Abstract
The function of the streptococcal cytoplasmic membrane lipoprotein, LppC, was identified with isogenic Streptococcus equisimilis H46A and Escherichia coli JM109 strain pairs differing in whether they contained [H46A and JM109(pLPP2)] or lacked (H46A lppC::pLPP10 and JM109) the functional lppC gene for comparative phosphatase determinations under acidic conditions. lppC-directed acid phosphatase activity was demonstrated zymographically and by specific enzymatic activity assays, with whole cells or cell membrane preparations as enzyme sources. LppC acid phosphatase showed optimum activity at pH 5, and the enzyme activity was unaffected by Triton X-100, l-(+)-tartaric acid, or EDTA. Database searches revealed significant structural homology of LppC to the Streptococcus pyogenes LppA, Flavobacterium meningosepticum OplA, Helicobacter pylori HP1285, and Haemophilus influenzae Hel [e (P4)] proteins. These results suggest a possible function for these proteins and establish a novel function of streptococcal cell membrane lipoproteins.
In a previous study from this laboratory, we reported the cloning and nucleotide sequence of a novel Streptococcus equisimilis chromosomal gene, designated lppC, which encodes a 32.4-kDa lipoprotein associated with the streptococcal cytoplasmic membrane or the outer membrane of Escherichia coli when expressed in this organism (5). The lppC gene is located immediately 3′ to and is transcribed independently of the unrelated gapC gene that codes for glyceraldehyde-3-phosphate dehydrogenase (4). As revealed by Southern, Northern, and Western analyses, homologs of lppC (and gapC) are conserved and also expressed in Streptococcus pyogenes (5). Database searches performed at that time found homology of LppC only to the hel gene-encoded outer-membrane antigen e (P4) from Haemophilus influenzae (6), to which it exhibits 58% sequence similarity. The biological function of e (P4) has remained elusive until very recently, when it was reported to be involved in the uptake of hemin as a source of porphyrin, an essential growth factor for H. influenzae when grown aerobically (9). Our attempts to provide evidence for a role of lppC in hemin uptake failed as, unlike the hel gene, lppC was unable to complement hemA mutants of E. coli for growth on hemin as the sole porphyrin source in aerobic conditions. Furthermore, S. equisimilis H46A, the source of lppC, was incapable of hemin binding or of growing on this compound in iron-limited medium (5).
Sequence database searching was continued at regular intervals for additional homologs of LppC and revealed weak structural similarity at low quality (quality score, 92.3) to the aphA gene product of E. coli MG1655 (sequence identity and similarity between LppC and AphA, 20.3 and 46.7%, respectively). Thaller et al. (16) had cloned and sequenced the aphA gene in the meantime and functionally characterized its product as an acid phosphatase. Sequence similarity between LppC and AphA prompted me to explore the possibility that the streptococcal protein has similar enzymatic activity. Here I provide biochemical, serological, and genetic evidence that the LppC protein does function as an acid phosphatase.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Two pairs of strains, with or without functional lppC, were used to identify the function of this gene. S. equisimilis H46A, a human serogroup C strain, contained wild-type lppC, whereas H46A lppC::pLPP10 was an erythromycin-resistant chromosomal insertion mutant carrying lppC interrupted at codon 144 by pLPP10 (5). Similarly, E. coli JM109 (21) was free of lppC, and JM109(pLPP2) contained plasmid-located lppC together with its promoter and terminator as a 1,152-bp fragment in the EcoRV site of pACYC184 (5). The streptococcal strains were grown without agitation at 37°C in brain-heart infusion medium (Difco) in ambient air. E. coli strains were cultured aerobically at 37°C in Luria-Bertani medium (11). If appropriate, the media contained erythromycin (2.5 μg/ml) and chloramphenicol (35 μg/ml) to select for plasmids.
Zymographic detection of phosphatase activity.
Whole-cell protein preparations were examined for phosphatase activity by the zymogram technique essentially as described by Thaller et al. (14). Briefly, cells from 10-ml overnight cultures were disrupted by sonication, and the unheated sonicates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (5). After treatment with renaturation buffer containing 1% Triton X-100, the gels were incubated overnight at 37°C in 100 mM sodium acetate buffer (pH 5.5) containing the phosphatase substrate (0.25 mM) 5-bromo-4-chloro-3-indolyl-phosphate (BCIP; Sigma). The appearance of blue bands indicated the presence of phosphatase activity. Western immunoblot analysis of the protein extracts performed in parallel with monospecific LppC antibodies served to compare the migration distances of the reactive protein bands in the two detection systems. The LppC antibodies had an enzyme-linked immunosorbent assay titer of >1,000 and were raised in rabbits as described previously (5). Western blots incubated with the primary antibodies were subsequently reacted with peroxidase-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad) as the secondary antibody by standard procedures.
Phosphatase assays.
Whole cells or subcellular cell fractions were assayed for phosphatase activity with disodium p-nitrophenyl phosphate (pNPP; Sigma) as the substrate by measuring the released p-nitrophenol (pNP) colorimetrically at 415 nm in the linear range of the calibration curves. Under standard conditions, the assays were performed in a volume of 1.2 ml in 37.5 mM citrate–4.16 mM chloride buffer (pH 4.8) containing 7.6 mM pNPP. The reactions were initiated by addition of the preparations to be tested for enzyme activity. Incubation was at 37°C for 30 min before the reactions were terminated by the addition of 5 ml of 0.1 N NaOH. Phosphatase inhibition tests were performed in standard reaction mixtures in the presence of 2% Triton X-100, 16.7 mM l-(+)-tartaric acid, or 15 mM EDTA. The pH dependence of the phosphatase activity was determined in citrate buffer with pNPP as substrate.
For whole-cell phosphatase assays, bacteria were harvested from 16-h cultures, washed twice in physiological NaCl solution, and used at 1.6 U of optical density at 600 nm (OD600) (corresponding to ∼3.2 × 109 cells) in the reaction mixtures. The bacteria were removed by centrifugation before absorbance was recorded. Cell membrane fractions were prepared as described previously (5). Briefly, E. coli cells were spheroplasted by lysozyme treatment in the presence of 1 M sucrose and lysed in Tris-HCl buffer containing 2% Triton X-100 and DNase. Triton X-100 specifically solubilizes the proteins of the inner membrane (7, 12). The lysate was centrifuged for 1 h at 40,000 × g in an SW50 rotor, and the precipitate containing the outer membrane was washed three times in distilled water before being frozen for further use. Streptococcal cells were protoplasted by combined treatment with lysozyme and mutanolysin in the presence of 66% sucrose (5). The protoplasts were lysed by three freeze-thaw cycles, followed by sonication to shear the DNA. Subsequent centrifugation at 40,000 × g yielded the pelleted cytoplasmic membrane fraction, which was washed and stored as described above.
The protein content of the bacterial membrane fractions was determined by the bicinchoninic acid method with bovine serum albumin as the standard, following the procedure recommended by the supplier of the assay kit (Sigma). Specific acid phosphatase activities in the whole-cell and the cell membrane assay were expressed on a per-OD600 unit basis and a per-milligram of membrane protein basis, respectively.
RESULTS
Zymographic detection of lppC-directed phosphatase activity.
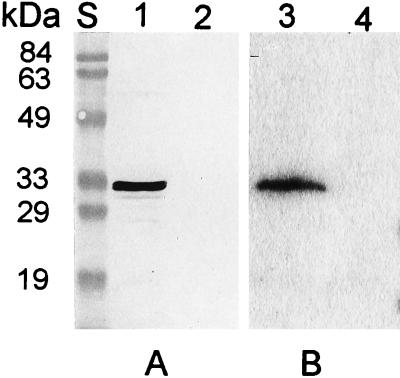
The lppC gene is efficiently expressed under its natural promoter in E. coli JM109(pLPP2) as demonstrated previously by reactivity of a novel ∼32-kDa protein with monospecific anti-LppC antibodies on Western blots (5). Whole-cell protein preparations from strains JM109 and JM109(pLPP2) were therefore tested for phosphatase activity by zymography. As shown in Fig. 1, the JM109(pLPP2) extract gave rise to a prominent band in the zymogram that migrated at the same rate as the anti-LppC reactive band in the Western blot run in parallel. Neither of the reactive bands was detected in the pLPP2-free control strain. This result indicated that lppC specified enzymatic activity that releases phosphate from BCIP under acidic conditions. Zymography with BCIP as the phosphatase substrate was too insensitive to detect the enzymatic activity of LppC in whole-cell protein preparations of H46A, presumably due to much lower relative LppC amounts in the latter cells compared to those of E. coli JM109(pLPP2) that overproduced the protein (5).
FIG. 1.
Detection of LppC protein in E. coli JM109(pLPP2) by Western blotting (A) and zymography (B). The results were obtained from sodium dodecyl sulfate–12% polyacrylamide gel electrophoretograms of whole-cell extracts (equivalent to ∼6 × 108 cells/slot) from JM109(pLPP2) (lanes 1 and 3) and JM109 (plasmid-free control, lanes 2 and 4) reacted, after blotting, with a 1:1,000 dilution of affinity-purified polyclonal antibodies to LppC (A) or, without blotting, with phosphatase substrate, BCIP (B). Lane S contained marker proteins, with molecular masses indicated on the left.
Acid phosphatase activity of whole cells and membrane fractions from S. equisimilis and E. coli.
To substantiate and expand the zymographic observation, a whole-cell assay was performed to determine whether the LppC protein functions as an acid phosphatase capable of releasing pNP from the standard substrate, pNPP. As shown in Table 1, the enzyme activity produced by wild-type H46A cells exceeded that elaborated by cells of the isogenic lppC insertion mutant by a factor of 7.9. An even-greater difference between the enzyme activities of cells with or without functional lppC was seen in the heterologous E. coli strain pair, in which JM109(pLPP2) was 14.8 times more active than the plasmid-free control strain.
TABLE 1.
Specific acid phosphatase activity of whole cells and cell membranes of the indicated S. equisimilis and E. coli strains as measured by the release of pNP from p-nitrophenyl phosphate
| Strain | pNP release froma:
|
|
|---|---|---|
| Whole cells | Membrane proteins | |
| H46A | 3.23 ± 0.20 | 9.90 ± 0.33 |
| H46A lppC::pLPP10 | 0.41 ± 0.05 | 0.36 ± 0.10 |
| JM109 (pLPP2) | 11.72 ± 0.16 | 119.43 ± 4.13 |
| JM109 | 0.79 ± 0.08 | 0.56 ± 0.11 |
Data are the mean values and standard errors from three to six independent experiments. For whole cells, values are expressed as {pNP [micromole milliliter−1 minute−1 (OD600)−1]}, and for membrane proteins, values are expressed as [pNP (micromole milliliter−1 minute−1 milligram−1)].
Since previous data show that the LppC protein is located in the streptococcal cytoplasmic membrane or in the E. coli (pLPP2) outer membrane (5), the corresponding membrane fractions were also assayed for phosphatase activity (Table 1). The specific enzyme activities in terms of total membrane protein were 27.5-fold and more than 200-fold higher in the preparations from H46A and JM109(pLPP2), respectively, than the corresponding activities seen in the membranes of the control strains. In fact, the activities of the latter ranged close to the borderline of measurability. In both the whole-cell and the membrane assay, the specific phosphatase activities detected in JM109(pLPP2) exceeded those observed in H46A by factors of about 3.6 and 12.1, respectively. Presumably, the greater activities in JM109(pLPP2) reflected the higher dose or more efficient expression of lppC, or both, in the heterologous strain.
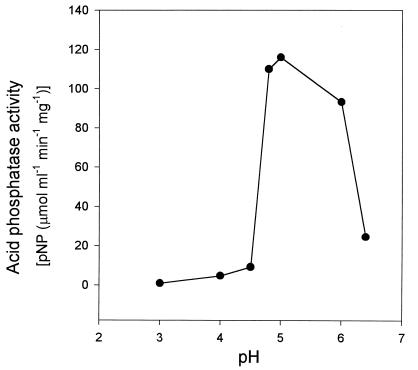
The E. coli JM109(pLPP2) outer-membrane preparation was used as a source of LppC phosphatase to study some basic properties of the enzyme. Optimum acid phosphatase activity of LppC was observed in a relatively sharp peak at pH 5, with activities at pH 4.5 and 6.4 amounting to only about 8 and 21%, respectively, of the activity seen at optimum pH (Fig. 2). Enzyme activity was resistant to 2% Triton X-100 in the reaction mixture. Furthermore, the enzyme could not be inhibited by tartrate and EDTA at concentrations of 16.7 and 15 mM, respectively (data not shown). Regarding the pH optimum of the enzyme produced in the homologous organism, the phosphatase activity of S. equisimilis membrane preparations also showed an optimum at about pH 5.
FIG. 2.
lppC-directed specific p-nitrophenyl phosphate-hydrolyzing activity of E. coli JM109(pLPP2) outer-membrane protein in citrate buffer of different pH values.
Identification of LppC homologs by sequence comparisons.
When initially described, LppC was reported to exhibit significant sequence similarity only to the Hel [e (P4)] protein of H. influenzae (5). In the meantime, the expanded sequence databases allowed the additional identification of putative structural homologs of LppC in S. pyogenes [LppA (12a)], Flavobacterium meningosepticum (OlpA; EMBL database accession no. Y12759), Helicobacter pylori (HP1285 [18]) and, as mentioned in the introduction, E. coli (AphA [16]). When ranked on the basis of optimized homology scores (8) relative to LppC, these proteins fell into the order LppC > LppA > OlpA > Hel > HP1285 > AphA (data not shown). By using Monte Carlo statistical analysis, based on the guidelines of Lipman and Pearson (8), to evaluate the significance of homology, LppC was found to be significantly homologous to all of the above proteins except AphA, to which, however, homology is still probable (data not shown). Multiple alignment of the amino acid sequences of the above-mentioned proteins (Fig. 3) revealed the greatest degree of diversion at both termini of the proteins. The 13-amino-acid region corresponding to LppC coordinates 97 to 109 was detected as the longest region with the highest degree of sequence similarity, followed by a 4- and a 6-amino-acid region of similar quality toward the C-terminal ends. However, none of these regions or any other part of the sequences exhibited the conserved RHG triad of the high-molecular-mass acid phosphatases. This sequence motif has been proposed to contain the histidine residue used in the phosphoryl transfer reaction that may proceed through a transient phosphohistidine enzyme intermediate (2, 19). Of the six proteins shown in Fig. 3, data are available only for AphA (16) and, as shown here, LppC that establish their functioning as acid phosphatases. Furthermore, evidence for the lipoprotein nature of the proteins has been published only for the Hel (6), LppC, and LppA proteins (5). The N-terminal sequences of HP1285 and AphA, although containing a cysteine residue, differ strongly from the consensus [(L, V) (A, S, T) (G, A) ↓ C] of the lipoprotein signal sequence cleavage site (13, 20). Besides, AphA can be readily released from the periplasmic space (16), suggesting that, unlike lipoproteins, it is not tightly associated with the cell envelope.
FIG. 3.
Gapped sequence alignment (17) of the S. equisimilis LppC (5), S. pyogenes LppA (12a), F. meningosepticum OlpA (EMBL database accession no. Y12759), H. pylori HP1285 (18), H. influenzae Hel (6), and E. coli AphA (16) proteins. Amino acids conserved in at least two-thirds of the proteins are in inverse font and were used to formulate a consensus sequence. Different degrees of shading indicate aligned amino acids with similar contributions to secondary structure. Note that the LppA sequence is still preliminary and incomplete at the N terminus.
DISCUSSION
As a group of enzymes, phosphatases are diverse and have widely different properties (19). The present results characterize the LppC enzyme as a cell membrane lipoprotein acid phosphatase that functions in the presence of tartrate and EDTA. EDTA resistance of its functional activity suggests that, similar to the class A acid phosphatases of the enteric bacteria (14–16), the enzyme does not require metal ion cofactors. The LppC enzyme has resistance to the inhibitory action of tartrate, as do the enteric class B acid phosphatases which, however, are EDTA susceptible and appear not to accept BCIP as a substrate (15, 16). With regard to size, the molecular mass of the LppC polypeptide (∼32 kDa) lies between that of the high- (40 to 60 kDa) and the low-molecular-mass (14 to 18 kDa) acid phosphatases (10, 19). It remains to be investigated whether the active form of the streptococcal protein is a homo-oligomer, as are, e.g., the enterobacterial NapA (15) and AphA (16) phosphatases.
Of considerable interest is the fact that the functional identification of lppC establishes a novel role for cytoplasmic membrane-associated lipoproteins of the streptococci, if not of the gram-positive bacteria in general. The various functions attributed to these proteins in recent years have hitherto not included phosphatase activity (for a review, see reference 13). Although the number of LppC homologs proposed here is still small, it is remarkable that those recognized occur in pathogenic or potentially pathogenic species (Fig. 3). Most of them, including LppC, require further functional characterization to provide insight into their possible physiological role. It remains to be seen whether this role is limited to serving nutritional and metabolic regulatory functions by scavenging organic phosphoesters (16, 19) or extends to pathogenetic functions. As a matter of fact, acid phosphatases from several bacterial species have recently been recognized as virulence factors that support intracellular survival by inhibiting the respiratory burst (1, 3, 10). In this connection, hydrolysis of phosphate esters, particularly when localized to cell surface structures, may be linked to cellular signal transduction processes. It is thus important to know whether or not LppC also exhibits phosphotransferase activity, as shown for the NapA (15) and AphA (16) phosphatases. A more specific issue that is raised by the functional identification of lppC relates to the primary function of the H. influenzae Hel [e (P4)] protein. On the basis of the present results, it can be speculated that this protein is an acid phosphatase and thus requires reevaluation with respect to its functional role.
ACKNOWLEDGMENTS
I thank Ulrike Wrazidlo for outstanding technical assistance. Accessibility of the database of the Streptococcus pyogenes genome sequencing project at the University of Oklahoma is gratefully acknowledged.
This work was supported by grants from the Thuringian Ministry of Science, Research and Arts; the German Research Association; and the Fonds of the Chemical Industry to H.M.
REFERENCES
- 1.Baca O G, Roman M J, Glew R H, Christner R F, Buhler J E, Aragon S A. Acid phosphatase activity in Coxiella burnetti: a possible virulence factor. Infect Immun. 1993;61:4232–4239. doi: 10.1128/iai.61.10.4232-4239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bazan J F, Fletterick R J, Pilkis S J. Evolution of a bifunctional enzyme: 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase. Proc Natl Acad Sci USA. 1989;86:9642–9646. doi: 10.1073/pnas.86.24.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chhatwal G S, Walker M J, Yan H, Timmis K N, Guzmán C A. Temperature dependent expression of an acid phosphatase by Bordetella bronchiseptica: role in intracellular survival. Microb Pathog. 1997;22:257–264. doi: 10.1006/mpat.1996.0118. [DOI] [PubMed] [Google Scholar]
- 4.Gase K, Gase A, Schirmer H, Malke H. Cloning, sequencing and functional overexpression of the Streptococcus equisimilis H46A gapC gene encoding a glyceraldehyde-3-phosphate dehydrogenase that also functions as plasmin(ogen)-binding protein: purification and biochemical characterization of the protein. Eur J Biochem. 1996;239:42–51. doi: 10.1111/j.1432-1033.1996.0042u.x. [DOI] [PubMed] [Google Scholar]
- 5.Gase K, Liu G, Bruckmann A, Steiner K, Ozegowski J, Malke H. The lppC gene of Streptococcus equisimilis encodes a lipoprotein that is homologous to the e (P4) outer membrane protein from Haemophilus influenzae. Med Microbiol Immunol. 1997;186:63–73. doi: 10.1007/s004300050047. [DOI] [PubMed] [Google Scholar]
- 6.Green B A, Farley J E, Quinn-Dey T, Deich R A, Zlotnick G W. The e (P4) outer membrane protein of Haemophilus influenzae: biologic activity of anti-e serum and cloning and sequencing of the structural gene. Infect Immun. 1991;59:3191–3198. doi: 10.1128/iai.59.9.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 8.Lipman D J, Pearson W R. Rapid and sensitive similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 9.Reidl J, Mekalanos J J. Lipoprotein e (P4) is essential for hemin uptake by Haemophilus influenzae. J Exp Med. 1996;183:621–629. doi: 10.1084/jem.183.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reilly T J, Baron G S, Nano F E, Kuhlenschmidt M S. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J Biol Chem. 1996;271:10973–10983. doi: 10.1074/jbc.271.18.10973. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T E. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 12.Schnaitman C A. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J Bacteriol. 1971;108:545–552. doi: 10.1128/jb.108.1.545-552.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Streptococcal Genome Sequencing Project. 1997. http://www.genome.ou.edu/strep.html.
- 13.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thaller M C, Berlutti F, Schippa S, Iori P, Passariello C, Rossolini G M. Heterogeneous patterns of acid phosphatases containing low-molecular-mass polypeptides in members of the family Enterobacteriaceae. Int J Syst Bacteriol. 1995;45:255–261. [Google Scholar]
- 15.Thaller M C, Lombardi G, Berlutti F, Schippa S, Rossolini G M. Cloning and characterization of the NapA acid phosphatase/phosphotransferase of Morganella morganii: identification of a new family of bacterial acid-phosphatase-encoding genes. Microbiology. 1995;141:147–154. doi: 10.1099/00221287-141-1-147. [DOI] [PubMed] [Google Scholar]
- 16.Thaller M C, Schippa S, Bonci A, Cresti S, Rossolini G M. Identification of the gene (aphA) encoding the class B acid phosphatase/phosphotransferase of Escherichia coli MG1655 and characterization of its product. FEMS Microbiol Lett. 1997;146:191–198. doi: 10.1111/j.1574-6968.1997.tb10192.x. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 19.Vincent J B, Crowder M W, Averill B A. Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem Sci. 1992;17:105–110. doi: 10.1016/0968-0004(92)90246-6. [DOI] [PubMed] [Google Scholar]
- 20.Von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 21.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]