Abstract
Reproducibility has been identified as an outstanding challenge in science, and the field of synthetic biology is no exception. Meeting this challenge is critical to allow the transformative technological capabilities emerging from this field to reach their full potential to benefit the society. We discuss the current state of reproducibility in synthetic biology and how improvements can address some of the central shortcomings in the field. We argue that the successful adoption of reproducibility as a routine aspect of research and development requires commitment spanning researchers and relevant institutions via education, incentivization and investment in related infrastructure. The urgency of this topic pervades synthetic biology as it strives to advance fundamental insights and unlock new capabilities for safe, secure and scalable applications of biotechnology.
Graphical Abstract
Keywords: synthetic biology, engineering biology, reproducibility, standardization, IV&V
1. Introduction
The core of synthetic biology is applying the rigor and practice of engineering disciplines to biological systems, which has captured the imagination and led to the popularization of this new, transformative science well beyond the halls of academia (1, 2). Now in its third decade, there has been significant technical progress and synthetic biology products are indeed impacting a variety of industries from healthcare to manufacturing (3). However, given the financial, experimental and intellectual resources that have been made available to the basic and applied synthetic biology research enterprise, we argue that the efficiency and success rate of such transitions are far lower than anticipated. Some explanatory power for this observation is provided by revisiting a seminal 2010 news feature, aptly titled ‘Five hard truths for synthetic biology’ (4), which succinctly identified challenges in the field. While there has been significant progress, many of those same challenges still resonate more than a decade later. As trusted agents responsible for the transition of synthetic biology–based products to specific stakeholders, our experience suggests that a lack of attention to reproducibility in synthetic biology contributes to each of these challenges, playing an outsized role in limiting the field’s ability to transition technologies and products.
Like many fields of science, synthetic biology faces a reproducibility challenge, although the issue is considered so rarely that the full scope of the problem remains unknown. We argue that addressing reproducibility will mitigate or even resolve the outstanding ‘hard truths’ in the field. Beginning with shared definitions of related terms, we describe the value of reproducibility, especially with respect to these core challenges, and examine the efforts to explicitly measure reproducibility. We call on individuals and institutions engaged in synthetic biology to balance emphasis on innovation and reproducibility. Even a modest shift toward supporting reproducibility studies would have a substantial impact on the long-term success of the field, enabling synthetic biology to finally meet our lofty expectations for transformative applications (Figure 1).
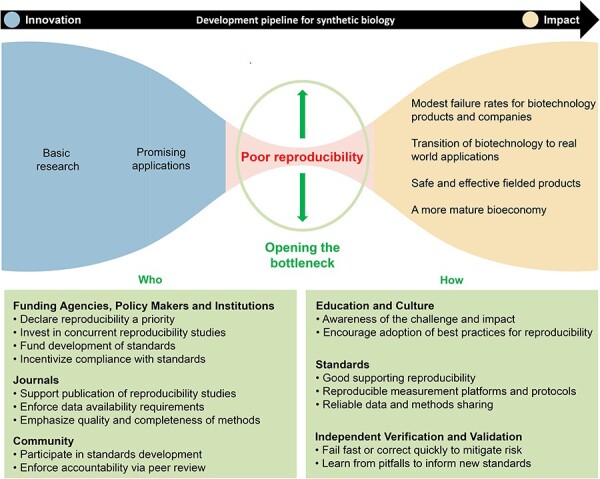
Figure 1.
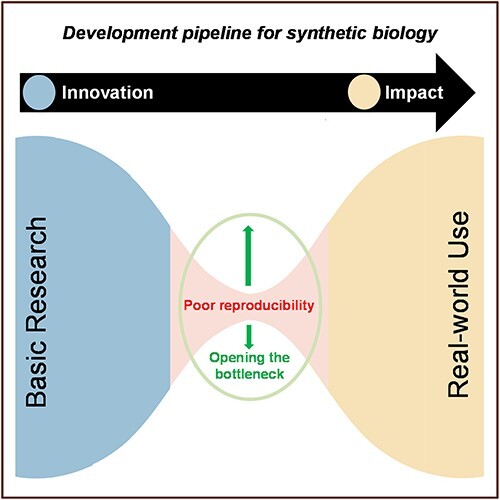
Reproducibility challenges introduce a bottleneck in the development pipeline for synthetic biology. Despite considerable public and private investments, we posit that reproducibility in synthetic biology is the most immediate process bottleneck that limits the flow of synthetic biology products to market—regardless of the merits or financial ‘push’—and reduces confidence in the overall enterprise, reducing stakeholder ‘pull’. Improving reproducibility is also critical to making biology more engineerable. The recommended strategic and tactical investments (boxed text) to address reproducibility needs can widen the bottleneck and increase the flow of impactful synthetic biology products.
2. The challenge of reproducibility
Reproducibility in scientific research has drawn increasing attention in recent years (5–7). While a lack of reproducibility is widely acknowledged as an issue across science, the scope of the problem has only rarely been measured (8). The Reproducibility Project: Cancer Biology (RPCB) is arguably the most significant effort to examine this issue in life sciences. Shockingly, it reported that 0 of 193 experiments from 53 selected papers had sufficient details to attempt reproduction without contacting the authors of the original studies. After 8 years and with $1 524 640 spent, only 26% of the studies could be attempted experimentally (9). Of those experiments, at least 85% of the quantitative effects were smaller than reported and only 46% of binary effects were reproduced (10). This poses a significant problem for early-stage researchers attempting to build on published results, as well as developers trying to transform these discoveries into products for real-world impact. Such findings also reduce confidence in the overall enterprise, dissuading investment from funding agencies and reducing stakeholder ‘pull’. Indeed, the RPCB was motivated by the high failure rates reported by the pharmaceutical industry when attempting to translate basic research findings to the clinic (11–13). The challenge spans beyond wet laboratory experimentation and also impacts modeling efforts, as recent studies have found that about half of systems biology models are not reproducible (14).
The central aim of synthetic biology to transform biology into an engineering discipline should position the field at the vanguard of improving reproducibility in the life sciences; however, this has not been the case. Far from an expansive effort similar to RPCB, only a handful of studies directly measuring interlaboratory reproducibility can be found in the synthetic biology literature (15–18), despite the rapid growth of interest and investment in synthetic biology research globally (19). While many calls have been made to improve standardization in design, method description, measurement, characterization and information sharing (20–29), the central role of reproducibility warrants more explicit attention. Even with all these calls to action, this Special Issue represents the most substantial collection of efforts to shed light on the current state of reproducibility in synthetic biology to date (30–38).
Contrasting the state of reproducibility in synthetic biology with electrical engineering offers an apt, if overused, analogy. Careful design of electrical components to meet well-defined specifications results in reliable electrical circuit implementation. The afforded predictability enables design by allowing the abstraction of function to increasingly greater device complexity, yielding the sweeping impact of the modern digital age. While most of the focus is directed toward component function, a thorough understanding of how context influences reproducibility is just critical: an electrical resistor will work as expected when inserted into any bench-top breadboard, while producing a microprocessor requires a controlled cleanroom. This example highlights the concept of robustness, which can only be established from the starting point of a reproducible experiment to understand the bounds of variability. Without understanding and improving reproducibility in synthetic biology as we move toward genome-scale engineering (39), we will fall short of the predictability needed for abstraction and the full impact of biotechnologies.
One challenge with improving reproducibility is a lack of clearly defined and widely accepted terminology. Definitions of relevant terms can vary by discipline (40), creating problems for the interdisciplinary field of synthetic biology. In recognition of the importance of a common terminology, a recent Executive Order in the USA (41) includes a call to establish such a lexicon for the larger US bioeconomy. We offer here specific definitions of reproducibility and some common associated terms (Box 1).
Box 1.
A lexicon for reproducibility.
Synthetic biology requires a shared lexicon of important terms to facilitate progress toward reproducibility. Here, we propose definitions for a set of relevant terms.
Reproducibility: Measurement precision under conditions of measurement that include different locations, operators, measuring systems and on the same or similar objects and protocols.
Repeatability: Measurement precision under conditions of measurement that include the same location, operators, protocols, measuring systems and on the same or similar objects and protocols.
Robustness: Quantitative capacity of a measurement to remain unaffected within the given conditions of measurement.
Variability: Uncertainty quantified from replicate measurements and attributable to specific parameters of measurement.
Technical replicate measurement: One of the multiple measurements intended to resolve variability in measurement results attributable to the measuring system, objects and/or protocol.
Biological replicate measurement: One of the multiple measurements intended to resolve variability in measurement results attributable to relevant biological processes.
Comparability: Suitability of quantitative comparison between measurements obtained under different conditions.
Reproducibility study: Study designed with measurements to quantify the reproducibility of measurements.
Independent Verification and Validation: Confirmation, typically by an unbiased third party, that measurements are reproducible, the technology meets the requisite specifications and the stakeholder’s operational needs are satisfied.
3. The ‘hard truths’ of synthetic biology are still hard and true
Synthetic biology has long hosted discussions identifying aspects of biology that confound our ability to engineer living systems, an endeavor undertaken without full knowledge of first principles and seemingly working to counter millions to billions of years of evolution (42, 43). In revisiting Roberta Kwok’s cogent and impactful summary of the ‘five hard truths for synthetic biology’ (4), it is implicitly assumed that the distinguishing features of biological systems exist in opposition to the goals of engineering. Since then, the field of synthetic biology has evolved. Attempting to engineer biological systems has led to many lessons learned about how to work within and even exploit the constraints of biology. Further progress now requires meeting the challenge of reproducibility directly to provide a path forward through the persistent ‘hard truths’ of synthetic biology.
3.1. Hard truth #1—many of the parts are undefined
In 2010, this first challenge was viewed as a lack of engineered genetic and biomolecular components or parts. Many researchers envisioned libraries of well-characterized and modular parts whose function could be ported into new designs (20, 21). The field of synthetic biology now acknowledges this oversimplification of the role of context, beginning with interdependence of function for neighboring parts. Even simply defining the sequence boundaries of parts, such as between ribosome binding sites and downstream coding sequences or promoter sequences beyond the +1 site and the corresponding transcribed 5ʹ untranslated region, makes assigning functional data to a part nontrivial (24, 44). Parts that insulate function from genetic context exist (45, 46), although they are not commonly used, and models have made strides in predicting function that accounts for local genetic context (47–51).
The experimental context further complicates context-dependent function. The influence of small changes in conditions, e.g. between experimenters, instruments or laboratories, is poorly understood, making it difficult or impossible to deconvolute the causes of variability when an attempt to reproduce a published result fails. Sequence-based models and insulating parts do not account for differences in experimental conditions. By probing reproducibility explicitly, key information missing in published methods can be identified to more confidently attribute function to specific aspects of context.
3.2. Hard truth #2—the circuitry is unpredictable
The second truth referred to the challenge of scaling from parts to genetic circuits with increasingly sophisticated functions in a predictive way. A pioneering study in synthetic biology took 3 years to combine just two expression cassettes into a toggle switch (4, 52). Considerable effort has shown steady improvement (53) leading to genetic circuits with up to 14 interacting genes (54), built using design automation and taking advantage of insulating parts (55, 56). While these accomplishments have been substantial, far more progress in synthetic biology could have been expected by now considering the frequent comparisons with electrical engineering, where Moore’s Law has described a doubling of the number of transistors per chip roughly every 2 years for decades (57). The fact that the ability to read and write DNA has progressed in a manner similar to or faster than Moore’s Law since the first toggle switch (58) means that the limitations are not in our ability to build genetic circuits but rather to design them reliably at higher levels of abstraction.
Abstraction requires predictive understanding of the function and variability of each underlying layer. The fact that synthetic biology still lacks the fundamental knowledge and engineering capability to support abstraction stems in part from a lack of emphasis on reproducibility. Reproducibility studies would inform the development of tools to understand and troubleshoot the function of parts individually and integrated with each other across different contexts. These should be measured at many points beyond simply the final output to optimize performance and reduce variability. Reproducibility studies can help reveal bounds on how to achieve predictable and reproducible part function in context.
3.3. Hard truth #3—the complexity is unwieldy
The third truth highlighted the challenge of building and testing genetic circuits as the increased scale leads to massive combinatorial design spaces. With significant advances in DNA synthesis and assembly, costs have fallen dramatically, and biofoundries have been established around the world to provide assembly services (59). The ability to build a large genetic construct is more accessible than ever; however, the fact that the underlying components often interact with each other and with the host means that the design space to explore rapidly becomes unwieldy. To account for this challenge, biotechnology companies like Amyris, Inc. and Ginkgo Bioworks have invested heavily in platforms to build and screen genetic design variants with impressive throughput. Scaling up requires infrastructure beyond the instrumentation itself, such as thoroughly defined methods, software to capture relevant data and metadata and careful metrology. Although much of these resources and knowledge remain confined to industry, community-adopted measurement (30, 60–62) and data standards (63–69), along with reproducibility studies, can allow for building upon work distributed across laboratories and institutions, effectively enabling scale-up for the field without costly centralized infrastructure.
3.4. Hard truth #4—many parts are incompatible
The idea of an incompatible part refers to how the function of parts often changes in other hosts. While progress has been made toward the goal of orthogonal parts that function across hosts, this challenge highlights more broadly the complex relationship between engineered parts, the host organism and the surrounding environment. Numerous factors can influence this interplay. Even within a single host, the utilization of cellular resources, or cellular burden, can play a major role in the function of a genetic circuit, potentially leading a cell to evolve away from the intended function (70). Cells in different growth phases have different expression profiles that can dramatically influence performance (71). Moving to a new host complicates these issues further in ways that are poorly understood.
Experimental variability is deeply enmeshed in interactions between the host and engineered parts. Eventually, a more complete picture of host cell function will emerge and lead to predictions of these interactions. Until then, a more practical approach is to design genetic circuits that are robust to particular host and environmental contexts, ensuring their wider compatibility (72). Doing so necessitates defining the boundaries within which the engineered system is intended to function; defining these boundaries requires measurement standards and detailed descriptions of metadata to thoroughly capture the experimental context. The first step to evaluating the robustness of an implementation across various conditions is to establish the reproducibility for a single condition (15–18), again highlighting the need for rigorous reproducibility studies.
3.5. Hard truth #5—variability crashes the system
Noise arising from biological processes—referred to as ‘variability’ in the study by Kwok (2010)—has primarily been seen as a challenge to overcome or engineer away. However, an important shift increasingly seeks to embrace this source of variability. Many efforts have looked to better understand the role of noise in biological systems, with some even identifying noise as a necessary component to achieving a particular function (73, 74). While it is certainly true that noisy behavior can disrupt the intended function of a design, this truth speaks more to a lack of understanding and control of the underlying system than noise as a problem per se. We know that naturally evolved living systems often operate well despite noise or even because of noise, which points to a need for new design approaches that explicitly account for this reality. Doing so requires a clear understanding of what portion of observed variability stems from the conditions of the measurement, error in the measurement and noise in the underlying system. Achieving such understanding likely also requires improvements in measurement technologies. For example, single-molecule measurements sensitive to stochastic noise also tend to have low throughput, making measurements to characterize the performance of an implementation across time and conditions impractical or unobtainable. Nonetheless, with an improved grasp of noise profiles for the system, it becomes far more feasible to design systems that are robust to stochastic noise in biological systems or other sources of variability.
A theme throughout the five hard truths is understanding or circumventing the limitations of engineering in biology. However, the long-term value of these efforts is undercut if the results are different under ostensibly the same conditions. A focus on reproducibility will clarify such confounding problems, leading to more direct resolution of the design rules of biology.
4. Advancing reproducibility
An intrinsic tension exists between rigor and innovation in science. Pseudoscientific ‘innovations’ can be churned out rapidly by ignoring sound logic, good scientific practice and uncertainty analysis, while an exhaustive examination of every alternative explanation of a result halts innovation. Current practices often value innovation over rigor. A greater emphasis on reproducibility would enforce greater rigor. While this shift may appear to slow innovation initially, increased rigor naturally speeds subsequent innovation by reducing the time spent trying to replicate the initial findings. Detailed reports on the reproducibility of past studies are needed to assess how dire the need for reproducibility studies may be in synthetic biology. If the results of the RPCB are any indication, synthetic biology—and biological sciences more broadly—has the cause for serious concern. Yet, other fields provide paths forward. For example, a recent study of imaging literature found that method reporting was poor (75), motivating a Special Issue on improving standards for reporting data and metadata toward improving reproducibility in microscopy (76).
4.1. An approach to formal reproducibility studies
Formal reproducibility studies provide a path to elucidate the state of reproducibility in synthetic biology. In realizing that synthetic biology technologies will require the same level of scrutiny as any other engineered system, the US Defense Advanced Research Projects Agency (DARPA) Biological Technologies Office has incorporated concurrent Independent Verification and Validation (IV&V) into their programs toward improving the likelihood of transitioning early-stage research into real products (77). These programs take a holistic approach to reproducibility in which the program manager, grantee and IV&V partner work together closely and in real time to determine whether the work product meets the required specifications (verification) and does what it is supposed to do in its intended operational environment to satisfy the needs of the stakeholders (validation). Performing IV&V concurrently mitigates risk by identifying failure quickly and enabling quick correction before errors are propagated. This ensures that the IV&V process is fit-for-purpose and effective, despite the absence of standard practices for IV&V in the biosciences. Importantly, in an IV&V approach, validating software and data analysis methodology are just as critical as validating hardware and experimental protocols. Beyond validating an implementation, the IV&V process brings into stark relief the gaps in key details that must be communicated and clear definitions of intent in a design. DARPA’s Information Innovation Office (I2O) approached the challenge of reproducibility slightly less directly with its Synergistic Discovery and Design (SD2) program, which endeavored to accelerate design in synthetic biology by making the description of an intended design, implementation and evaluation as seamless as possible—even for tasks distributed across different laboratories and researchers. Valuable and instructive examples of IV&V and work under SD2 are published in this Special Issue and elsewhere (33–35, 37, 38, 63, 65, 78–80).
What is a realistic model for immediately implementing some or all of the elements of IV&V studies into synthetic biology research? Unfortunately, extensive, real-time efforts to examine reproducibility fall outside the scope of current projects and resourcing. Until this is implemented, retroactive reproducibility studies remain critical to build trust in and advance previous results. Eventually, we envision the emergence of an ecosystem of laboratories skilled at providing IV&V consultation and services in synthetic biology, whether housed in industry, academia or government organizations. However, the success of an IV&V partner working closely with the original researchers and laboratory does not ensure wider reproducibility. The field of synthetic biology must learn from both retroactive and concurrent reproducibility efforts to define acceptable metrics for reproducibility across its various biotechnologies.
The fact that reproduction occurred concurrently with the original research in the DARPA programs is important. In the retroactive RPCB, lack of engagement by the original authors was one of the most significant factors prohibiting successful reproducibility studies of published work. This result is hardly surprising: researchers fail to adequately document their work; researchers change positions; materials are discontinued, compromised, or lost and, perhaps most importantly, researchers have no immediate incentive to devote time and resources to assisting reproducibility studies. Any action on the part of the original researchers is motivated largely by professional courtesy and not aligned with current incentives of funding or publishing.
In contrast, by integrating concurrent IV&V into research projects, researchers are directly incentivized to ensure that their results are reproducible. All aspects of a study can be carefully designed at the outset to maximize rigor and reproducibility. Poor study design or other problems can be identified and mitigated rapidly, rather than resorting to triaging the data after the conclusion of a study. For DARPA, the potential of concurrent IV&V to identify technology transition failure points earlier has justified the upfront investment. By publishing IV&V studies, the community can benefit from the lessons learned and improve all aspects of sharing research results, from descriptions of materials and protocols to measurements and data. Over time, this virtuous cycle of increased transparency and rigor can facilitate further retroactive reproducibility studies and, in turn, accelerate innovation and facilitate technology transition.
4.2. Role of policymakers, funding agencies and institutions
Emphasizing reproducibility as a routine part of research workflows in synthetic biology requires a cultural shift that must be supported by adequate infrastructure and incentivization. Policymakers, funding agencies and research institutions can play critical roles in declaring the importance of reproducibility, probing the current state of the problem, supporting the development of infrastructure and incentivizing researchers to value reproducibility. When developing policy documents, the costs of poor reproducibility in early-stage research should be considered and ideally measured, especially at a time when significant attention is being paid to synthetic biology and its major role in the emerging bioeconomy. Synthetic biology does not have to wait for damning articles from industry bemoaning a lack of reproducibility to motivate its own RPCB; action can be taken now.
While it is clear that scientists (8) and funding agencies (81) recognize reproducibility in science as a problem generally, understanding of the specific drivers and consequences of failing to address reproducibility in synthetic biology is still lacking. Even measuring the scale of the problem for a particular field, as the RPCB did for cancer biology, is rare. In synthetic biology, the problem of reproducibility is largely unmeasured. Funding agencies could invest in understanding the scope of the problem by directly funding reproducibility studies. Ideally, these investments would include concurrent IV&V to immediately highlight problems and solutions, sidestepping some of the challenges with retroactive reproduction.
Meeting infrastructure needs for reproducibility requires support from funding agencies and research institutions. Many previous articles have advocated for increased development and adoption of standards in design, method description, measurement, characterization and information sharing (20–29), and several examples of such standards exist (63–69, 82, 83). While supporting the trend toward automation may improve reproducibility (26), measurement assurance and standards are also important to advance reproducibility for more common laboratory tools. Fluorescent plate readers, for example, are a workhorse instrument, yet important instrument parameters and control measurements are often not reported (84). Only in the last few years have standards for plate reader measurements in synthetic biology emerged (30, 60–62). Inclusion of experts in metrology and instrumentation, not just users, is important. The increasing engagement of standards organizations on standards in biotechnology relevant to synthetic biology, such as the International Organization for Standards Technical Committee 276 on Biotechnology, is a necessary and encouraging step forward. Development of these standards can be accelerated dramatically.
Funding agencies and institutions can also effect a culture shift toward better reproducibility by encouraging and incentivizing good practices (85). The UK Reproducibility Network provides a model to improve reproducibility at a national level and has spawned similar national efforts around the globe (86). The Berlin Institute of Health offers an example of how institutions can prioritize good practices (87). Proposal calls could require adoption of community standards in measurement and data reporting. Program reviews could be made to enforce such requirements. Similarly, proposals could require engagement with an IV&V partner, whether the partners are identified by the proposers or matched by the funding agency. Ultimately, improving reproducibility requires an investment of time, effort and materials by researchers and funding priorities to accommodate and incentivize those investments.
4.3. Responsibilities of journals
Journals can similarly do more to encourage improved reproducibility. Editors could request reviewers to pay closer attention to methods, statistics, data sharing and likelihood of reproducibility in general. Such factors could have explicit sections to address in the evaluation systems. Many journals have taken steps to influence authors, including introducing data sharing requirements, publishing community calls for better practices (88), advocating for publication of methods before results (89) and encouraging adoption of standards (90). Extending these initiatives further will be important. A common refrain by researchers is that attempts to reproduce a result, failed or successful, are difficult to publish. The situation has improved, with several journals now explicitly accepting manuscripts that do not have a perceived impact or innovation or that report negative results; however, Special Issues, such as this one, or article types explicitly for reproducibility studies would undoubtedly help. In addition to the advantage of allowing others to learn from these efforts, the chance of publication could improve the rigor of attempts to reproduce results from the outset.
4.4. Responsibilities of researchers
Scientists have a long history of self-policing research quality. Indeed, a 2016 survey on reproducibility scored researcher-driven approaches as more likely to be effective than incentivization (8). As peer review is one of the core drivers of this process, reviewers can address reproducibility by paying attention to the completeness of methods; the availability of sequences, data and materials and participation in community standards, such as the Synthetic Biology Open Language (63, 66, 67, 69, 82). Similarly, when preparing a manuscript, extra attention can be paid to these aspects. Becoming involved in the development of community standards can help advance these causes, especially because when and how to apply standards is open to debate (91). Participation in research coordination communities, such as Build-A-Cell (https://www.buildacell.org/), pushes interlaboratory reproducibility forward. When attempting to reproduce published results, researchers can be more rigorous in documenting and disseminating those efforts, whether in a journal or a preprint service such as bioRχiv. More broadly, research can be performed with reproducibility in mind. As an experiment is being planned, consideration of how aspects of the protocol might influence reproducibility can lead to better defined methods and potentially explain unexpected results. In addition to funding agencies and journals, professional societies or conference organizers could provide incentives by creating awards for reproducibility. Ultimately, researchers perform the work they are incentivized to do, and reproducibility is under-incentivized. Nonetheless, the scientific tradition of holding one another to the expectation of performing and reporting rigorous work can be applied to improving reproducibility in synthetic biology.
5. A cause for optimism
While the field of synthetic biology has not fully overcome its hard truths, major progress offers evidence for optimism. A range of successful products based on advances in our capabilities to engineer living systems has begun to impact people’s daily lives (3). The complexity of engineered genetic circuits has climbed, for example, with the creation of a digital display using genetic circuits engineered into seven strains using 63 transcription factors (54). The capacity to synthesize DNA at ever higher scales and lower costs continues to progress rapidly. Bibliometric assessments show the continued expansion of the field, in terms of both funding and global participation (19, 92). Communities dedicated to open sharing of information thrive and grow. Maintaining this trajectory requires a sustained commitment to reproducibility and a willingness to share what works—and just importantly, what does not work—with the community at large.
Trends in other technical and engineering fields offer further reasons for optimism. The reproducibility challenges in cancer biology hold lessons for synthetic biology. The US National Institutes of Health’s (NIH) Brain Research Through Advancing Innovative Neurotechnologies Initiative explicitly emphasizes data sharing and reproducibility, along with funding to support this work. Programs that focus on implementing data standards to enable artificial intelligence, such as DARPA’s SD2 and NIH’s Bridge2AI programs, implicitly require greater reproducibility to achieve their goals. Beyond biology, data transparency is often visible in physics, where raw data shared at large scales advance collaborations like the Sloan Digital Sky Survey and the Conseil Européen pour la Recherche Nucléaire Large Hadron Collider. It is not too late to learn from these efforts and adapt them to synthetic biology, hopefully leading to accelerated research and translation to application in the long term.
The promise of synthetic biology remains as tantalizing as ever to deliver safe, effective biotechnological products and services that improve every aspect of our lives, from our inner microbiome to environmental security. The ability to reliably engineer biological systems could indeed still change the world, beyond recent breakthroughs in vaccine technology: the production of novel molecules and materials impossible or impractical by current means; green, petroleum-free biomanufacturing from diverse feedstocks; medicines with unprecedented efficacy and precision; new sustainable approaches to bioremediation, agriculture and nutrition; synthetic cells and many others. The ability to reproduce studies, and the measurements that support IV&V, can directly inform effective regulation of synthetic biology products. Even as the societal transformations brought about by early computers were impossible to predict, projections about the impact of synthetic biology are likely to be wildly wrong. Such progress in electronics was enabled by reproducible results at ever higher levels of abstraction; to reach its lofty potential, synthetic biology must do the same. Guided by ethics, security and policy, innovation in synthetic biology should only be limited by imagination—not a lack of reproducibility.
Acknowledgments
The authors thank the synthetic biology community for insightful conversations regarding reproducibility. The opinions and assertions contained herein are those of the authors and are not to be construed as those of the National Institute of Standards and Technology, US Army, US Navy, military service at large or US Government. Certain commercial equipment, instruments or materials are identified in this paper to foster understanding. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the stated purpose.
Contributor Information
Matthew W Lux, Research & Operations Directorate, U.S. Army Combat Capabilities Development Command Chemical Biological Center, APG, MD 21010, USA.
Elizabeth A Strychalski, Cellular Engineering Group, National Institute of Standards and Technology, Gaithersburg, MD 20899, USA.
Gary J Vora, Center for Bio/Molecular Science & Engineering, U.S. Naval Research Laboratory, Washington, DC 20375, USA.
Data Availability Statement
No data were collected in this work.
Material Availability Statement
No materials were used in this work.
Conflict of interest statement.
No potential conflict of interest was reported by the authors.
References
- 1. Ananthaswamy A. (2014) Rewiring nature with synthetic biology. Disco. Mag. [Google Scholar]
- 2. Morton O. (2019) Redesigning life: The promise of synthetic biology. The Economist., 3–12. [Google Scholar]
- 3. Voigt C.A. (2020) Synthetic biology 2020-2030: six commercially-available products that are changing our world. Nat. Commun., 11, 6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwok R. (2010) Five hard truths for synthetic biology. Nature, 463, 288–290. [DOI] [PubMed] [Google Scholar]
- 5. Munafo M.R., Nosek B.A., Bishop D.V.M., Button K.S., Chambers C.D., du Sert N.P., Simonsohn U., Wagenmakers E.J., Ware J.J. and Ioannidis J.P.A. (2017) A manifesto for reproducible science. Nat. Hum. Behav., 1, 0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Begley C.G. and Ioannidis J.P. (2015) Reproducibility in science: improving the standard for basic and preclinical research. Circ. Res., 116, 116–126. [DOI] [PubMed] [Google Scholar]
- 7. Nichols J.D., Oli M.K., Kendall W.L. and Boomer G.S. (2021) Opinion: a better approach for dealing with reproducibility and replicability in science. Proc. Natl. Acad. Sci. U.S.A., 118, e2100769118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baker M. (2016) 1,500 scientists lift the lid on reproducibility. Nature, 533, 452–454. [DOI] [PubMed] [Google Scholar]
- 9. Errington T.M., Denis A., Perfito N., Iorns E. and Nosek B.A. (2021) Challenges for assessing replicability in preclinical cancer biology. Elife, 10, e67995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Errington T.M., Mathur M., Soderberg C.K., Denis A., Perfito N., Iorns E. and Nosek B.A. (2021) Investigating the replicability of preclinical cancer biology. Elife, 10, e71601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Begley C.G. and Ellis L.M. (2012) Drug development: raise standards for preclinical cancer research. Nature, 483, 531–533. [DOI] [PubMed] [Google Scholar]
- 12. Prinz F., Schlange T. and Asadullah K. (2011) Believe it or not: how much can we rely on published data on potential drug targets?. Nat. Rev. Drug Discov., 10, 712. [DOI] [PubMed] [Google Scholar]
- 13. Begley C.G. (2013) An unappreciated challenge to oncology drug discovery: pitfalls in preclinical research. Am Soc. Clin. Oncol. Educ. Book, 466–468. [DOI] [PubMed] [Google Scholar]
- 14. Tiwari K., Kananathan S., Roberts M.G., Meyer J.P., Sharif Shohan M.U., Xavier A., Maire M., Zyoud A., Men J., Ng S. et al. (2021) Reproducibility in systems biology modelling. Mol. Syst. Biol., 17, e9982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelly J.R., Rubin A.J., Davis J.H., Ajo-Franklin C.M., Cumbers J., Czar M.J., de Mora K., Glieberman A.L., Monie D.D. and Endy D. (2009) Measuring the activity of BioBrick promoters using an in vivo reference standard. J Biol. Eng., 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beal J., Haddock-Angelli T., Gershater M., de Mora K., Lizarazo M., Hollenhorst J., Rettberg R. and Jones D.D. and iGEM Interlab Study Contributors . (2016) Reproducibility of fluorescent expression from engineered biological constructs in E. coli. PLoS One., 11, e0150182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cole S.D., Beabout K., Turner K.B., Smith Z.K., Funk V.L., Harbaugh S.V., Liem A.T., Roth P.A., Geier B.A., Emanuel P.A. et al. (2019) Quantification of interlaboratory cell-free protein synthesis variability. ACS Synth. Biol., 8, 2080–2091. [DOI] [PubMed] [Google Scholar]
- 18. Beal J., Farny N.G., Haddock-Angelli T., Selvarajah V., Baldwin G.S., Buckley-Taylor R., Gershater M., Kiga D., Marken J., Sanchania V. et al. (2020) Robust estimation of bacterial cell count from optical density. Commun Biol., 3, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shapira P., Kwon S. and Youtie J. (2017) Tracking the emergence of synthetic biology. Scientometrics, 112, 1439–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Canton B., Labno A. and Endy D. (2008) Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol., 26, 787–793. [DOI] [PubMed] [Google Scholar]
- 21. Lucks J.B., Qi L., Whitaker W.R. and Arkin A.P. (2008) Toward scalable parts families for predictable design of biological circuits. Curr. Opin. Microbiol., 11, 567–573. [DOI] [PubMed] [Google Scholar]
- 22. Lux M.W., Bramlett B.W., Ball D.A. and Peccoud J. (2012) Genetic design automation: engineering fantasy or scientific renewal?. Trends Biotechnol., 30, 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Lorenzo V. and Schmidt M. (2018) Biological standards for the knowledge-based bioeconomy: what is at stake. N Biotechnol., 40, 170–180. [DOI] [PubMed] [Google Scholar]
- 24. Decoene T., De Paepe B., Maertens J., Coussement P., Peters G., De Maeseneire S.L. and De Mey M. (2018) Standardization in synthetic biology: an engineering discipline coming of age. Crit. Rev. Biotechnol., 38, 647–656. [DOI] [PubMed] [Google Scholar]
- 25. Beal J., Haddock-Angelli T., Farny N. and Rettberg R. (2018) Time to get serious about measurement in synthetic biology. Trends Biotechnol., 36, 869–871. [DOI] [PubMed] [Google Scholar]
- 26. Jessop-Fabre M.M. and Sonnenschein N. (2019) Improving reproducibility in synthetic biology. Front Bioeng. Biotechnol., 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beal J., Goni-Moreno A., Myers C., Hecht A., de Vicente M.D.C., Parco M., Schmidt M., Timmis K., Baldwin G., Friedrichs S. et al. (2020) The long journey towards standards for engineering biosystems: are the molecular biology and the biotech communities ready to standardise?. EMBO Rep., 21, e50521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Glykofrydis F. and Elfick A. (2022) Exploring standards for multicellular mammalian synthetic biology. Trends Biotechnol., 40, 1299–1312. [DOI] [PubMed] [Google Scholar]
- 29. Beal J., Teague B., Sexton J.T., Castillo-Hair S., DeLateur N.A., Samineni M., Tabor J.J. and Weiss R. and the Calibrated Flow Cytometry Study Consortium . (2022) Meeting measurement precision requirements for effective engineering of genetic regulatory networks. ACS Synth. Biol., 11, 1196–1207. [DOI] [PubMed] [Google Scholar]
- 30. Beal J., Telmer C.A., Vignoni A., Boada Y., Baldwin G.S., Hallett L., Lee T., Selvarajah V., Billerbeck S., Brown B. et al. (2022) Multicolor plate reader fluorescence calibration. Synth. Biol., 7, ysac010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ross D., Tonner P.D. and Vasilyeva O.B. (2022) Method for reproducible automated bacterial cell culture and measurement. Synth. Biol., 7, ysac013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Romantseva E., Alperovich N., Ross D., Lund S.P. and Strychalski E.A. (2022) Effects of DNA template preparation on variability in cell-free protein production. Synth. Biol., 7, ysac015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rhea K.A., McDonald N.D., Cole S.D., Noireaux V., Lux M.W. and Buckley P.E. (2022) Variability in cell-free expression reactions can impact qualitative genetic circuit characterization. Synth. Biol., 7, ysac011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McDonald N.D., Rhea K.A., DaviesJ.P. Jr, Zacharko J.L., Berk K.L. and Buckley P.E. (2022) Evaluating the persistence and stability of a DNA-barcoded microbial system in a mock home environment. Synth. Biol., 7, ysac016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goldman R.P., Moseley R., Roehner N., Cummins B., Vrana J.D., Clowers K.J., Bryce D., Beal J., DeHaven M., Nowak J. et al. (2022) Highly-automated, high-throughput replication of yeast-based logic circuit design assessments. Synth. Biol., 7, ysac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia B.J., Urrutia J., Zheng G., Becker D., Corbet C., Maschhoff P., Cristofaro A., Gaffney N., Vaughn M., Saxena U. et al. (2022) A toolkit for enhanced reproducibility of RNASeq analysis for synthetic biologists. Synth. Biol., 7, ysac012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robitaille M.C., Byers J.M., Christodoulides J.A. and Raphael M.P. (2023) Automated cell segmentation for reproducibility in bioimage analysis. Synth. Biol., 8, ysad001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cummins B., Vrana J., Moseley R.C., Eramian H., Deckard A., Fontanarrosa P., Bryce D., Weston M., Zheng G., Nowak J. et al. (2022) Robustness and reproducibility of simple and complex synthetic logic circuit designs using a DBTL loop. Synth. Biol., 8, ysad005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bartley B.A., Beal J., Karr J.R. and Strychalski E.A. (2020) Organizing genome engineering for the gigabase scale. Nat. Commun., 11, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Plesser H.E. (2017) Reproducibility vs. replicability: a brief history of a confused terminology. Front. Neuroinform., 11, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Biden J. (2022) Executive order on advancing biotechnology and biomanufacturing innovation for a sustainable, safe, and secure American bioeconomy.
- 42. Endy D. (2005) Foundations for engineering biology. Nature, 438, 449–453. [DOI] [PubMed] [Google Scholar]
- 43. Knight T.F. (2005) Engineering novel life. Mol. Syst. Biol., 1, 0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Smit M.H. and van Duin J. (1990) Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. U.S.A., 87, 7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lou C., Stanton B., Chen Y.J., Munsky B. and Voigt C.A. (2012) Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol., 30, 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ali T., Renkawitz R. and Bartkuhn M. (2016) Insulators and domains of gene expression. Curr. Opin. Genet. Dev., 37, 17–26. [DOI] [PubMed] [Google Scholar]
- 47. Salis H.M., Mirsky E.A. and Voigt C.A. (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol., 27, 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bonde M.T., Pedersen M., Klausen M.S., Jensen S.I., Wulff T., Harrison S., Nielsen A.T., Herrgard M.J. and Sommer M.O. (2016) Predictable tuning of protein expression in bacteria. Nat. Methods, 13, 233–236. [DOI] [PubMed] [Google Scholar]
- 49. Cetnar D.P. and Salis H.M. (2021) Systematic quantification of sequence and structural determinants controlling mrna stability in bacterial operons. ACS Synth. Biol., 10, 318–332. [DOI] [PubMed] [Google Scholar]
- 50. Hollerer S. and Jeschek M. (2023) Ultradeep characterisation of translational sequence determinants refutes rare-codon hypothesis and unveils quadruplet base pairing of initiator tRNA and transcript. Nucleic Acids Res., 51, 2377–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. LaFleur T.L., Hossain A. and Salis H.M. (2022) Automated model-predictive design of synthetic promoters to control transcriptional profiles in bacteria. Nat. Commun., 13, 5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gardner T.S., Cantor C.R. and Collins J.J. (2000) Construction of a genetic toggle switch in Escherichia coli. Nature, 403, 339–342. [DOI] [PubMed] [Google Scholar]
- 53. Purnick P.E. and Weiss R. (2009) The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol., 10, 410–422. [DOI] [PubMed] [Google Scholar]
- 54. Shin J., Zhang S., Der B.S., Nielsen A.A. and Voigt C.A. (2020) Programming Escherichia coli to function as a digital display. Mol. Syst. Biol., 16, e9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nielsen A.A., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D. and Voigt C.A. (2016) Genetic circuit design automation. Science, 352, aac7341. [DOI] [PubMed] [Google Scholar]
- 56. Jones T.S., Oliveira S.M.D., Myers C.J., Voigt C.A. and Densmore D. (2022) Genetic circuit design automation with Cello 2.0. Nat. Protoc., 17, 1097–1113. [DOI] [PubMed] [Google Scholar]
- 57. Schaller R.R. (1997) Moore’s law: past, present and future. IEEE Spectr., 34, 52–59. [Google Scholar]
- 58. Stephens Z.D., Lee S.Y., Faghri F., Campbell R.H., Zhai C., Efron M.J., Iyer R., Schatz M.C., Sinha S. and Robinson G.E. (2015) Big data: astronomical or genomical?. PLoS Biol., 13, e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Farzaneh T. and Freemont P.S. (2021) Biofoundries are a nucleating hub for industrial translation. Synth. Biol., 6, ysab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beal J., Haddock-Angelli T., Baldwin G., Gershater M., Dwijayanti A., Storch M., de Mora K., Lizarazo M. and Rettberg R. and iGEM Interlab Study Contributors . (2018) Quantification of bacterial fluorescence using independent calibrants. PLoS One., 13, e0199432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fedorec A.J.H., Robinson C.M., Wen K.Y. and Barnes C.P. (2020) FlopR: an open source software package for calibration and normalization of plate reader and flow cytometry data. ACS Synth. Biol., 9, 2258–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Csibra E. and Stan G.B. (2022) Absolute protein quantification using fluorescence measurements with FPCountR. Nat. Commun., 13, 6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McLaughlin J.A., Beal J., Misirli G., Grunberg R., Bartley B.A., Scott-Brown J., Vaidyanathan P., Fontanarrosa P., Oberortner E., Wipat A. et al. (2020) The synthetic biology open language (sbol) version 3: simplified data exchange for bioengineering. Front. Bioeng. Biotechnol., 8, 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schreiber F., Gleeson P., Golebiewski M., Gorochowski T.E., Hucka M., Keating S.M., Konig M., Myers C.J., Nickerson D.P., Sommer B. et al. (2021) Specifications of standards in systems and synthetic biology: status and developments in 2021. J Integr. Bioinform., 18, 20210026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Martinez-Garcia E., Goni-Moreno A., Bartley B., McLaughlin J., Sanchez-Sampedro L., Pascual Del Pozo H., Prieto Hernandez C., Marletta A.S., De Lucrezia D., Sanchez-Fernandez G. et al. (2020) SEVA 3.0: an update of the Standard European Vector Architecture for enabling portability of genetic constructs among diverse bacterial hosts. Nucleic Acids Res., 48, D1164–D1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Misirli G., Hallinan J., Pocock M., Lord P., McLaughlin J.A., Sauro H. and Wipat A. (2016) Data integration and mining for synthetic biology design. ACS Synth. Biol., 5, 1086–1097. [DOI] [PubMed] [Google Scholar]
- 67. Sainz de Murieta I., Bultelle M. and Kitney R.I. (2016) Toward the first data acquisition standard in synthetic biology. ACS Synth. Biol., 5, 817–826. [DOI] [PubMed] [Google Scholar]
- 68. Walsh D.I. 3rd, Pavan M., Ortiz L., Wick S., Bobrow J., Guido N.J., Leinicke S., Fu D., Pandit S., Qin L. et al. (2019) Standardizing automated DNA assembly: best practices, metrics, and protocols using robots. SLAS Technol., 24, 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baig H., Fontanarossa P., McLaughlin J., Scott-Brown J., Vaidyanathan P., Gorochowski T., Misirli G., Beal J. and Myers C. (2021) Synthetic biology open language visual (SBOL visual) version 3.0. J Integr. Bioinform., 18, 20210013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grob A., Di Blasi R. and Ceroni F. (2021) Experimental tools to reduce the burden of bacterial synthetic biology. Curr. Opin. Syst. Biol., 28, 100393. [Google Scholar]
- 71. Zilberzwige-Tal S., Fontanarrosa P., Bychenko D., Dorfan Y., Gazit E. and Myers C.J. (2022) Investigating and modeling the factors that affect genetic circuit performance. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Frei T., Cella F., Tedeschi F., Gutierrez J., Stan G.B., Khammash M. and Siciliano V. (2020) Characterization and mitigation of gene expression burden in mammalian cells. Nat. Commun., 11, 4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eldar A. and Elowitz M.B. (2010) Functional roles for noise in genetic circuits. Nature, 467, 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Eling N., Morgan M.D. and Marioni J.C. (2019) Challenges in measuring and understanding biological noise. Nat. Rev. Genet., 20, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Marques G., Pengo T. and Sanders M.A. (2020) Imaging methods are vastly underreported in biomedical research. Elife, 9, e55133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.(2021) Minding microscopy metadata. Nat. Methods, 18, 1411. [DOI] [PubMed] [Google Scholar]
- 77. Raphael M.P., Sheehan P.E. and Vora G.J. (2020) A controlled trial for reproducibility. Nature, 579, 190–192. [DOI] [PubMed] [Google Scholar]
- 78. Crowther M., Grozinger L., Pocock M., Taylor C.P.D., McLaughlin J.A., Misirli G., Bartley B.A., Beal J., Goni-Moreno A. and Wipat A. (2020) ShortBOL: a language for scripting designs for engineered biological systems using synthetic biology open language (SBOL). ACS Synth. Biol., 9, 962–966. [DOI] [PubMed] [Google Scholar]
- 79. Bryce D., Goldman R.P., DeHaven M., Beal J., Bartley B., Nguyen T.T., Walczak N., Weston M., Zheng G., Nowak J. et al. (2022) Round Trip: an automated pipeline for experimental design, execution, and analysis. ACS Synth. Biol., 11, 608–622. [DOI] [PubMed] [Google Scholar]
- 80. Hasnain A., Balakrishnan S., Joshy D.M., Smith J., Haase S.B. and Yeung E. (2022) Learning perturbation-inducible cell states of novel compounds from observability analysis of transcriptome dynamics. Nat. Commun. 14, 3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Collins F.S. and Tabak L.A. (2014) Policy: NIH plans to enhance reproducibility. Nature, 505, 612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Galdzicki M., Clancy K.P., Oberortner E., Pocock M., Quinn J.Y., Rodriguez C.A., Roehner N., Wilson M.L., Adam L., Anderson J.C. et al. (2014) The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology. Nat. Biotechnol., 32, 545–550. [DOI] [PubMed] [Google Scholar]
- 83. Schreiber F., Sommer B., Czauderna T., Golebiewski M., Gorochowski T.E., Hucka M., Keating S.M., Konig M., Myers C., Nickerson D. et al. (2020) Specifications of standards in systems and synthetic biology: status and developments in 2020. J Integr. Bioinform., 17, 20200022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chavez M., Ho J. and Tan C. (2017) Reproducibility of high-throughput plate-reader experiments in synthetic biology. ACS Synth. Biol., 6, 375–380. [DOI] [PubMed] [Google Scholar]
- 85. Diaba-Nuhoho P. and Amponsah-Offeh M. (2021) Reproducibility and research integrity: the role of scientists and institutions. BMC Res. Notes, 14, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. U.K. Reproducibility Network Steering Committee . (2021) From grassroots to global: a blueprint for building a reproducibility network. PLoS Biol., 19, e3001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Strech D., Weissgerber T. and Dirnagl U. (2020) Improving the trustworthiness, usefulness, and ethics of biomedical research through an innovative and comprehensive institutional initiative. PLoS Biol., 18, e3000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Peccoud J., Anderson J.C., Chandran D., Densmore D., Galdzicki M., Lux M.W., Rodriguez C.A., Stan G.B. and Sauro H.M. (2011) Essential information for synthetic DNA sequences. Nat. Biotechnol., 29, 22. [DOI] [PubMed] [Google Scholar]
- 89.(2020) The method comes first. Nat. Methods, 17, 1169. [DOI] [PubMed] [Google Scholar]
- 90. Hillson N.J., Plahar H.A., Beal J. and Prithviraj R. (2016) Improving synthetic biology communication: recommended practices for visual depiction and digital submission of genetic designs. ACS Synth. Biol., 5, 449–451. [DOI] [PubMed] [Google Scholar]
- 91. Tas H., Amara A., Cueva M.E., Bongaerts N., Calvo-Villamanan A., Hamadache S. and Vavitsas K. (2020) Are synthetic biology standards applicable in everyday research practice?. Microb. Biotechnol., 13, 1304–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Meyer C., Nakamura Y., Rasor B.J., Karim A.S., Jewett M.C. and Tan C. (2021) Analysis of the innovation trend in cell-free synthetic biology. Life, 11, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were collected in this work.


