Abstract
Background
Inflammatory bowel disease (IBD) is a multifactorial chronic inflammatory disease resulting from dysregulation of the mucosal immune response and gut microbiota. Crohn's disease (CD) and ulcerative colitis (UC) are difficult to distinguish, and differential diagnosis is essential for establishing a long-term treatment plan for patients. Furthermore, the abundance of mucosal bacteria is associated with the severity of the disease. This study aimed to differentiate and diagnose these two diseases using the microbiome and identify specific biomarkers associated with disease activity.
Results
Differences in the abundance and composition of the microbiome between IBD patients and healthy controls (HC) were observed. Compared to HC, the diversity of the gut microbiome in patients with IBD decreased; the diversity of the gut microbiome in patients with CD was significantly lower. Sixty-eight microbiota members (28 for CD and 40 for UC) associated with these diseases were identified. Additionally, as the disease progressed through different stages, the diversity of the bacteria decreased. The abundances of Alistipes shahii and Pseudodesulfovibrio aespoeensis were negatively correlated with the severity of CD, whereas the abundance of Polynucleobacter wianus was positively correlated. The severity of UC was negatively correlated with the abundance of A. shahii, Porphyromonas asaccharolytica and Akkermansia muciniphilla, while it was positively correlated with the abundance of Pantoea candidatus pantoea carbekii. A regularized logistic regression model was used for the differential diagnosis of the two diseases. The area under the curve (AUC) was used to examine the performance of the model. The model discriminated UC and CD at an AUC of 0.873 (train set), 0.778 (test set), and 0.633 (validation set) and an area under the precision-recall curve (PRAUC) of 0.888 (train set), 0.806 (test set), and 0.474 (validation set).
Conclusions
Based on fecal whole-metagenome shotgun (WMS) sequencing, CD and UC were diagnosed using a machine-learning predictive model. Microbiome biomarkers associated with disease activity (UC and CD) are also proposed.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-023-03084-5.
Keywords: Gut microbiome, Inflammatory bowel disease, Crohn’s disease, Ulcerative colitis, Machine learning, Whole metagenome shotgun (WMS) sequencing
Introduction
Inflammatory bowel disease (IBD) is a multifactorial disease that results in chronic intestinal inflammation due to the dysregulation of immune responses. More than 3.5 million patients in the United States and Europe have IBD, which is becoming more common worldwide [1].
The human gut microbiome plays an important role in nutrient metabolism, pathogen protection, and immune system development. It is generally accepted that the development and progression of IBD are more closely associated with the gut microbiome [2–6]. In IBD, genetic and environmental factors such as an altered gut microbiome and enhanced intestinal permeability play roles in deregulating intestinal immunity, eventually leading to gastrointestinal damage [7, 8]. IBD includes Crohn's disease (CD) and ulcerative colitis (UC). Although the distribution, location, and histology of the inflammatory site vary between these two diseases, approximately 10–15% of patients exhibit the same pattern [9, 10]. Moreover, it is difficult to classify the state of the disease because the clinical and pathological characteristics overlap [9]. A clear differentiation between these two disorders must be made to develop an effective treatment plan.
As CD and UC are progressive conditions, determining their severity is essential for selecting the best course of treatment [11, 12]. According to Swidsinski et al., the concentration of mucosal-associated bacteria was related to the severity of the disease [13, 14]. Potential variations in the microbial composition related to disease activity may serve as markers for disease monitoring. This study investigated biomarkers for differential diagnosis and identified distinctive microbiomes associated with disease activity.
Results
Clinical statistics of the study population
In this study, 482 samples were obtained from 50 healthy controls (HC), 173 patients with CD, and 259 patients with UC. The demographic and clinical characteristics of the participants are presented in Table 1. There were differences in sex, age, and body mass index (BMI) between CD and UC patients, but no differences in family history. Table 2 shows the basic characteristics of patients with CD according to disease activity groups. Of the 173 patients, we excluded 82 who received biologics, 4 who underwent surgery, and 18 who underwent both biologics and surgery. Of the 69 patients with CD, 49 had mild CD, 19 moderate CD, and one severe CD. There were no differences in sex, age, BMI, family history, or smoking status between the mild and moderate patient groups. Table 3 shows the basic characteristics of the patients with UC according to their disease activity. Of the 259 patients, 61 who received biologics were excluded. Of 198 patients with UC, 162 had mild UC, 25 had moderate UC, and 11 had severe UC. Sex, age, BMI, and smoking status were not significantly different in the mild, moderate, and severe groups of patients, but there was a significant difference in family history (Table 3).
Table 1.
Clinicopathological characteristics used in this study
| Group | HC | CD | UC | p-value |
|---|---|---|---|---|
| Number | 50 | 173 | 259 | |
| Gender | < 0.001 | |||
| Male | 17 (34.0%) | 133 (76.9%) | 155 (59.8%) | |
| Female | 33 (66.0%) | 40 (23.1%) | 104 (40.2%) | |
| Age, median years (range) | 38 (19—70) | 30 (14—83) | 49 (19—84) | < 0.001 |
| BMI, median BMI (range) | 23.7 (16.2—32.8) | 20.4 (12.4—36.5) | 22.68 (14.06—38.2) | < 0.001 |
| Family history | - | 0.493 | ||
| Yes | 6 (3.5%) | 15 (5.8%) | ||
| No | 149 (86.1%) | 221(85.3%) | ||
| Un-known | 18 (10.4%) | 23 (8.9%) | ||
| Smoking history | < 0.001 | |||
| Never | 36 (72.0%) | 102 (59.0%) | 125 (48.3%) | |
| Past smoker | 11 (22.0%) | 26 (15.0%) | 96 (37.1%) | |
| Current smoker | 3 (6.0%) | 26 (15.0%) | 18 (6.9%) | |
| Un-known | 0 (0.0%) | 19 (11.0%) | 20 (7.7%) |
Table 2.
A basic characteristic of CD patients according to disease activity groups
| Group | Mild | Moderate | p-value |
|---|---|---|---|
| Number | 49 | 19 | |
| Gender | 0.153 | ||
| Male | 36 (73.5%) | 17 (89.5%) | |
| Female | 13 (26.5%) | 2 (10.5%) | |
| Age, median years (range) | 30 (19–83) | 29 (14–56) | 0.341 |
| BMI, median BMI (range) | 22.2 (14.5–36.5) | 20.1 (13.1–31.5) | 0.110 |
| Family history | 0.446 | ||
| Yes | 2 (4.1%) | 0 (0.0%) | |
| No | 41 (83.7%) | 18 (94.7%) | |
| Un-known | 6 (12.2%) | 1 (5.3%) | |
| Smoking history | 0.774 | ||
| Never | 27 (55.1%) | 13 (68.4%) | |
| Past smoker | 7 (14.3%) | 2 (10.5%) | |
| Current smoker | 9 (18.4%) | 2 (10.5%) | |
| Un-known | 6 (12.2%) | 2 (10.5%) |
Table 3.
A basic characteristic of UC patients according to disease activity groups
| Group | Mild | Moderate | Severe | p-value |
|---|---|---|---|---|
| Number | 162 | 25 | 11 | |
| Gender | 0.520 | |||
| Male | 102 (63.0%) | 13 (52.0%) | 6 (54.5%) | |
| Female | 60 (37.0%) | 12 (48.0%) | 5 (45.5%) | |
| Age, median years (range) | 49 (19–84) | 43 (21–71) | 47 (24–67) | 0.396 |
| BMI, median BMI (range) | 22.8 (14.5–38.2) | 23.9 (16.9–30) | 19.7 (17.6–26.4) | 0.176 |
| Family history | 0.020 | |||
| Yes | 8 (4.9%) | 0 (0.0%) | 3 (27.3%) | |
| No | 138 (85.2%) | 22 (88.0%) | 7 (63.6%) | |
| Un-known | 16 (9.9%) | 3 (12.0%) | 1 (9.1%) | |
| Smoking history | 0.806 | |||
| Never | 77 (47.5%) | 14 (56.0%) | 4 (36.4%) | |
| Past smoker | 59 (36.4%) | 7 (28.0%) | 4 (36.4%) | |
| Current smoker | 13 (8.0%) | 1 (4.0%) | 1 (9.1%) | |
| Un-known | 13 (8.0%) | 3 (12.0%) | 2 (18.2%) |
Association of the enterotypes with IBD
Enterotypes are used to categorize people according to their gut microbiomes and as potential biomarkers of the healthy human intestine. Enterotypes have recently attracted attention in predicting their relationships with diseases [15]. So, we examined the relationships between enterotypes and the health status of our cohort. Principal Coordinate Analysis (PCoA) of the gut microbiome of 50 HC and 432 IBD patients revealed three groups (Fig. S1A). The abundance of Bifidobacterium was similar between enterotype 2 and 3, although that of Bacteroides was higher in enterotype 2 than in enterotype 1, and that of Faecalibacterium was similar between enterotype 1 and 3 (Fig. S1B). 42.5% of the samples were grouped into the enterotype 2 cluster (Faecalibacterium), followed by 32.1% of the enterotype 1 cluster (Bifidobacterium) and 25.3% of the enterotype 3 cluster (Bacteroides). The analysis showed that their distribution in HC and IBD was significantly different, as shown in Table S1 (Fisher’s exact test, p < 0.05). The enterotype with a Faecalibacterium predominance was underrepresented in the HC (74% vs 37% vs 40%).
Differences in the taxonomic composition of the gut microbiome in patients with CD and UC
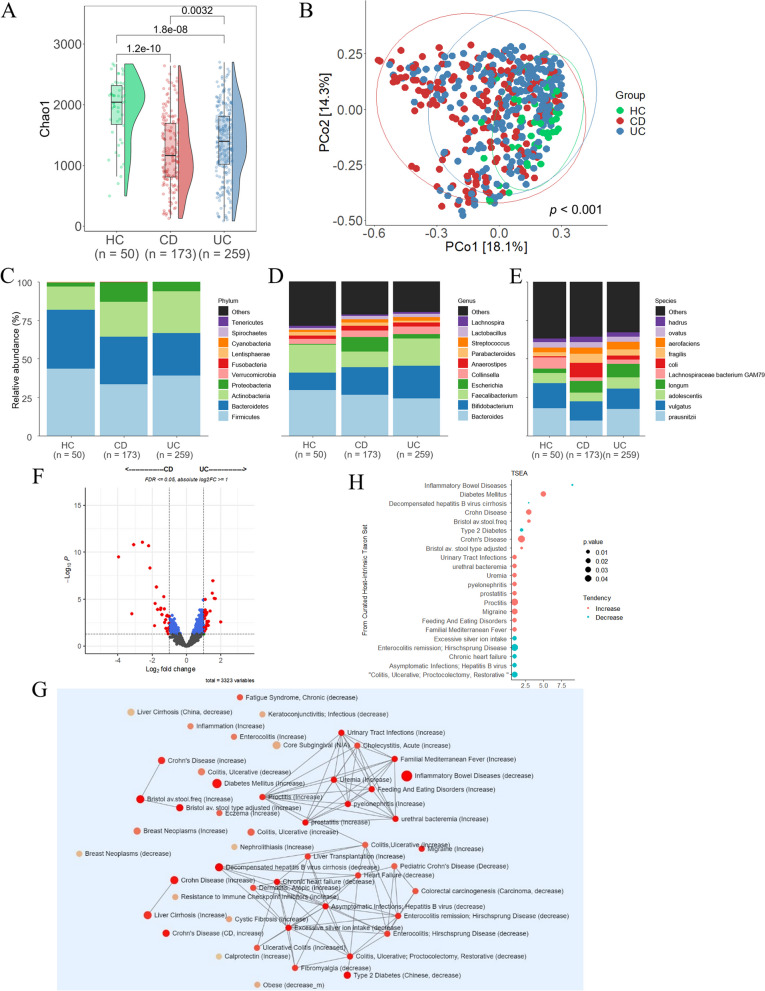
The abundance tables were rarefied to 1,222,561 sequences per sample by random subsampling using the phyloseq R package [16]. To evaluate the alpha diversity, we calculated the Chao1 diversity index, which considers the number, uniformity, and abundance of taxa observed in each sample. The alpha diversity indices were lower in patients with IBD than healthy controls. The Chao1 diversity index was significantly lower in patients with UC and CD than in HC and was significantly lower in patients with CD than in UC patients (Fig. 1A). Beta diversity was measured as the Bray–Curtis distance, which was visualized using principal coordinate analysis (PCoA) (Fig. 1B). The relative abundance of the gut microbiome was compared among the groups (HC, CD, and UC). The following percentages in parentheses are in the order of HC > CD > UC. Compared to HC, IBD patients had significantly higher abundances of phylum Actinobacteria (15.22, 22.77, and 27.13%). It is also more abundant in patients with UC than those with CD. In contrast, Firmicutes (43.60, 33.75, and 39.14%) decreased in patients with IBD compared to HC and were more reduced in patients with CD than in UC patients (Fig. 1C and Fig. S2A). At the genus level, Escherichia (0.64, 9.48, and 2.67%) increased in patients with IBD compared to HC and was significantly higher in patients with CD than in UC patients. In contrast, Faecalibacterium (18.13, 10.04, and 17.54%) was reduced in patients with IBD compared to that in HC; it decreased significantly in patients with CD compared to that in UC patients (Fig. 1D and Fig. S2B). At the species level, Lachnospiraceae bacterium GAM79 (7.53, 2.37, and 2.97%) was lower in patients with IBD than in HC, and patients with CD than in UC patients. E. coli (0.67, 9.45, and 2.68%) was more abundant in patients with IBD and significantly more abundant in CD patients (Fig. 1E and Fig. S2C). Linear models for differential abundance analysis (LinDA) was used to compare the predominance of communities between groups, and significant differences in microbial abundance were observed between the CD and UC groups. Sixty-eight microbiotas were identified at the species level, among which 28 were over-represented in the CD group and 40 were over-represented in the UC group (Fig. 1F, FDR < = 0.05, absolute log2FC > = 1). Shigella dysenteriae, Erysipelotrichaceae bacterium|46, E. coli, and Escherichia marmotae were more abundant in the CD group, whereas Bacillus vallismortis, Lactobacillus ruminis, Alicycliphilus dentirficans, and Lachnospira eligens were more abundant in the UC group (Table S2). Taxon set enrichment analysis (TSEA) was performed to identify the taxon sets associated with host intrinsic factors. We identified 23 disease-related terms that were significant in the curated set of host intrinsic factors, including decompensated hepatitis B virus cirrhosis, IBD, type 2 diabetes, chronic heart failure, excessive silver ion intake, and familial Mediterranean fever (Fig. 1G and H).
Fig. 1.
Diversity and distribution of microbiomes in healthy control (HC) and IBD (CD and UC) patient groups. A Alpha diversity was calculated as the Chao1 index; (B) Beta diversity was calculated as Bray–Curtis dissimilarity. PCoA was used to display the microbiome space among the groups. C, D, E Comparison of relative taxa abundance among HC, CD, and UC at the phylum (C), genus (D), and species (E) levels. The taxonomic biomarkers were identified using LinDA (FDR < = 0.05, |log2FC|> = 1). F Volcano plot of the composition of the intestinal microbial community at the species level. G Taxon Set Enrichment Analysis (TSEA) results showing the network modules of enriched terms. H Taxon set terms representing the association of human diseases with the known composition of the groups (p < = 0.05)
Metabolic functions of the different gut microbiome
The HUMAnN3 tool was used to explore the potential functions of the microbial community. Subsequently, we compared the differentially enriched MetaCyc pathways and KEGG Orthology (KO) terms using LinDA. A significant difference in the MetaCyc pathway and KO terms was observed between patients with CD and UC. Sixty-eight differentially abundant MetaCyc pathways were identified. Among these, 66 pathways (phytate degradation I, mannosylglycerate biosynthesis I, and 2-methylcitrate cycle I etc.) were enriched in CD patients. In comparison, only two pathways (super pathways of polyamine biosynthesis II and superpathway ofmenaquinol-8 biosynthesis II) were enriched in UC patients (FDR < = 0.05 and |log2FC|> = 1) (Table S3). In the KOs, 1,242 differentially enriched KO terms were found, and 1153 KO terms (FKBP-type peptidyl-prolyl cis–trans isomerase SlpA, biopolymer transport protein ExbD, and trehalose 6-phosphate phosphatase [EC:3.1.3.12]) were enriched in CD patients. In contrast, 89 KO terms (hypothetical protein, stage V sporulation protein AB, and multidrug/hemolysin transport system permease protein) were enriched in UC patients (FDR < = 0.05 and |log2FC|> = 1) (Table S4).
Alteration in gut microbiome according to the activity of CD
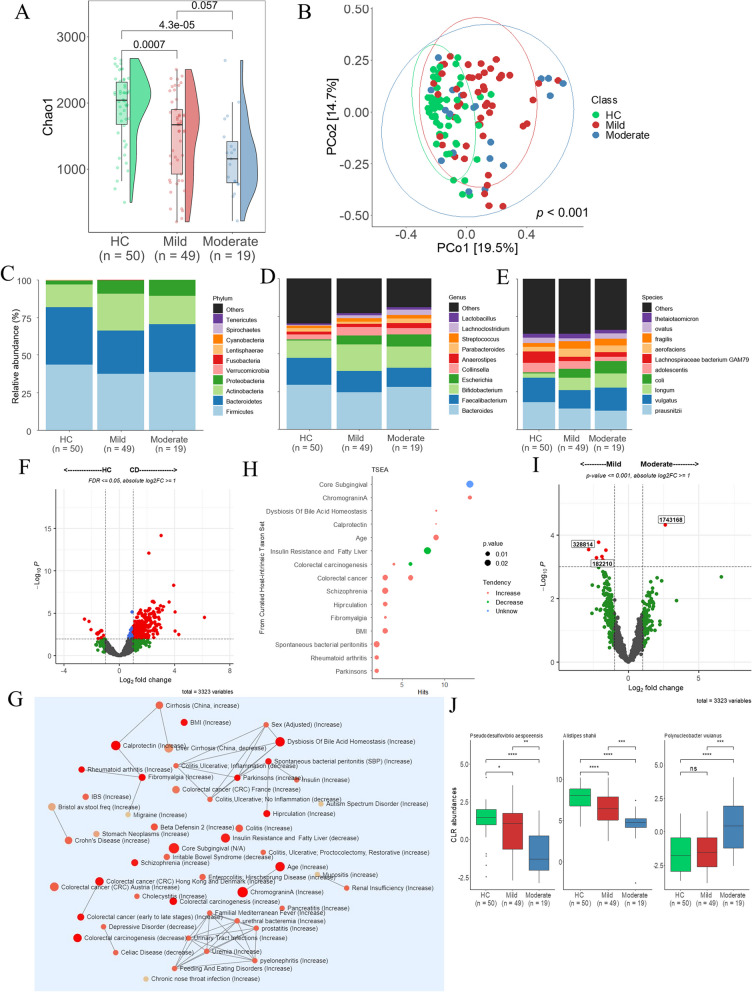
The analysis was performed using the same pipeline as in the previous analysis. The alpha diversity index of the mild and moderate CD groups decreased more than that of the HC group. However, the difference between the mild and moderate groups was not significant. By analyzing beta diversity using Bray–Curtis and PCoA, we found significant differences in the microbial communities among the HC, mild, and moderate groups (Fig. 2A and B). The microbial composition of relative abundance differed between HC and CD but was similar in the mild and moderate groups. The following percentages in parentheses are in the order of HC > mild > moderate groups. Verruciomicrobia (0.27, 0.05, and 0.01%) and Cyanobacteria (0.04, 0.03, and 0.02%) at the phylum level were lower in the mild and moderate CD groups than in the HC group (Fig. 2C and Fig. S3A). At the genus level, Escherichia coli (0.64, 5.97, and 8.03%) increased in the mild and moderate groups compared to the HC group, but the difference between the mild and moderate groups was not significant (Fig. 2D and Fig. S3B). At the species level, E. coli (0.67, 5.99, and 8.07%) increased in the mild and moderate groups compared with the HC group, but the difference between the mild and moderate groups was not significant (Fig. 2E and Fig. S3C). Subsequently, differential abundance analysis was performed. First, the microbial compositions of the HC and CD groups were compared using LinDA. Differential abundance tests revealed that 554 species were differentially abundant between HC and CD groups. Fifty microbiotas were over-represented in HC, whereas 504 microbiotas were over-represented in CD (Fig. 2F, FDR < = 0.05 and |log2FC|> = 1) (Table S5). Then, TSEA was performed and identified numerous terms related to colorectal cancer and 17 other terms (Fig. 2G and H). Second, in patients with CD, the mild and moderate groups were compared at the species level. Eight bacteria (Fig. 2I, p < = 0.001 and |log2FC|> = 1) were identified (Table S6), but among them, bacteria that did not differ between the HC and CD groups were excluded. Three bacterial species were identified. Alistipes shahii and Pseudodesulfovibrio aespoeensis were over-represented in the mild group, whereas Polynucleobacter wuianus was over-represented in the moderate group (Fig. 2J). Third, a correlation between the CD activity and bacterial abundance was identified (Table S7). The abundance of A. shahii (R = -0.57, p = 1.41E-11) and P. aespoeensis (R = -0.46, p = 1.59E-07) were negatively correlated with CD activity. In contrast, the abundance of Polynucleobacter wianus (R = 0.35 and p = 8.77E-05) showed a positive correlation.
Fig. 2.
Diversity and distribution of microbiomes in CD subgroups. A Measurement of the Chao1 index of community richness. B PCoA showing differences in microbial composition by the Bray–Curtis index. C, D, E Taxonomic composition of the gut microbiome in the levels of CD-active group at the phylum (C), genus (D), and species (E). F Difference in microbial abundances between HC and CD (FDR < = 0.05, |log2FC|> = 1). G TSEA network modules displayed by TSEA. H The TSEA results show 17 host-intrinsic terms (p < = 0.05). I Comparison of the microbial composition between mild and moderate (p < = 0.001, absolute log2FC > = 1) at the species level. J Signatures of the gut microbiota related to CD activity. Box plots represent the median and interquartile range. The significances are represented as follows: ****p < = 0.0001; ***p < = 0.001; **p < = 0.01; *p < = 0.05; and ns, p > 0.05
Alteration in gut microbiome according to the activity of UC
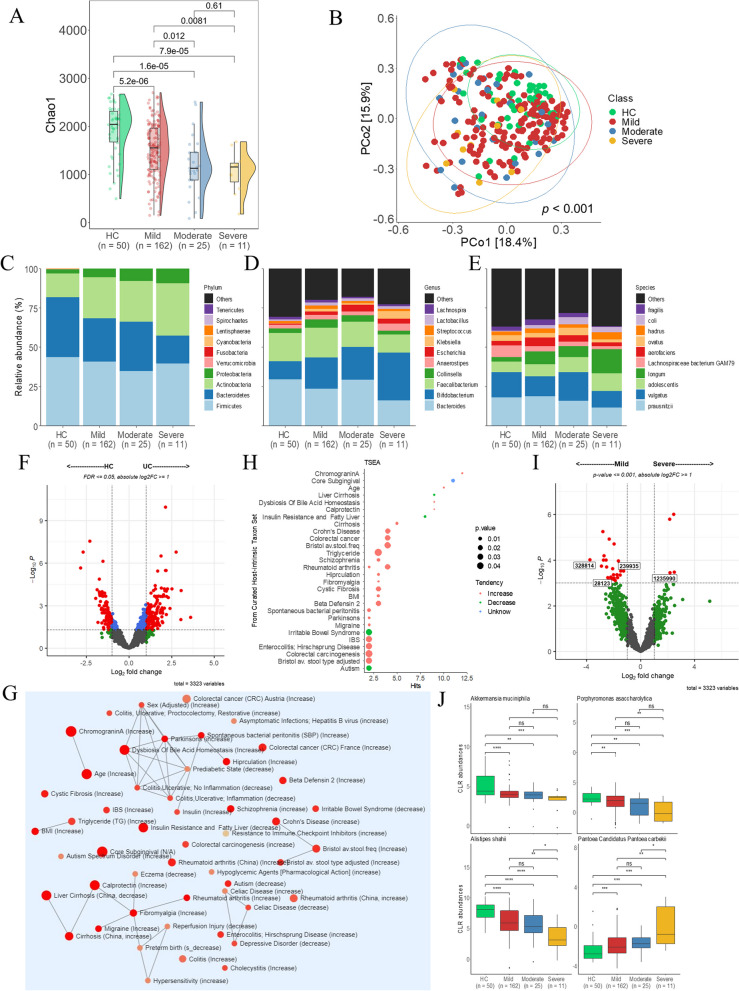
The alpha diversity index decreased in patients with mild, moderate, and severe disease compared to the HC group. Furthermore, the alpha diversity in patients with mild disease differed significantly from those with moderate or severe disease. In contrast, there were no differences in the Chao1 index between the patients with moderate and severe disease. PCoA based on the Bray–Curtis dissimilarity index identified significant differences in microbial composition among the HC, mild, moderate, and severe groups (Fig. 3A and B). The composition of relatively abundant microbiomes differed between the HC and UC groups. Among patients with UC, the mild and moderate groups were similar, but the mild and severe groups differed. The following percentages are in the order of HC > mild > moderate > severe groups. At the phylum level, Actinobacteria (15.22, 26.00, 25.97, and 33.16%) increased in the mild and severe groups compared with the HC group. However, Spirochaetes (0.03, 0.02, 0.01, and 0.01%) and Cynobacteria (0.04, 0.03, 0.02, and 0.02%) decreased (Fig. 3C and Fig. S4A). At the genus level, Bifidobacterium (11.42, 19.80, 20.84, and 30.44%) increased in patients with mild, moderate, and severe UC, respectively, compared to HC. However, there were no significant differences between the patients with UC (Fig. 3D and Fig. S4B). At the species level, the abundance of the Lachnospiraceae bacterium GAM79 (7.53, 3.47, 2.45, and 2.19%) was lower in the UC group than in the HC group, but the difference between the moderate and severe groups was insignificant (Fig. 3E and Fig. S4C). Differential abundance between HC and UC was then compared using LinDA. Between the HC and UC groups, 332 species were differentially abundant. In the HC group, 170 bacteria were over-represented, whereas 162 were over-represented in the UC group (Fig. 3F, FDR < 0.05 and |log2FC|> = 1). (Table S8). These bacteria are associated with many diseases, including irritable bowel syndrome (IBS), colorectal cancer, enterocolitis, and UC (Fig. 3G and H). Second, we compared patients with mild and severe UC at the species level and identified 30 bacterial species. Among them, bacteria that did not change between patients with HC and UC were omitted (Fig. 3I, p < 0.001 and |log2FC|> = 1) (Table S9). Therefore, a total of four microbial communities were discovered. Pantoea Candidatus Pantoea carbekii increased in the severe group, whereas Porphyromonas asaccharolytica, Akkermansia muciniphila, and A. shahii decreased (Fig. 3J). Third, a correlation between UC disease activity and species abundance was identified (Table S10). UC activity was negatively correlated with the abundance of A. shahii (R = -0.42, p = 2.97E-12), P. asaccharolytica (R = -0.30, p = 9.08E-07), and A. muciniphila (R = -0.37, p = 2.00E-09). However, P. candidatus Pantoea carbekii exhibited a positive correlation (R = 0.31, p = 4.51E-07).
Fig. 3.
Diversity and distribution of microbiomes in the subgroups of UC according to disease severity. A Alpha diversity measured by the Chao1 index. B Bray–Curtis-based Beta diversity. Relative abundance of the gut microbiome of the levels of UC activity groups at the phylum (C), genus (D) and species (E) levels. F The difference in microbial abundances between HC and UC (FDR < = 0.05, |log2FC|> = 1). G Network modules produced by the microbiota. H The microbiota between HC and UC have been implicated in various human diseases (p < = 0.05). I Comparison of the microbial composition between the mild and severe groups at the species level (p < = 0.001, log2FC|> = 1). J Biomarkers of bacteria associated with the severity of UC. Box plots represent the median and interquartile range. The significances are represented as follows: ***p < = 0.0001; ***p < = 0.001; **p < = 0.01; *p < = 0.05; and ns, p > 0.05
Classification of IBD by machine learning with microbiome
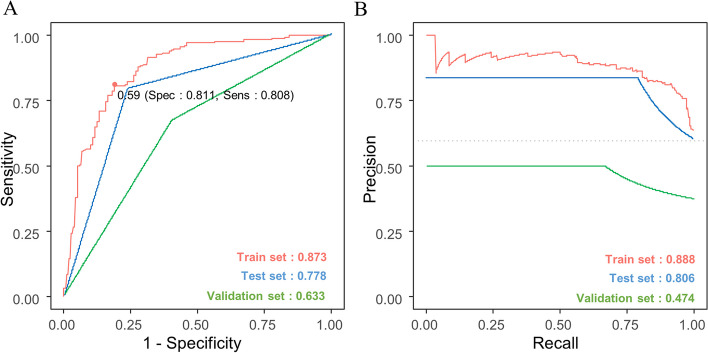
After establishing that CD and UC have different microbial compositions, we attempted to develop a microbiota-based diagnostic model to distinguish between these two diseases. Lazy predict was used to compare classification models to get the best-performing method. Regarding the classification accuracy for CD and UC, ridge logistic regression had the highest accuracy, F1 score and area under the curve (AUC), followed by LinearSVC, RidgeClassifiter, and LinearDiscriminantAnalysis (Fig. S5A, B). Therefore, a penalized (regularized) logistic ridge regression model was used with tenfold cross-validation to select the best method. The optimal lambda (log [λ] = -2.016529) and accompanying regression coefficients were selected as the location where the mean square error (MSE) was the lowest (Table S11 and Fig. S6A). Using the ROC analysis, criterion-related cut-off values were generated to predict IBD. Based on the ideal cut-off threshold (0.59) of the training set, sensitivity and specificity were determined for each data set. We obtained the AUCs of 0.873 and 0.778 for the training and test sets. The model was validated in an independent cohort of 50 IBD patients based on the logistic regression coefficients of the selected predictors. As a result, an AUC of 0.633 was confirmed in the validation cohort (Fig. 4A). PRAUC of 0.888, 0.806 and 0.474 were achieved for the training, test and validation sets (Fig. 4B). In a test set of 128 IBD, the accuracy, sensitivity, specificity, precision, and F1 scores were 0.781, 0.765, 0.792, 0.709, and 0.736. In a validation cohort of 50 IBD, the accuracy, sensitivity, specificity, precision, and F1 scores were 0.625, 0.600, 0.667, 0.750, and 0.667 (Table 4).
Fig. 4.
The performance of discriminating CD from UC using 68 microbiome biomarkers using a ridge regression model. A Receiver operating characteristic (ROC) curve. B Precision-recall (PR) curve
Table 4.
A summary of the diagnostic performance of the microbiome-based biomarker panel in the test set and validation cohorts
| Data set | Accuracy | Sensitivity | Specificity | Precision | F1 score |
|---|---|---|---|---|---|
| Test set |
0.781 95% CI: (0.700, 0.850) |
0.765 | 0.792 | 0.709 | 0.736 |
| Validation set |
0.625 95% CI: (0.510, 0.730) |
0.600 | 0.667 | 0.750 | 0.667 |
Finally, when we compared the 68 microbes with those in previous studies, we found three overlapping species (Morganella morganii, Faecalibacterium prausnitzii, and Haemophilus parainfluenzae) [17] (Fig. S6B).
Discussion
In recent decades, considerable progress has been made in the diagnosis and treatment of IBD and our knowledge of the gut microbiome [18]. The gut microbiome contains many potential biomarkers associated with disease activity and treatment efficacy in patients with IBD. Using machine learning classification based on gut microbiome analysis of fecal samples, we provide a non-invasive approach to IBD diagnosis that is practical and effective compared to invasive procedures like colonoscopy.
We identified 68 microbiomes from 3,323 species using LinDA to compare the dominance of each community in CD and UC. Of the 68 microbiomes, 6 species belonged to Citrobacter, 4 species belonged to Escherichia, and 3 species belonged to Shigella. The microbiomes known to cause inflammation were abundant in CD. Especially, the abundance of E. coli and S. dysenteriae was significantly higher in CD. Adherent invasive E. coli can invade and attach to intestinal epithelial cells and proliferate inside macrophages [19]. On the other hand, in UC, anti-inflammatory bacteria were more abundant. Eight species were identified as belonging to Prevotella and 2 species belonging to Comamonas. Prevotella, through its antibacterial effect, can prevent the proliferation of other pathogens, significantly alleviating the severity of IBD [20]. Furthermore, three species of the gut microbiome, P. mirabilis, M. morganii, and C. amalonaticus, showed a significant increase in patients with CD and UC compared to that in HC. This result is consistent with previous literature [21–23]. The metabolic activity of M. morganii produces indolimins that cause DNA damage. M. morganii increased intestinal permeability and exacerbated colon tumorigenesis in gnotobiotic mice [21].
In previous studies, a machine learning model for IBD diagnosis was trained using information from genomic databases [24], the gut microbiome [25, 26], and histological and endoscopic findings [27]. We applied machine learning methods to the microbiome data to distinguish between CD and UC. Our results showed that the model performed better than previous models. For instance, a previous study in which ML models were trained with data from bacterial genus and operational taxonomic unit (OTU) data from only 20 patients with CD and 19 with UC, achieved 0.79 AUC and 0.72 AUC, respectively [28]. Our study achieved predictive performance with 0.873 AUC in the train set and 0.778 AUC in the test set. Additionally, when we validated our model using an independent cohort, we attained performance of an AUC of 0.633. We suggest that the AUC of 0.633 in an independent cohort is reasonably good, considering the differences in cohorts (Korean vs. American) and sequencing platforms (MGI vs. Illumina), which may cause batch effects.
We identified patterns in which some bacteria changed continuously as the disease progressed in patients with CD and UC. This result suggests that assessing the severity of a disease utilizing specific microbial biomarkers (anti- and pro-inflammatory bacteria) may help predict and monitor the effectiveness of an intervention [29]. By analyzing their association with CD activity, A. shahii, P. aespoeensis and P. wuianus were identified. In contrast, the microbiota associated with disease activity in UC includes P. candidatus pantoea carbekii, A. shahii, A. muciniphila, and P. asaccharolytica. Interestingly, A. shahii was common in both diseases. The depletion of Alistipes is an important indicator of a gut microbiome imbalance, although A. shahii has a less clearly defined metabolic role in the gut microbiota [30]. Recent research has highlighted the significant function of A. shahii as an inhibitory regulator of tumor development and its protective activity against various disorders [31, 32]. Like A. shahii, A. muciniphila acts as an immunomodulator in the intestine through regulatory T cells that produce IL-10 [33]. A. muciniphila reduces inflammatory cytokines and chemokines (TNF-, IL1-, IL6, IL12A, MIP-1A, G-CSF, and KC) in the serum and tissues [34]. The known colitis-causing bacteria Clostridium difficile, Ruminococcus gnavus, Bacteroides fragilis and E. coli, were not associated with disease activity in any of the diseases.
Most previous studies on various gut microbiomes, including IBD, have used 16S ribosomal RNA gene sequencing, which can accurately identify microbiomes at the phylum level but is limited at the species level [35]. In contrast, the whole-metagenome shotgun (WMS) sequencing method allows taxonomy to be defined more accurately at the species level and identify specific genes [36, 37]. Our study is more accurate than previous studies in identifying and comparing species levels using WMS sequencing. Another strength was the large sample size. IBD research using WMS technology has been carried out on a small scale, but our study used a large sample size, resulting in a better performance of the diagnostic model.
The present study had a few limitations. First, confounders, including BMI, age, drug use, lifestyle, and enterotypes, were not controlled for, which may affect the microbiome. Second, while we validated our model in an independent cohort of 80 Western patients, another validation in a larger cohort will strengthen the usefulness of our microbiome markers. Nonetheless, identifying fecal microbiomes associated with disease phenotypes and activity at the species level will advance our knowledge in the field.
Conclusions
CD and UC had significant differences in microbial diversity and gut microbial community composition. We classified the two diseases using shotgun metagenomics and machine learning approaches. A potential biomarker for predicting disease progression has been proposed by comparing the activity of each disease. These analyses could aid in developing novel prognostic and therapeutic strategies for CD and UC using the gut microbiome.
Materials and methods
Data collection
Two hospitals (Chungnam National University Hospital and Daejeon St. Mary’s Hospital) participated in this study. The biospecimen and data used in this study were provided by the Biobank of Chungnam National University Hospital, a member of the Korea Biobank Network. We collected the clinical data from newly diagnosed and monitored patients with CD and UC). Patients with CD and UC were divided into three subgroups according to disease activity (mild, moderate, or severe). To characterize the microbiome associated with disease severity in each group, we excluded patients who used drugs that could affect the gut microbiome and those who underwent disease-related surgery.
Public data processing
Metagenomics data and clinical data generated through the Inflammatory Bowel Disease Multiomics Database website (IBDMDB http://ibdmdb.org) were downloaded from the Sequence Read Archive (SRA). Among them, the Shotgun metagenome sequencing data was downloaded by default, and the microbial abundance file was used as an alternative to the original Shotgun metagenome sequencing fastq file. If the host_id was the same, samples from the initial fecal collection date were used. Thus, CD (n = 50) and UC (n = 30) were used to validate the machine learning model. MBECS (https://github.com/rmolbrich/MBECS) was used to correct for the batch effect between the two studies.
DNA shotgun sequencing
Fecal samples were collected between 2019 and 2020, and DNA was extracted from the fecal sample and stored at -80 °C until use. gDNA (1 mg) was sheared using an S220 Ultra Sonicator (Covaris, Woburn, MA, USA). Library preparation was performed using the MGIEasy DNA Library Prep Kit (MGI, China), according to the manufacturer’s instructions. Briefly, after size selection of the fragmented gDNA using AMPure XP magnetic beads, the fragmented gDNA was end-repaired and A-tailed at 37 °C for 30 min and 65 °C for 15 min. The index adapter was ligated to the ends of the DNA fragments at 23 °C for 60 min. After the adapter-ligated DNA was removed, PCR was performed to enrich the DNA fragments with adapter molecules. Thermocycler conditions were as follows: 95 °C for 3 min, 7 cycles at 98 °C for 20 s, 60 °C for 15 s, and 72 °C for 30 s, with a final extension at 72 °C for 10 min. The double-stranded library was quantified using the QauntiFluor ONE dsDNA system (Promega, Madison, WI, USA). The library was circularized at 37 °C for 30 min and then digested at 37 °C for 30 min, followed by cleanup of the circularization product. The library was incubated at 30 °C for 25 min using the DNA nanoball (DNB) enzyme to make the DNB. Finally, the library was quantified using the QuantiFluor ssDNA System (Promega). The prepared DNB was sequenced on the MGIseq system (MGI) with 150 bp paired end reads at a single run.
Processing of sequence data
Quality control of the raw sequences of each sample was performed using the FastQC software (ver. 0.11.9). Primer sequences were removed using Cutadapt, and low-quality sequences were trimmed using Trim Galore (ver. 0.6.4). Human host reads were subtracted by mapping them to the human reference genome (GRCh38) using the Burrows-Wheeler Aligner (BWA) (ver. 0.7.17). After mapping, the FASTQ-paired files were sorted using Samtools Sort (ver. 1.10). Taxonomic classification was performed using the standard taxonomic sequence classification tool Kraken2 (ver. 1.1.1), and relative abundance was estimated using Bayesian Reestimation of Abundance with KrakEN (Bracken) (ver. 2.6.2). After removing low-abundance reads, the data was normalized in several ways (rarefaction and centered log ratio) using R (ver. 4.1.1). Based on the MetaCyc database, HMP Unified Metabolic Analysis Network 3 (HUMAnN3) (ver. 3.0.0) was used to describe the metabolic potential of individuals within a microbial population. The data were then combined using a join table script. An unstratified table was obtained using the split_stratified_table script. LinDA was used to compare variations in the MetaCyc pathway abundance. The regrouped_table script was used to translate the gene table to uniref90_ko. The KO term was renamed using the rename_table script. Then, using the same procedure as above, an unstratified table was created, and the abundances of KO were compared.
Enterotypes identification using sample clustering between healthy control and IBD individuals
The enterotype classification was performed using the genus of relative abundance table based on the Jensen-Shannon branch clustering of samples published by the MetaHIT consortium in 2011 in the enterotyping pipeline (https://enterotype.embl.de/#) [38]. Clustering results were visualized in Principal Coordinates Analysis (PCoA) plots using the ade4 package.
Statistical analysis of metagenome data
R software (ver. 4.1.1) was used for statistical analyses and visualization. Chi-square and Fisher’s exact tests were used to assess the association between clinical characteristics and microbiome composition. The Wilcoxon rank-sum test and Student’s t-test were used to evaluate the significance of differences in microbiomes between groups. The abundance table was rarefied to 1,222,561 sequences per sample by random subsampling to analyze microbial diversity in R. Permutation analysis of variance (PERM3ANOVA) was performed using the adonis2 function of the vegan package in R (ver. 4.1.3). LinDA were used to identify significant differences in relative abundance between the two groups at the species level [39]. The analysis codes are provided at https://github.com/kangdy9801/Diagnosis-of-Crohn-s-Disease-and-Ulcerative-Colitis-Using-the-Microbiome.
From a given list of taxa of interest, TSEA examines whether any patterns exist that are biologically or ecologically significant. Each node represents a taxon set with color based on the p-value and size based on the number of hits. An edge connects two taxa if the shared hits are > 20% of the combined taxa [40]. Spearman's rank correlation analysis was used to evaluate the correlation between microbiota and disease activity.
Supervised machine learning: Classification
Utilizing Lazy Predict (https://github.com/shankarpandala/lazypredict), 27 classification models were trained to diagnose diseases based on the microbiota identified by differential abundance analysis. All hyperparameters were set to default. Samples were divided at random into 70% and 30%. The model was trained on 70% of the total sample, and the remaining 30% comprised the test set. The reported results used a ridge logistic regression model and performed the best. After selecting the most optimal model, the performance of the supervised machine learning model utilizing only the training samples was evaluated during the training phase using tenfold cross-validation. Data shuffling and splitting were performed in ten independent iterations. ROC curve analysis was performed to establish the ideal cut-off threshold, sensitivity, and specificity. To overcome the class imbalance problem, we conducted the precision-recall (PR) curve [41, 42]. Seven performance indicators, including accuracy, sensitivity, specificity, precision, AUC, PRAUC and F1 score, were used to assess the models’ performance when tested on the dataset.
Supplementary Information
Additional file 1: Fig. S1. Enterotypes identified in individuals with HC and IBD. (A) Using principal coordinate analysis (PCoA), participants were divided into three enterotypes, with Bacteroides, Faecalibacterium or Bifidobacterium as the primary distinguishing factors. (B) Three bacteria most abundant among the three enterotypes (Wilcoxon test, p < 0.05). Fig. S2. Comparison of the relative abundance of microbiota among HC, CD, and UC in phylum (A), genus (B) and species (C) based on the Kruskal-Wallis test. Each figure is shown in the order of HC, CD and UC. Fig. S3. Comparison of the relative abundance of microbiota between disease stages (HC, mild, and moderate) in CD patients at the phylum (A), genus (B) and species (C) levels. The numbers are listed in the following order: HC, mild and moderate. Fig. S4. Comparison of the relative abundance of microbiota between disease stages in UC patients at the levels of phylum (A), genus (B), and species (C) levels. The numbers are listed in the following order: HC, mild, moderate, and severe. Fig. S5. Comparison of 27 supervised machine learning models for diagnosing IBD subtypes using differential abundance. (A) F1 score; (B) ROC AUC. Fig. S6. (A) Parameter adjustment of a supervised machine learning model to classify CD and UC individuals using gut microbiomes. Ten-fold cross-validation was performed to select the optimal lambda. The point where the mean square error (MSE) was minimized was designated as the best lambda (log [λ] = -2.016529). (B) Venn diagram comparing our microbiome list with that of Clonney et al.. Fig. S7. Graphical abstract
Additional file 2: Supplementary Table 1. Association between enterotypes and IBD occurrence.
Additional file 3: Supplementary Table 2. Differences in the taxonomic composition of the gut microbiome in patients with CD and UC. Supplementary Table 3. Metacyc pathway of the different gut microbiomes (CD VS UC). Supplementary Table 4. KEGG Orthology of the different gut microbiomes (CD VS UC). Supplementary Table 5. Differences in the taxonomic composition of the gut microbiome in HC and CD. Supplementary Table 6. Differences in the taxonomic composition of the gut microbiome in mild and Moderate patients with CD. Supplementary Table 7. Correlation between microbes and severity of CD. Supplementary Table 8. Differences in the taxonomic composition of the gut microbiome in HC and UC. Supplementary Table 9. Differences in the taxonomic composition of the gut microbiome in mild and Severe patients with UC. Supplementary Table 10. Correlation between microbes and the severity of UC. Supplementary Table 11. Microbial coefficients used for modeling
Acknowledgements
Not Applicable.
Authors’ contributions
BD and healthy control cohort samples were collected by Min-Kyung Yeo, Sang-Bum Kang, Jin-Man Kim and Ju-Seok Kim. Experimental design was performed by Ju-Seok Kim, Da-Yeon Kang, Jong-Lyul Park and Seon-Young Kim. Data analysis, machine learning pipeline and figure preparation were performed by Da-Yeon Kang. The manuscript was written by Da-Yeon Kang, Ju-Seok Kim and Seon-Young Kim. All authors have read and approved the final manuscript.
Funding
This research was supported by Systemic Industrial Infrastructure Projects through the Ministry of Trade, Industry and Energy (MOTIE) (P0009796, 2019) and National Research Foundation (NRF) grant (NRF-2020M3A9I6A01036057) funded through the Ministry of Science and ICT (MOST).
Availability of data and materials
The data sets utilized in this research are available from NCBI’s Sequence Read Archive (SRA) (Accession ID: PRJNA945504) and Korean Nucleotide Archive (KoNA) (Accession ID: PRJKA220304) (https://www.kobic.re.kr/kona/).
Declarations
Ethics approval and consent to participate
This study was approved by the Chungnam National University Hospital Internal Review Board (IRB number: CNUH 2019–06-034) with informed consent from the participants. Studies involving human subjects were conducted in accordance with the Declaration of Helsinki. Minors (under the age of 16) have obtained informed consent to participate from their parents and/or legal guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Da-Yeon Kang, Jong-Lyul Park and Min-Kyung Yeo contributed equally to this work.
Contributor Information
Ju Seok Kim, Email: showsik@hanmail.net.
Seon-Young Kim, Email: kimsy@kribb.re.kr.
References
- 1.Miyoshi J, Chang EB. The gut microbiota and inflammatory bowel diseases. Transl Res. 2017;179:38–48. doi: 10.1016/j.trsl.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sartor RB. Microbial influences in inflammatory bowel Diseases. Gastroenterology. 2008;134(2):577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 3.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9(10):599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 4.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel Disease. Semin Immunopathol. 2015;37(1):47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajca S, Grondin V, Louis E, Vernier-Massouille G, Grimaud JC, Bouhnik Y, et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s Disease. Inflamm Bowel Dis. 2014;20(6):978–986. doi: 10.1097/MIB.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 6.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel Disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartor RB. Mechanisms of Disease: pathogenesis of Crohn’s Disease and ulcerative Colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3(7):390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 8.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel Disease. Nature. 2007;448(7152):427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 9.Yantiss RK, Odze RD. Diagnostic difficulties in inflammatory bowel Disease pathology. Histopathology. 2006;48(2):116–132. doi: 10.1111/j.1365-2559.2005.02248.x. [DOI] [PubMed] [Google Scholar]
- 10.Yu CS, Pemberton JH, Larson D. Ileal pouch-anal anastomosis in patients with indeterminate colitis: long-term results. Dis Colon Rectum. 2000;43(11):1487–1496. doi: 10.1007/BF02236726. [DOI] [PubMed] [Google Scholar]
- 11.Ooi CJ, Hilmi I, Banerjee R, Chuah SW, Ng SC, Wei SC, et al. Best practices on immunomodulators and biologic agents for ulcerative colitis and crohn’s disease in Asia. Intest Res. 2019;17(3):285–310. doi: 10.5217/ir.2019.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe K. Clinical management for small bowel of crohn’s disease in the treat-to-target era: now is the time to optimize treatment based on the dominant lesion. Intest Res. 2020;18(4):347–354. doi: 10.5217/ir.2020.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel Disease. Gastroenterology. 2002;122(1):44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 14.Basset C, Holton J, Bazeos A, Vaira D, Bloom S. Are Helicobacter species and enterotoxigenic bacteroides fragilis involved in inflammatory bowel disease? Dig Dis Sci. 2004;49(9):1425–1432. doi: 10.1023/b:ddas.0000042241.13489.88. [DOI] [PubMed] [Google Scholar]
- 15.Tap J, Derrien M, Tornblom H, Brazeilles R, Cools-Portier S, Dore J, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152(1):111–23e8. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 16.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clooney AG, Eckenberger J, Laserna-Mendieta E, Sexton KA, Bernstein MT, Vagianos K, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut. 2021;70(3):499–510. doi: 10.1136/gutjnl-2020-321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361(21):2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darfeuille-Michaud A. Adherent-invasive Escherichia coli: a putative new E. Coli pathotype associated with Crohn’s Disease. Int J Med Microbiol. 2002;292(3–4):185–193. doi: 10.1078/1438-4221-00201. [DOI] [PubMed] [Google Scholar]
- 20.Azimirad M, Jo Y, Kim MS, Jeong M, Shahrokh S, Asadzadeh Aghdaei H, et al. Alterations and prediction of functional profiles of gut microbiota after fecal microbiota transplantation for iranian recurrent Clostridioides difficile Infection with underlying inflammatory bowel Disease: a pilot study. J Inflamm Res. 2022;15:105–116. doi: 10.2147/JIR.S338212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y, Oh J, Xue M, Huh WJ, Wang J, Gonzalez-Hernandez JA, et al. Commensal microbiota from patients with inflammatory bowel Disease produce genotoxic metabolites. Science. 2022;378(6618):eabm3233. doi: 10.1126/science.abm3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peloquin JM, Nguyen DD. The microbiota and inflammatory bowel disease: insights from animal models. Anaerobe. 2013;24:102–106. doi: 10.1016/j.anaerobe.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khorsand B, Asadzadeh Aghdaei H, Nazemalhosseini-Mojarad E, Nadalian B, Nadalian B, Houri H. Overrepresentation of Enterobacteriaceae and Escherichia coli is the major gut microbiome signature in Crohn’s Disease and ulcerative Colitis; a comprehensive metagenomic analysis of IBDMDB datasets. Front Cell Infect Microbiol. 2022;12:1015890. doi: 10.3389/fcimb.2022.1015890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Z, Wang W, Bradfield J, Li J, Cardinale C, Frackelton E, et al. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel Disease. Am J Hum Genet. 2013;92(6):1008–1012. doi: 10.1016/j.ajhg.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh M, Zhang L. DeepMicro: deep representation learning for Disease prediction based on microbiome data. Sci Rep. 2020;10(1):6026. doi: 10.1038/s41598-020-63159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douglas GM, Hansen R, Jones CMA, Dunn KA, Comeau AM, Bielawski JP, et al. Multi-omics differentially classify Disease state and treatment outcome in pediatric Crohn’s Disease. Microbiome. 2018;6(1):13. doi: 10.1186/s40168-018-0398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mossotto E, Ashton JJ, Coelho T, Beattie RM, MacArthur BD, Ennis S. Classification of paediatric inflammatory bowel Disease using machine learning. Sci Rep. 2017;7(1):2427. doi: 10.1038/s41598-017-02606-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forbes JD, Chen CY, Knox NC, Marrie RA, El-Gabalawy H, de Kievit T, et al. A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome. 2018;6(1):221. doi: 10.1186/s40168-018-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He XX, Li YH, Yan PG, Meng XC, Chen CY, Li KM, et al. Relationship between clinical features and intestinal microbiota in Chinese patients with ulcerative Colitis. World J Gastroenterol. 2021;27(28):4722–4737. doi: 10.3748/wjg.v27.i28.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The Genus Alistipes: gut Bacteria with emerging implications to inflammation, Cancer, and Mental Health. Front Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the Tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li YD, He KX, Zhu WF. Correlation between invasive microbiota in margin-surrounding mucosa and anastomotic healing in patients with Colorectal cancer. World J Gastrointest Oncol. 2019;11(9):717–728. doi: 10.4251/wjgo.v11.i9.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Round JL, Mazmanian SK. Inducible Foxp3 + regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, et al. Administration of Akkermansia muciniphila ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in mice. Front Microbiol. 2019;10:2259. doi: 10.3389/fmicb.2019.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;469(4):967–977. doi: 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35(9):833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 37.Durazzi F, Sala C, Castellani G, Manfreda G, Remondini D, De Cesare A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep. 2021;11(1):3030. doi: 10.1038/s41598-021-82726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H, He K, Chen J, Zhang X. LinDA: linear models for differential abundance analysis of microbiome compositional data. Genome Biol. 2022;23(1):95. doi: 10.1186/s13059-022-02655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15(3):799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 41.Linden A. Measuring diagnostic and predictive accuracy in Disease management: an introduction to receiver operating characteristic (ROC) analysis. J Eval Clin Pract. 2006;12(2):132–139. doi: 10.1111/j.1365-2753.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 42.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE. 2015;10(3):e0118432. doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Enterotypes identified in individuals with HC and IBD. (A) Using principal coordinate analysis (PCoA), participants were divided into three enterotypes, with Bacteroides, Faecalibacterium or Bifidobacterium as the primary distinguishing factors. (B) Three bacteria most abundant among the three enterotypes (Wilcoxon test, p < 0.05). Fig. S2. Comparison of the relative abundance of microbiota among HC, CD, and UC in phylum (A), genus (B) and species (C) based on the Kruskal-Wallis test. Each figure is shown in the order of HC, CD and UC. Fig. S3. Comparison of the relative abundance of microbiota between disease stages (HC, mild, and moderate) in CD patients at the phylum (A), genus (B) and species (C) levels. The numbers are listed in the following order: HC, mild and moderate. Fig. S4. Comparison of the relative abundance of microbiota between disease stages in UC patients at the levels of phylum (A), genus (B), and species (C) levels. The numbers are listed in the following order: HC, mild, moderate, and severe. Fig. S5. Comparison of 27 supervised machine learning models for diagnosing IBD subtypes using differential abundance. (A) F1 score; (B) ROC AUC. Fig. S6. (A) Parameter adjustment of a supervised machine learning model to classify CD and UC individuals using gut microbiomes. Ten-fold cross-validation was performed to select the optimal lambda. The point where the mean square error (MSE) was minimized was designated as the best lambda (log [λ] = -2.016529). (B) Venn diagram comparing our microbiome list with that of Clonney et al.. Fig. S7. Graphical abstract
Additional file 2: Supplementary Table 1. Association between enterotypes and IBD occurrence.
Additional file 3: Supplementary Table 2. Differences in the taxonomic composition of the gut microbiome in patients with CD and UC. Supplementary Table 3. Metacyc pathway of the different gut microbiomes (CD VS UC). Supplementary Table 4. KEGG Orthology of the different gut microbiomes (CD VS UC). Supplementary Table 5. Differences in the taxonomic composition of the gut microbiome in HC and CD. Supplementary Table 6. Differences in the taxonomic composition of the gut microbiome in mild and Moderate patients with CD. Supplementary Table 7. Correlation between microbes and severity of CD. Supplementary Table 8. Differences in the taxonomic composition of the gut microbiome in HC and UC. Supplementary Table 9. Differences in the taxonomic composition of the gut microbiome in mild and Severe patients with UC. Supplementary Table 10. Correlation between microbes and the severity of UC. Supplementary Table 11. Microbial coefficients used for modeling
Data Availability Statement
The data sets utilized in this research are available from NCBI’s Sequence Read Archive (SRA) (Accession ID: PRJNA945504) and Korean Nucleotide Archive (KoNA) (Accession ID: PRJKA220304) (https://www.kobic.re.kr/kona/).