation regulatory sensing of uncharged tryptophan tRNA. Science 301:211-213. [DOI] [PubMed] [Google Scholar]
- 7.Chen, G., and C. Yanofsky. 2004. Features of a leader peptide coding region that regulate translation initiation for the anti-TRAP protein of B. subtilis. Mol. Cell 13:703-711. [DOI] [PubMed] [Google Scholar]
- 8.Creighton, T. E., and C. Yanofsky. 1969. Chorismate to tryptophan (Escherichia coli)-anthranilate synthetase, PR transferase, PRA isomerase, InGP synthetase, tryptophan synthetase. Methods Enzymol. 17:365-386. [Google Scholar]
- 9.Deikus, G., P. Babitzke, and D. H. Bechhofer. 2004. Recycling of a regulatory protein by degradation of the RNA to which it binds. Proc. Natl. Acad. Sci. USA 101:2747-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du, H., and P. Babitzke. 1998. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis. J. Biol. Chem. 273:20494-20503. [DOI] [PubMed] [Google Scholar]
- 11.Du, H., R. Tarpey, and P. Babitzke. 1997. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J. Bacteriol. 179:2582-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gollnick, P., P. Babitzke, E. Merino, and C. Yanofsky. 2002. Aromatic amino acid metabolism in Bacillus subtilis, p. 233-244. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 13.Gollnick, P., S. Ishino, M. I. Kuroda, D. J. Henner, and C. Yanofsky. 1990. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:8726-8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundy, F. J., and T. M. Henkin. 2003. The T box and S box transcription termination control systems. Front. Biosci. 8:d20-d31. [DOI] [PubMed] [Google Scholar]
- 15.Henkin, T. M., and C. Yanofsky. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700-707. [DOI] [PubMed] [Google Scholar]
- 16.Henner, D., and C. Yanofsky. 1993. Biosynthesis of aromatic amino acids, p. 269-280. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 17.Lee, A. I., J. P. Sarsero, and C. Yanofsky. 1996. A temperature-sensitive trpS mutation interferes with TRAP regulation of trp gene expression in Bacillus subtilis. J. Bacteriol. 178:6518-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCabe, B. C., and P. Gollnick. 2004. Cellular levels of trp RNA-binding attenuation protein in Bacillus subtilis. J. Bacteriol. 186:5157-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merino, E., P. Babitzke, and C. Yanofsky. 1995. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 177:6362-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otridge, J., and P. Gollnick. 1993. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc. Natl. Acad. Sci. USA 90:128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarsero, J. P., E. Merino, and C. Yanofsky. 2000. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J. Bacteriol. 182:2329-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarsero, J. P., E. Merino, and C. Yanofsky. 2000. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc. Natl. Acad. Sci. USA 97:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 24.Sekiguchi, J., N. Takada, and H. Okada. 1975. Genes affecting the productivity of α-amylase in Bacillus subtilis Marburg. J. Bacteriol. 121:688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevtsov, M. B., Y. Chen, P. Gollnick, and A. A. Antson. 2004. Anti-TRAP protein from Bacillus subtilis: crystallization and internal symmetry. Acta Crystallogr. D 60:1311-1314. [DOI] [PubMed] [Google Scholar]
- 26.Shudershana, S., H. Du, M. Mahalanabis, and P. Babitzke. 1999. A 5′ RNA stem-loop participates in the transcription attenuation mechanism that controls expression of the Bacillus subtilis trpEDCFBA operon. J. Bacteriol. 181:5742-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder, D., J. Lary, Y. Chen, P. Gollnick, and J. L. Cole. 2004. Interaction of the trp RNA-binding attenuation protein (TRAP) with anti-TRAP. J. Mol. Biol. 338:669-682. [DOI] [PubMed] [Google Scholar]
- 28.Steinberg, W. 1974. Temperature-induced derepression of tryptophan biosynthesis in a tryptophanyl-transfer ribonucleic acid synthetase mutant of Bacillus subtilis. J. Bacteriol. 117:1023-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinberg, W., and C. Anagnostopoulos. 1971. Biochemical and genetic characterization of a temperature-sensitive tryptophanyl-transfer ribonucleic acid synthetase mutant of Bacillus subtilis. J. Bacteriol. 105:6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valbuzzi, A., and C. Yanofsky. 2001. Inhibition of the B. subtilis regulatory protein TRAP by the TRAP-inhibition protein, AT. Science 293:2057-2059. [DOI] [PubMed] [Google Scholar]
- 31.Valbuzzi, A., and C. Yanofsky. 2002. Zinc is required for assembly and function of the anti-trp RNA-binding attenuation protein, AT. J. Biol. Chem. 277:48574-48578. [DOI] [PubMed] [Google Scholar]
- 32.Valbuzzi, A., P. Gollnick, P. Babitzke, and C. Yanofsky. 2002. The anti-trp RNA-binding attenuation protein (anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J. Biol. Chem. 277:10608-10613. [DOI] [PubMed] [Google Scholar]
- 33.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 34.Yakhnin, H., H. Zhang, A. V. Yakhnin, and P. Babitzke. 2004. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J. Bacteriol. 186:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, M., A. de Saizieu, A. P. van Loon, and P. Gollnick. 1995. Translation of trpG in Bacillus subtilis is regulated by the trp RNA-binding attenuation protein (TRAP). J. Bacteriol. 177:4272-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanofsky, C. 2004. The different roles of tryptophan transfer RNA I regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 20:367-374. [DOI] [PubMed] [Google Scholar]