Abstract
Current data-dependent acquisition (DDA) approaches select precursor ions for MS/MS characterization based on their abundance, known as a TopN approach. Low abundant species may not be identified as biomarkers in a TopN approach. Herein, a new DDA approach is proposed, DiffN, which uses the differential abundance of ions between two samples to selectively target species undergoing the largest fold-changes for MS/MS. Using a dual nano-electrospray (nESI) ionization source which allows samples contained in separate capillaries to be analyzed in parallel, the DiffN approach was developed and validated with well-defined lipid extracts. The dual nESI source and DiffN DDA approach was applied to quantify the differences in lipid abundance between two colorectal cancer cell lines. The SW480 and SW620 lines represent a matched pair from the same patient: the SW480 cells from a primary tumor and the SW620 cells from a metastatic lesion. A comparison of the TopN and DiffN DDA approaches on these cancer cell samples highlights the ability of DiffN to increase the likelihood of biomarker discovery and the decreased probability of TopN to efficiently select lipid species that undergo large fold changes. The ability of the DiffN approach to efficiently select precursor ions of interest makes it a strong candidate for lipidomic analyses. This DiffN DDA approach may also apply to other molecule classes (e.g., other metabolites or proteins) that are amenable to shotgun analyses.
Graphical Abstract
Introduction
In biomarker discovery, prognostic molecular signatures distinguish a phenotype of interest from a control, typically a disease-related state vs. a healthy state. Qualitative assessments rely on the presence or absence of a unique set of biomolecules, such as unique oligonucleotide sequences, antigens, or host-generated antibodies that result from a bacterial or viral infection. Many disease-related states, however, are identified by subtle changes in metabolite pools.1–4 Detecting these changes requires accurate quantitative measures capable of identifying and measuring multiple species simultaneously. Coupling a separation technique such as liquid chromatography (LC) to mass spectrometry (MS) is a powerful tool for biomarker discovery,5,6 providing molecular identification and quantification. However, due to instrumental limitations, not every species eluting under an LC peak can be targeted and identified with tandem mass spectrometry (MS/MS). Data-dependent acquisitions (DDAs) prioritize which m/z values are selected for MS/MS, allowing the mass spectrometer to focus on a subset of species. In a TopN approach, an N number of the most abundant ions in the mass spectrum are selected for MS/MS identification or quantification.7,8 Typical TopN approaches target the most abundant 10 – 15 ions to be selected as precursor ions, which can limit extensive sample coverage. Two TopN DDA approaches are typically used, dynamic exclusion and iterative exclusion. In dynamic exclusion, precursor ions selected in the first MS survey scan are placed on a “dynamic” list for a certain amount of time. The next set of topmost abundant precursors are selected in the next survey scan and this process can be repeated. Dynamic exclusion can typically target a few hundred species efficiently, but often misses low abundance peaks. Alternatively, an iterative exclusion approach can be used. In iterative exclusion, a single sample is repeatedly analyzed, and precursor ions selected in the previous analysis (i.e., the most abundant precursors) are placed on an exclusion list for the next analysis. This process can be repeated for N number of analyses. Iterative exclusion allows for identification of much lower abundance species compared to dynamic exclusion, but the need for repeat analyses significantly reduces the throughput and efficiency of the discovery process and can require large sample volumes.9–11
Direct infusion, or so-called “shotgun” analyses, introduce the sample to the MS without prior separation.12,13 Shotgun analyses offer a simpler setup and reduced analysis times, and can require less sample than chromatographic DDA methods where repeat injections are performed. A disadvantage of shotgun approaches is lower overall sample coverage due to matrix effects and/or ionization suppression from highly abundant species. TopN approaches are also used in shotgun analyses and dynamic and iterative exclusion DDA approaches can increase sample coverage.14–16
The coverage obtained in chromatographic and shotgun analyses of complex mixtures using a TopN approach is limited, less likely to identify species that are low abundance but undergo large fold-changes between the phenotype of interest and control samples. The detection and quantification of fold changes across multiple species are particularly useful in biomarker discovery17–19 and is information that can be lost when focusing on high abundant, housekeeping species. Here, a new method of precursor ion selection for MS/MS analysis is demonstrated to identify and characterize species that undergo the largest changes between multiple samples, DiffN. In a DiffN approach (Fig. 1), fold-changes for every precursor ion contained in two (or more) separate samples are determined in real-time. Species that meet a differential abundance threshold are then targeted for MS/MS acquisition. Unlike TopN approaches, DiffN focuses on fold changes rather than intensity. It offers the potential to improve biomarker discovery by focusing on less abundant species that undergo large fold changes and reducing the amount of sample needed by limiting the extent of reanalysis. Additionally, two (or more) samples are analyzed in parallel, rather than sequentially, which minimizes the effect of instrumental fluctuations.
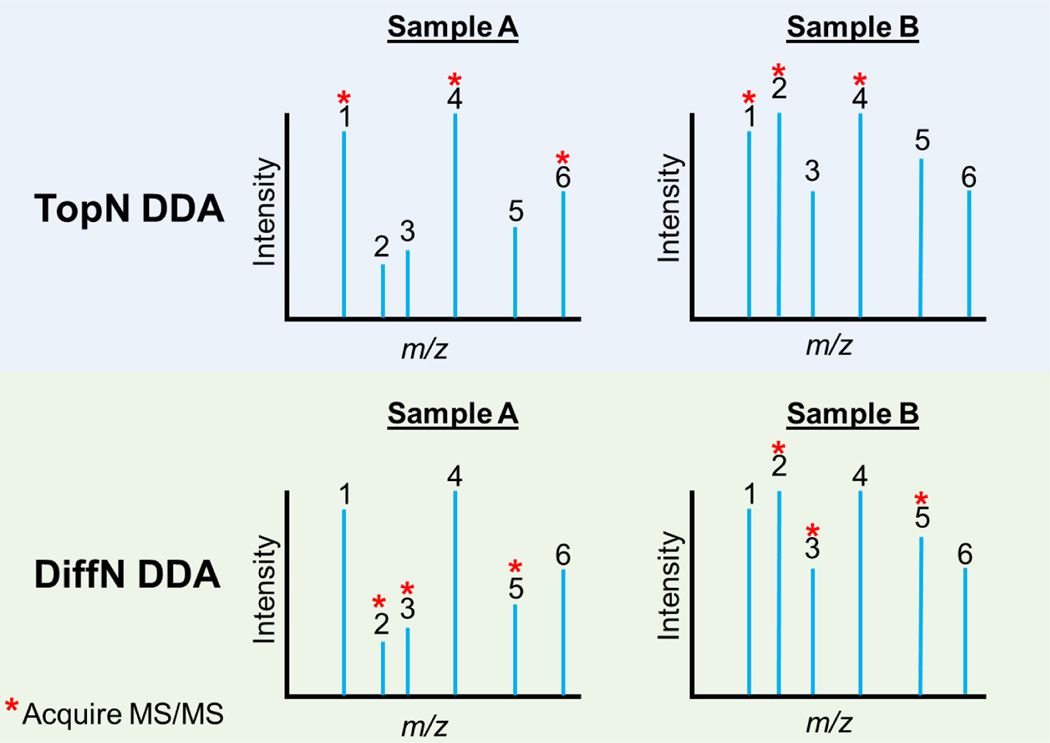
Figure 1.
Graphical representation of precursor ion selection for MS/MS analysis using (top) a TopN DDA approach or (bottom) the DiffN DDA approach. In TopN, Samples A and B are analyzed sequentially (e.g., separate chromatographic separations or separated direct infusions) and the most abundant species are selected for MS/MS (here, peaks 1, 4 and 6 in Sample A and 1, 2, and 4 in Sample B). TopN may miss low abundance species with higher fold changes (here, peaks 3 and 5). In DiffN, Samples A and B are analyzed in parallel. Species with the largest differential abundances are targeted for MS/MS (here, peaks 2, 3, and 5).
The DiffN approach to selecting precursor ions for MS/MS analysis does not alter current sample preparation, separation, or characterization workflows and thus should apply broadly to metabolomic and proteomic-based biomarker discovery. To demonstrate its utility, the DiffN DDA approach was applied to determine differences in lipid abundance between a matched pair of colorectal cancer cell lines obtained from a primary tumor (SW480 cells) and a metastasis (SW620 cells). Lipids were chosen because they are known biomarkers for distinguishing cancerous and healthy tissue.20–24 Altered lipid signatures also can distinguish drug responsive and resistant cancer cells.25–27
Materials and Methods
Reagents.
Unless otherwise stated, all reagents were used as received. Optima™ grade methyl tert-butyl ether (MTBE), dichloromethane (DCM), formic acid, methanol (MeOH), and water were purchased from Fisher Scientific. Ammonium acetate was purchased from Sigma Aldrich. Bovine liver extract (BLE) and the Differential Ion Mobility System Suitability Synthetic Standard Mix were purchased from Avanti Polar Lipids. Pyrex glass capillaries (1.5–1.8 × 90 mm) used for nESI were purchased from Fisher Scientific. All 3D- printed components used in this work were printed on a Form2 printer with Clear V4 resin (Formlabs).
Dual nESI Design.
The dual nano-electrospray ionization (dual nESI) source was based on a previous design.28,29 The two nESI emitters were controlled electrokinetically using a simple custom-built power supply and switching circuit, which allowed for the simultaneous collection and DiffN analysis of two samples. The circuit used to control the dual nESI source in this work used an EMCO C25N power module to apply −1.5 kV to each emitter, which initiated or stopped electrospray via two high voltage reed relay switches. The switching rate and duration of the applied voltage on a single emitter were controlled with a LabVIEW program. The constant voltage supplied by the EMCO module resulted in a switching time of 500 μs between emitters (Fig. S1B). This setup overcame the limitations of a previously described design,28,29 which required a 250 ms delay between emitter switching (Fig. S1A). This new level of control between emitters reduced crosstalk and allowed full utilization of the MS duty cycle. Figure S2 is a detailed diagram of the power supply used to control the dual nESI setup.
Dual nESI Parameters and Validation.
The electrospray solvent was 50/50 (v/v) dichloromethane/methanol containing 0.1% formic acid (by volume) and 5 mM ammonium acetate. The solvent also contained either 0.1 or 1.0 μg/mL of a synthetic lipid mixture (Table S1), which served as an internal standard. Each nESI emitter was loaded with approximately 30μL of the sample before analysis and placed 15 mm from the MS inlet. The spacing between emitter tips was set at 3.5 mm (Figure S3). The reproducibility and accuracy of the dual nESI setup were evaluated by measuring the fold change between two emitters, one containing 1.0 μg/mL and the other 2.0 μg/mL of a bovine liver extract (BLE) lipid standard. For each experiment, the two emitters were alternated (pulsed) on and off five times. Each emitter was on for 12 seconds per pulse during which time 10–12 mass spectra were acquired. As each emitter was on five times during the experiment, a total of 50–60 mass spectra were acquired per emitter. These mass spectra were then averaged to give an intensity value for each emitter. This same process was repeated for a total of three pairs of emitters, after which the average fold change, standard deviation, and %RSD were calculated for lipid species in BLE using the intensity values determined for the three emitter pairs.
MS and DIMS Parameters.
The dual nESI emitters were coupled to a Bruker HCT Ultra quadrupole ion trap mass spectrometer with a custom-built differential ion mobility spectrometer (DIMS) positioned on the front of the MS inlet capillary.30 The DIMS was operated at a compensation field (CF) of 67 V/cm and a dispersion field (DF) of 22 kV/cm. MS parameters used for all experiments were as follows: capillary voltage, 2.0 kV; skimmer voltage, 40.0 V; capillary exit voltage, 149.5 V; octopole 1 voltage, 8.00 V; octopole 2 voltage, 2.20 V; trap drive, 80.5; octupole RF, 200 Vpp; lens 1 voltage, −5.0 V; lens 2 voltage, −60.0 V; dry gas temperature setting, 200 °C; dry gas flow rate, 1 L/min; smart target, 100,000; maximum accumulation time, 200 ms; scan range, 400–1000 Da; spectral averages, 15.
The glass capillaries (Pyrex) used for nESI were pulled to an inner diameter of approximately 12.5 μm using a two-stage PC-10 heated capillary puller (Narishige) with the following temperature settings (arbitrary units): initial, 24.8; stage one, 60.0; stage two, 48.0. A 36-gauge stainless steel wire was positioned in the barrel of each capillary before they were mounted in a 3D printed holder.
Cell Culture and Lipid Extraction.
All cell culture medium and supplements were purchased from Gibco, except for fetal bovine serum (FBS, VWR). The SW480 and SW620 cell lines were purchased from the American Type Culture Collection and validated by short tandem repeating sequencing. Both cell lines were maintained as monolayers at 37 °C and 5% CO2 in phenol-free RPMI 1640 medium supplemented with 10% FBS, HEPES (12.5 mM), and penicillin-streptomycin (1%, v/v). The culture medium was exchanged every 2–3 days, and cells passed at 80–90% confluency with phenol-free TrypLE, following standard culture procedures.
The lipid extraction procedure was modified from a previously published protocol.31 Here, cells were detached from the plate and counted using the trypan blue exclusion method. Aliquots of 1.0×106 cells were pelleted by centrifugation (1000 xg, 5 min), washed with 1 mL of a 1X phosphate buffered saline solution, and re-pelleted to remove any residual culture medium. The cells were resuspended in 300 μL of ice-cold methanol (−20 °C) and vortexed for 30 s. Next, lipids were extracted into 1200 μL of MTBE. After 30 min of agitation on a reciprocating shaker, 300 μL of water was added, each sample was centrifuged (21,000 xg, 5 min), and 1000 μL of supernatant was collected. The supernatant was dried under vacuum and reconstituted in 1 mL of electrospray solvent containing 1 μg/mL of the internal standard lipid mixture.
Data Analysis.
Unless otherwise stated, values are the average and standard deviation of at least four samples prepared from separate plates of cells, loaded into separate pairs of capillaries, and placed in the dual nESI setup. To limit the biological variation associated with different cell passages, samples of the SW480 and SW620 cell lines were processed from a single cell pass. All plots and statistical analyses were made in GraphPad Prism version 9.3.1.
Results and Discussion
Validation of the DiffN Approach with Lipid Mixture Standards.
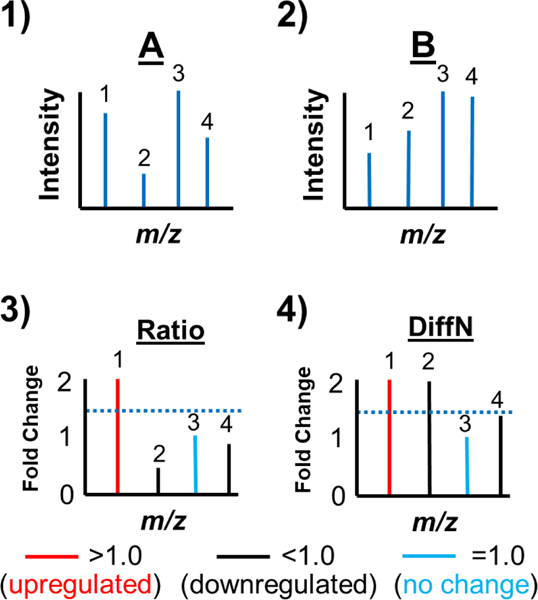
The DiffN approach selects precursor ions for MS/MS identification based on their differential abundance between two samples. In Figure 2 is the workflow for DiffN precursor ion selection, which depends on accurate relative quantification of each lipid species (fold change). In this experimental setup, equal concentrations of an internal standard were added to each sample and loaded into capillaries. The internal standards accounted for variations arising from differences in capillary tip geometry, spray instability, and ionization suppression. The sample-containing capillaries were aligned in front of the MS inlet, and a mass spectrum of each sample obtained (Fig. 2, steps 1 and 2). A Ratio spectrum was generated by dividing the internal standard-normalized intensities of each m/z value (Fig. 2, step 3). A DiffN spectrum was generated by plotting the reciprocal of ratios with a value of < 1.0 (Fig. 2, step 4). Finally, species with a DiffN value greater than a preset threshold (e.g., 1.5-fold, Fig. 2 dotted blue line) were selected for MS/MS identification (peaks 1 and 2).
Figure 2.
Workflow for DiffN MS/MS precursor ion selection. In steps 1 and 2, mass spectra for Samples A and B are acquired. Next, each peak in the two mass spectra is normalized to the internal standard intensity. In step 3 a Ratio Spectrum is generated where the normalized mass spectrum of Sample A is divided by the normalized mass spectrum of Sample B. Next, each value in the Ratio Spectrum with a fold change <1.0 is changed to its reciprocal value to generate a DiffN Spectrum (step 4). Finally, MS/MS spectra are acquired for each of the N selected peaks in the DiffN Spectrum.
The accuracy of the dual nESI source and the DiffN workflow was validated by comparing emitters loaded with 1.0 and 2.0 μg/mL BLE. This extract was chosen because it contains a high percentage of glycerophospholipids (GPs, Table S2), which are abundant in colorectal cancer cell lines.32,33 The DIMS parameters used for all experiments were determined empirically. A DF of 22 kV/cm and a CF of 67 V/cm were used because they allowed co-transmission of all lipid species into the MS while excluding background contaminant ions. To determine the optimal CF for lipid transmission, CF scans were performed. A CF of 67 V/cm was chosen because it was closest to the center of all lipid peaks transmitting through the DIMS. To ensure the DIMS CF chosen did not affect the fold change measurement accuracy, three CF values were tested: the optimal CF, 67, as well as 60, and 73 V/cm. The fold-changes for all lipid species were not statistically different for the three CFs (Fig. S5 A and B), as determined by a student’s t-test. Of the CFs tested, 67 V/cm gave the lowest average %error for all lipid species with a value of 10.5% (Fig. S5C) while maintaining good reproducibility (%RSD 11.4% across all lipids, Fig. S5D). Lipid classes could potentially be separated using a higher resolution DIMS34 or carrier gas modifiers,28 which could significantly enhance the capability of DiffN by separating isobars/isomers.
Figure 3 is a Ratio Spectrum of 25 species in the BLE sample that were confidently identified with MS/MS. (The Ratio and DiffN spectra are the same in this case as the samples are identical, the fold-change of each lipid species is due to a different concentration of the same BLE sample.) These values were determined by normalizing the raw signal intensities to either, 1) the sum of the signal of the internal standards, 2) the signal of the lipid class specific internal standard, or 3) the sum of the internal standard signal where the class specific species was excluded (e.g., when normalizing PC lipids, the PC class specific internal standard was excluded from the sum). All approaches were tested because the sum of the internal standards is most appropriate for unknown samples, as the identities of lipid species have yet to be determined. Additionally, the sum of all internal standards, class specific excluded, is relevant for when a class specific internal standard is not available. All normalization methods resulted in statistically indistinguishable ratios, as determined by Student’s t-tests for each lipid species, confirming the validity of each method of normalization. If a solvent modifier28 or high resolution DIMS34 is used to separate lipid classes, then the class specific internal standard would be the only one used.
Figure 3.
Comparison of the fold-changes of each lipid species between 2.0 μg/mL and 1.0 μg/mL BLE determined by normalizing to the sum of the internal standards (●), normalizing to each class-specific internal standard (), and normalizing to the sum of all internal standards (), excluding the class of the lipid being normalized (e.g., when normalizing PC lipids, the PC class specific IS was excluded from the sum). There is no statistical difference in fold changes for any method of normalization. Species are plotted in order of increasing abundance with the lowest abundance species on the left and the highest abundance species on the right.
Each data point is the average of fold changes determined from three replicates, where a new pair of nESI emitters were used for each replicate. The bias between the measured and expected fold-changes for all 25 of the identified lipid species was less than 20%. For 23 of the identified lipid species, the bias was less than 10% (Fig. S4).
The reproducibility of the fold changes between the three sets of nESI emitters was also acceptable with 22 of the lipid species having relative standard deviations (RSD) of less than 20%. Three species had RSD values greater than 20%, each a triglyceride (TG): 25.2% for TG 50:0, 43.2% for TG 48:0, and 21.6% for TG 48:1. The imprecision in the fold change measurements is likely due to a combination of low abundance and ion suppression by higher abundance phosphatidylcholines (PCs).
Applying the DiffN Workflow to Compare the SW480 and SW620 Cell Lines.
The DiffN DDA method was applied to select the precursor ions of lipid species extracted from the SW480 and SW620 colorectal cell lines. These two cell lines are a matched pair, obtained from the same patient. The SW480 cells were derived from the primary tumor and the SW620 cells from a lymph node metastasis. The lipid profiles of these cell lines were previously compared using a shotgun approach and a TopN DDA method, providing a good starting point to benchmark the DiffN workflow.
Lipids were extracted and analyzed from four samples, each containing 1×106 cells. The SW480 and SW620 cell lines were analyzed simultaneously in the dual nESI setup at a DIMS DV of 22 kV/cm and a static CV of 67 V/cm. The intensity of each precursor ion was normalized to the sum of the internal standard intensities. The fold-change between the SW620 and the SW480 peaks was determined from the analyte-to-internal standard ratios. Any fold changes with %RSD values > 30.0% were attributed to noise (e.g., signals near the instrument’s detection limit) and excluded. Additionally, fold changes between 0.7–1.3 were excluded to truncate the dataset and focus on lipids with larger differential abundances. The m/z list that met these criteria contained 118 peaks which were then characterized with MS/MS. Included in this list were peaks containing one or more 13C isotopes. Therefore, of the 118 peaks characterized by MS/MS, 40 of them were identified as a monoisotopic lipid peak.
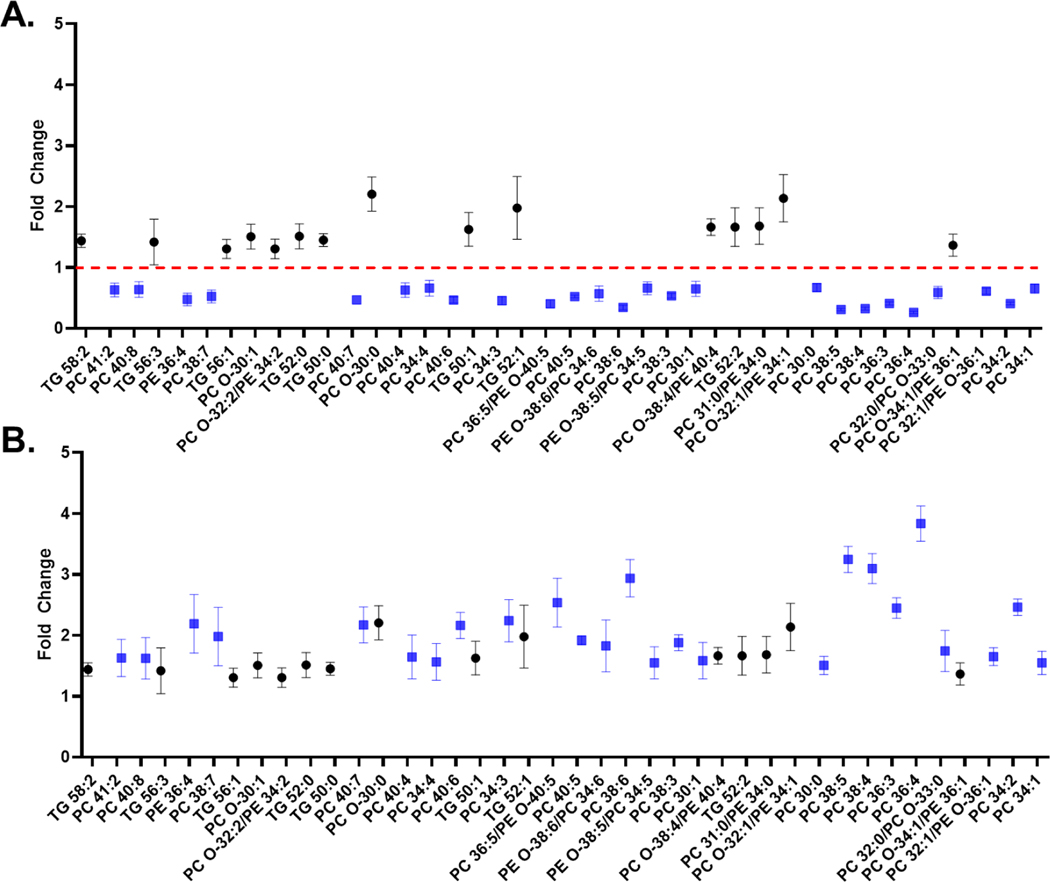
In Figure 4A the lipid fold changes between the SW620 and SW480 cells are depicted as a Ratio spectrum. Figure 4B is the corresponding DiffN spectrum, in which the reciprocal values for fold changes < 1.0 observed in the Ratio spectrum are plotted. For example, PC 36:4 has a fold change of 0.26 in the Ratio spectrum but an overall 3.84-fold difference in the DiffN spectrum. In Figures S6 and S7 the analyte-to-internal standard ratios and statistically significant differences in lipid abundance between the two cell lines are depicted.
Figure 4.
(A) Ratio spectrum displaying the fold-change of identified lipid species in the SW620 extract divided by the corresponding lipid species in the SW480 extract. (B) The DiffN spectrum, which plots the reciprocal of all fold changes in the ratio spectrum that were less than 1.0. All lipids were normalized to the sum of the internal standard intensities before determining the fold changes. Blue squares (■) represent lipids in the Ratio spectrum with a fold change <1.0, and black circles (●) represent lipids with a fold change >1.0. Lipids are ordered in terms of relative abundance with the lowest abundance species on the far left of the plot and the highest abundance species on the right of the plot.
Of the 15 upregulated lipid species in the SW620 cells, eight were TGs and seven GPs. Two of the GPs were confidently identified as phosphatidylcholines (PCs). The MS/MS spectra of the remaining four GPs contained product ions that corresponded to a PC phosphate headgroup (m/z = 184.1) and the neutral loss of a phosphatidyl ethanolamine (PE) phosphate headgroup (mass 141.1), indicating a pair of isobars at the same m/z value. Of the 25 downregulated lipids in the SW620 cells, 21 were confidently identified as PCs. The remaining four GPs also contained both PC and PE signatures. The species that gave product ion signatures of both a PC and PE originated from a pair of isobaric precursor ions. MS/MS was performed in an attempt to determine the fold-change of each of these isobaric species using the relative abundances of the m/z = 184.1 and ions with mass 141.1 losses. However, these values did not match the fold changes determined from the full scan mass spectra. The product ion signals were low for m/z 184.1 and the m/z resulting from neutral loss of 141.1 Da. The signal may have been suitable for qualitative identification but not for quantification. Given the discrepancies between the ratios from the full scan and MS/MS spectra, the fold change of the PC and PE isobars were reported as PC/PE in Figure 4. In the future, DIMS carrier gas modifiers could be used to potentially separate isobaric species to determine individual fold change contributions.28
With DiffN, fold changes are determined on an MS1 level. An added benefit of determining fold changes on an MS1 level is obtaining fold change values for the monoisotopic peak and each 13C-containing isotope detected. The isotopic distributions provide multiple measurements of the same molecule and allow for confirmation of the fold change measurement for the monoisotopic species. Two examples of using 13C-containing isotopes to confirm the fold change are depicted in Fig. S8. Depicted in Fig. S8A is a highly abundant peak, PC 34:1, which is 54% of the base peak, while depicted in Fig. S8B is a lower abundance peak, PC 38:4, which is only 5% of the base peak. For PC 34:1, three 13C-containing isotopes were detected, while only one was detected for PC 38:4. Nevertheless, the fold changes for the 13C-containing peaks were the same as the monoisotopic peak, confirming the correct fold change value was determined.
The lipids identified using the DiffN method were compared to those reported in literature. Previously, other groups have characterized the lipid profiles of the SW480 and SW620 cell lines using a modified Folch extraction, introducing the samples to the MS with a single nESI emitter and a PC 28:0 internal standard. PC, PE, and TG species were differentially abundant between the two cell lines in both datasets. Of the 40 lipids identified using DiffN, 29 (72.5%) were identified by others who used a TopN DDA approach. Of the 29 lipids identified in both studies, the fold-change direction (increase or decrease) was the same for 21 lipids. Of the eight lipids remaining, others found no statistical difference in abundance for four of them (all PCs) while the other four showed opposite fold changes (two TGs, one PC, and one PE). The discrepancies for those eight lipids are likely due to biological variation where different cell culture conditions and different passage numbers, leading to genetic variation, could affect lipid uptake, synthesis, and storage, and thereby change the observed abundance for certain lipid species.
To further illustrate the benefits of focusing on fold-changes between samples rather than abundances when selecting precursor ions for MS/MS, the m/z values selected using a TopN or DiffN approach for the same lipid extract samples from the SW480 and SW620 cell lines were compared. Table 1 lists the ten most abundant lipid species and the ten most differentially abundant species, the Top10 and Diff10 precursor ions across the two cell lines. The Top10 approach exhibited several limitations compared to Diff10. First, each lipid species selected by the DiffN approach underwent a fold change (> 2.0) while the seven lipid species unique to the Top10 list did not. Figures S6, S7, and S9 depict the analyte-to-internal standard ratios and statistical significance of each fold change for lipids in the Diff10 and Top10 lists. Nine out of ten species in the Top10 list had statistically significant fold changes based on a Student’s t-test (p-value <0.05) while all species in the Diff10 list showed statistically significant fold changes. PC 38:4 (fold change 1.11) in the Top10 list was not significant. Second, of all the lipids identified (43 lipids total including the 40 identified by DiffN and three unique lipids selected by Top10 and not by DiffN- PC 36:2, PC 36:1, and PE 38:4), the most abundant precursor ion, PC 34:1, ranked 31st in terms of fold change, indicating that intensity does not necessarily correlate with fold change.
Table 1.
Precursor ions detected in the SW480 and SW620 cell lysates selected using the Top10 or Diff10 approach. a,b,c
| Top10 | Diff10 | ||||
|---|---|---|---|---|---|
| Lipid ID | m/z | Fold Change | Lipid ID | m/z | Fold Change |
| PC 34:1 | 760.8 | 1.53 | PC 36:4 | 782.8 | 3.82 |
|
|
|
||||
| PC 34:2 | 758.8 | 2.45 | PC 38:5 | 808.7 | 3.23 |
|
|
|
||||
| PC 36:2 | 786.7 | 1.25 | PC 38:4 | 810.7 | 3.08 |
|
|
|
||||
| PC 32:1/PE O-36:1 | 732.8 | 1.64 | PC 38:6 | 806.7 | 2.91 |
|
|
|
||||
| PC 36:4 | 782.8 | 3.82 | PC 36:5/PE O-40:5 | 780.8 | 2.49 |
|
|
|
||||
| PC 36:1 | 788.8 | 1.18 | PC 34:2 | 758.8 | 2.45 |
|
|
|
||||
| PC O-34:1/PE 36:1 | 746.8 | 1.37 | PC 36:3 | 784.7 | 2.44 |
|
|
|
||||
| PC 32:0/PC O-33:0 | 734.8 | 1.70 | PC O-30:0 | 692.8 | 2.20 |
|
|
|
||||
| PE 38:4 | 768.7 | 1.11 | PC 34:3 | 756.7 | 2.20 |
|
|
|
||||
| PC 36:3 | 784.7 | 2.44 | PC 40:6 | 834.7 | 2.14 |
Values highlighted in blue were picked by the Top10 DDA approach
Values highlighted in green were picked by the Diff10 DDA approach
Values highlighted in black were picked by both the Top10 and Diff10 DDA approach
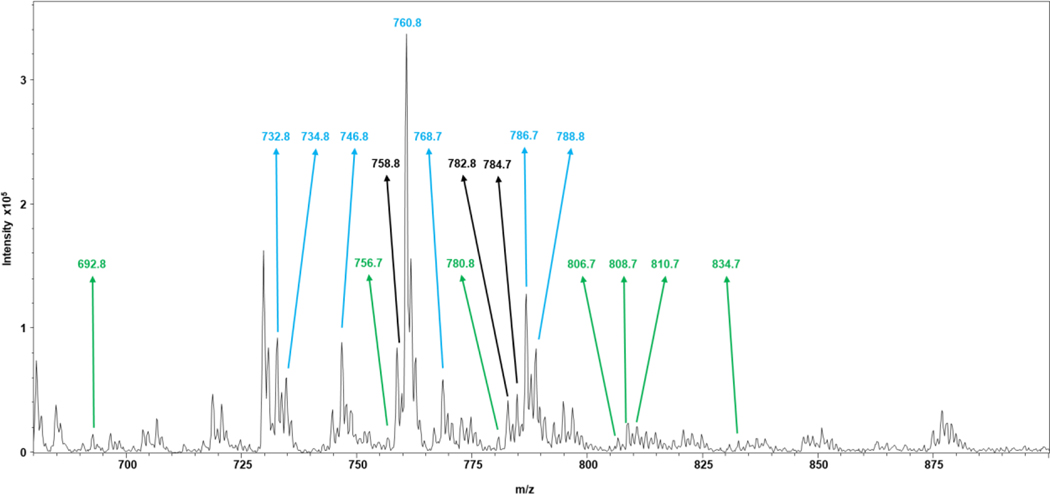
Figure 5 is a mass spectrum of the SW620 lipid extract (a mass spectrum of the SW480 lipid extract is shown in Fig. S10), with m/z values selected with the TopN and DiffN DDA approaches noted. These m/z values correspond to the lipid species in Table 1. This visualization of the data and the m/z selection process highlight the ability of the DiffN approach to target species that undergo large fold changes but are of low abundance. These species are targeted immediately and do not require many rounds of iterative exclusion needed in a TopN approach. This data also highlights the narrower mass range occupied by Top10 species, where the highly abundant ions are present, and shows the DiffN approach can target species across a wider mass range that are low abundance. A wider mass range could potentially lead to targeting a wider variety of lipid classes. With DiffN, the discovery of unique biomarker signatures is enabled with greater efficiency in terms of instrument duty cycle and experiment time.
Figure 5.
Mass spectrum obtained from a lipid extract of the SW620 cell line. The m/z values selected by the TopN approach are annotated in blue, the DiffN approach in green, and both approaches in black. Only lipid species in the cells were used in the peak picking process, with m/z values of the internal standards omitted from the selection.
Conclusion
A new method is described for selecting precursor ions for MS/MS characterization, the DiffN DDA approach. To demonstrate its utility, two samples were characterized with a dual nESI source coupled to DIMS-MS. Unlike the commonly used TopN approach, DiffN is not biased by absolute abundance but instead uses the differential abundance of species in the two samples to select MS/MS precursor ions undergoing the largest fold changes. A comparison of lipid species selected with the TopN and DiffN DDA approaches highlights that high abundance does not always correspond to a large fold change. Diff10 selected seven unique peaks with relatively low abundances, and all 10 peaks had fold changes > 2.0, while Top10 only selected three peaks with fold changes > 2.0. Differential abundance is important in biomarker discovery, where a change in one or more species indicates a cellular phenotype or disease state. While lipids were the biomolecule of choice for this work, the DiffN approach could be promising for many types of -omics analyses to identify biomarkers more efficiently.
Supplementary Material
Acknowledgments
This work was supported by funds provided by the National Institute of General Medical Sciences through Grant Award Number R35 GM128697.
Bruker Daltonics has licensed some UNC DIMS IP used in this work.
References
- (1).Hu C; van der Heijden R; Wang M; van der Greef J; Hankemeier T; Xu G. Analytical Strategies in Lipidomics and Applications in Disease Biomarker Discovery. Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences. September 15, 2009, pp 2836–2846. 10.1016/j.jchromb.2009.01.038. [DOI] [PubMed] [Google Scholar]
- (2).Cheng D; Jenner AM; Shui G; Cheong WF; Mitchell TW; Nealon JR; Kim WS; McCann H; Wenk MR; Halliday GM; Garner B. Lipid Pathway Alterations in Parkinson’s Disease Primary Visual Cortex. PLoS One 2011, 6 (2). 10.1371/journal.pone.0017299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Butler LM; Perone Y; Dehairs J; Lupien LE; de Laat V; Talebi A; Loda M; Kinlaw WB; Swinnen JV Lipids and Cancer: Emerging Roles in Pathogenesis, Diagnosis and Therapeutic Intervention. Adv. Drug Deliv. Rev 2020, 159, 245–293. 10.1016/j.addr.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Guijas C; Montenegro-Burke JR; Warth B; Spilker ME; Siuzdak G. Metabolomics Activity Screening for Identifying Metabolites That Modulate Phenotype. Nat. Biotechnol 2018, 36 (4), 316–320. 10.1038/nbt.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Dunn WB; Broadhurst DI; Atherton HJ; Goodacre R; Griffin JL Systems Level Studies of Mammalian Metabolomes: The Roles of Mass Spectrometry and Nuclear Magnetic Resonance Spectroscopy. Chem. Soc. Rev 2011, 40 (1), 387–426. 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- (6).Sobsey CA; Ibrahim S; Richard VR; Gaspar V; Mitsa G; Lacasse V; Zahedi RP; Batist G; Borchers CH Targeted and Untargeted Proteomics Approaches in Biomarker Development. Proteomics 2020, 20 (9), 1–15. 10.1002/pmic.201900029. [DOI] [PubMed] [Google Scholar]
- (7).Hilaire PB Saint; Rousseau K; Seyer A; Dechaumet S; Damont A; Junot C; Fenaille F. Comparative Evaluation of Data Dependent and Data Independent Acquisition Workflows Implemented on an Orbitrap Fusion for Untargeted Metabolomics. Metabolites 2020, 10 (4). 10.3390/metabo10040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kalli A; Smith GT; Sweredoski MJ; Hess S. Evaluation and Optimization of Mass Spectrometric Settings during Data-Dependent Acquisition Mode: Focus on LTQ-Orbitrap Mass Analyzers. J. Proteome Res 2013, 12 (7), 3071–3086. 10.1021/pr3011588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Koelmel JP; Kroeger NM; Gill EL; Ulmer CZ; Bowden JA; Patterson RE; Yost RA; Garrett TJ Expanding Lipidome Coverage Using LC-MS/MS Data-Dependent Acquisition with Automated Exclusion List Generation. J. Am. Soc. Mass Spectrom 2017, 28 (5), 908–917. 10.1007/s13361-017-1608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Rudomin EL; Carr SA; Jaffe JD Directed Sample Interrogation Utilizing an Accurate Mass Exclusion-Based Data-Dependent Acquisition Strategy (AMEx). J. Proteome Res 2009, 8 (6), 3154–3160. 10.1021/pr801017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kreimer S; Belov ME; Danielson WF; Levitsky LI; Gorshkov MV; Karger BL; Ivanov AR Advanced Precursor Ion Selection Algorithms for Increased Depth of Bottom-Up Proteomic Profiling. J. Proteome Res 2016, 15 (10), 3563–3573. 10.1021/acs.jproteome.6b00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lintonen TPII; Baker PRSS; Suoniemi M; Ubhi BK; Koistinen KM; Duchoslav E; Campbell JL; Ekroos K. Differential Mobility Spectrometry-Driven Shotgun Lipidomics. Anal. Chem 2014, 86 (19), 9662–9669. 10.1021/ac5021744. [DOI] [PubMed] [Google Scholar]
- (13).Han X; Gross RW Shotgun Lipidomics: Electrospray Ionization Mass Spectrometric Analysis and Quantitation of Cellular Lipidomes Directly from Crude Extracts of Biological Samples. Mass Spectrom. Rev 2005, 24 (3), 367–412. 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- (14).Schwudke D; Liebisch G; Herzog R; Schmitz G; Shevchenko A. Shotgun Lipidomics by Tandem Mass Spectrometry under Data-Dependent Acquisition Control. Methods in Enzymology. 2007, pp 175–191. 10.1016/S0076-6879(07)33010-3. [DOI] [PubMed] [Google Scholar]
- (15).Schwudke D; Oegema J; Burton L; Entchev E; Hannich JT; Ejsing CS; Kurzchalia T; Shevchenko A. Lipid Profiling by Multiple Precursor and Neutral Loss Scanning Driven by the Data-Dependent Acquisition. Anal. Chem 2006, 78 (2), 585–595. 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- (16).Nazari M; Muddiman DC Enhanced Lipidome Coverage in Shotgun Analyses by Using Gas-Phase Fractionation. J. Am. Soc. Mass Spectrom 2016, 27 (11), 1735–1744. 10.1007/s13361-016-1446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Polley MYC; Freidlin B; Korn EL; Conley BA; Abrams JS; McShane LM Statistical and Practical Considerations for Clinical Evaluation of Predictive Biomarkers. J. Natl. Cancer Inst 2013, 105 (22), 1677–1683. 10.1093/jnci/djt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Parker CE; Borchers CH Mass Spectrometry Based Biomarker Discovery, Verification, and Validation - Quality Assurance and Control of Protein Biomarker Assays. Mol. Oncol 2014, 8 (4), 840–858. 10.1016/j.molonc.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Grissa D; Pétéra M; Brandolini M; Napoli A; Comte B; Pujos-Guillot E. Feature Selection Methods for Early Predictive Biomarker Discovery Using Untargeted Metabolomic Data. Front. Mol. Biosci 2016, 3 (JUL), 1–15. 10.3389/fmolb.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Pakiet A; Kobiela J; Stepnowski P; Sledzinski T; Mika A. Changes in Lipids Composition and Metabolism in Colorectal Cancer: A Review. Lipids Health Dis. 2019, 18 (1), 1–21. 10.1186/s12944-019-0977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Deng J; Yang Y; Zeng Z; Xiao X; Li J; Luan T. Discovery of Potential Lipid Biomarkers for Human Colorectal Cancer by In-Capillary Extraction Nanoelectrospray Ionization Mass Spectrometry. Anal. Chem 2021, 93 (38), 13089–13098. 10.1021/acs.analchem.1c03249. [DOI] [PubMed] [Google Scholar]
- (22).Coleman O; Ecker M; Haller D. Dysregulated Lipid Metabolism in Colorectal Cancer. Curr. Opin. Gastroenterol 2022, 38 (2), 162–167. 10.1097/mog.0000000000000811. [DOI] [PubMed] [Google Scholar]
- (23).Silva CL; Perestrelo R; Sousa-Ferreira I; Capelinha F; Câmara JS; Petković M. Lipid Biosignature of Breast Cancer Tissues by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Breast Cancer Res. Treat 2020, 182 (1), 9–19. 10.1007/s10549-020-05672-9. [DOI] [PubMed] [Google Scholar]
- (24).Eberlin LS; Ferreira CR; Dill AL; Ifa DR; Cooks RG Desorption Electrospray Ionization Mass Spectrometry for Lipid Characterization and Biological Tissue Imaging. Biochim. Biophys. Acta 2011, 1811 (11), 946–960. 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chen X; Sun M; Yang Z. Single Cell Mass Spectrometry Analysis of Drug-Resistant Cancer Cells: Metabolomics Studies of Synergetic Effect of Combinational Treatment. Anal. Chim. Acta 2022, 1201, 339621. 10.1016/j.aca.2022.339621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kopecka J; Trouillas P; Gašparović AČ; Gazzano E; Assaraf YG; Riganti C. Phospholipids and Cholesterol: Inducers of Cancer Multidrug Resistance and Therapeutic Targets. Drug Resist. Updat 2020, 49 (February). 10.1016/j.drup.2019.100670. [DOI] [PubMed] [Google Scholar]
- (27).Nazari M; Muddiman DC Polarity Switching Mass Spectrometry Imaging of Healthy and Cancerous Hen Ovarian Tissue Sections by Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI). Analyst 2016, 141 (2), 595–605. 10.1039/c5an01513h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Keating JE; Glish GL Dual Emitter Nano-Electrospray Ionization Coupled to Differential Ion Mobility Spectrometry-Mass Spectrometry for Shotgun Lipidomics. Anal. Chem 2018, 90 (15), 9117–9124. 10.1021/acs.analchem.8b01528. [DOI] [PubMed] [Google Scholar]
- (29).Bushey JM; Kaplan DA; Danell RM; Glish GL; Kaplany DA; Daneil RM; Glish GL Pulsed Nano-Electrospray Ionization: Characterization of Temporal Response and Implementation with a Flared Inlet Capillary. Instrum. Sci. Technol 2009, 37 (3), 257–273. 10.1080/10739140902831313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Dharmasiri U; Isenberg SL; Glish GL; Armistead PM; Comprehensive L; Carolina N; Drive W; Hill C. Differential Ion Mobility Spectrometry Coupled to Tandem Mass Spectrometry Enables Targeted Leukemia Antigen Detection. J. Proteome Res 2014, 13 (10), 4356–4362. 10.1021/pr500527c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Matyash V; Liebisch G; Kurzchalia TV; Shevchenko A; Schwudke D. Lipid Extraction by Methyl-Terf-Butyl Ether for High-Throughput Lipidomics. In Journal of Lipid Research; 2008; Vol. 49, pp 1137–1146. 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Fhaner CJ; Liu S; Ji H; Simpson RJ; Reid GE Comprehensive Lipidome Profiling of Isogenic Primary and Metastatic Colon Adenocarcinoma Cell Lines. Anal. Chem 2012, 84 (21), 8917–8926. 10.1021/ac302154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Rombouts C; De Spiegeleer M; Van Meulebroek L; Vanhaecke L; De Vos WH Comprehensive Polar Metabolomics and Lipidomics Profiling Discriminates the Transformed from the Non-Transformed State in Colon Tissue and Cell Lines. Sci. Rep 2021, 11 (1), 1–12. 10.1038/s41598-021-96252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bowman AP; Abzalimov RR; Shvartsburg AA Broad Separation of Isomeric Lipids by High-Resolution Differential Ion Mobility Spectrometry with Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom 2017, 28 (8), 1552–1561. 10.1007/s13361-017-1675-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.