Abstract
Cementum, a thin layer of mineralized tissue covering tooth root surface, is recognized as the golden standard in periodontal regeneration. However, current efforts mainly focus on alveolar bone regeneration rather than cementum regeneration, and rarely take Porphyromonas gingivalis (Pg), the keystone pathogen responsible for periodontal tissue destruction, into consideration. Though M2 macrophage-derived exosomes (M2-EXO) show promise in tissue regeneration, the exosome-producing M2 macrophages are induced by exogenous cytokines with transitory and unstable effects, restricting the regeneration potential of M2-EXO. Here, exosomes derived from genetically engineered M2-like macrophages are constructed by silencing of casein kinase 2 interacting protein-1 (Ckip-1), a versatile player involved in various biological processes. Ckip-1 silencing is proved to be an effective gene regulation strategy to obtain permanent M2-like macrophages with mineralization-promoting effect. Further, exosomes derived from Ckip-1-silenced macrophages (sh-Ckip-1-EXO) rescue Pg-suppressed cementoblast mineralization and cementogenesis. Mechanismly, sh-Ckip-1-EXO delivers Let-7f-5p targeting and silencing Ckip-1, a negative regulator also for cementum formation and cementoblast mineralization. More deeply, downregulation of Ckip-1 in cementoblasts by exosomal Let-7f-5p activates PGC-1α-dependent mitochondrial biogenesis. In all, this study provides a new strategy of genetically engineered M2-like macrophage-derived exosomes for cementum regeneration under Pg-dominated inflammation.
Keywords: Casein kinase 2 interacting protein-1, M2 macrophage, Exosome, Cementum regeneration, Porphyromonas gingivalis
Graphical abstract
Highlights
-
•
Genetically engineered sh-Ckip-1-EXO is designed based on M2-EXO.
-
•
sh-Ckip-1-EXO delivers Let-7f-5p targeting and silencing Ckip-1 in cementoblasts.
-
•
PGC-1α-dependent mitochondrial biogenesis is activated in Ckip-1-silenced cementoblasts.
-
•
sh-Ckip-1-EXO benefits P. gingivalis-suppressed cementoblast mineralization.
1. Introduction
Periodontitis is a chronic inflammatory disease characterized by the collapse of periodontal supporting tissues, which eventually leads to tooth loss [1]. At present, nearly one billion people suffer from periodontitis worldwide, in which, severe periodontitis has become the sixth most prevalent disease in the world [2]. Among the supporting tissues, cementum is a thin layer of mineralized tissue that covers the surface of tooth root, and anchors the tooth in the alveolar socket by embedded fibers [3]. Given the critical role of cementum in maintaining the tooth stability and health, cementum regeneration is considered the golden standard of periodontal regeneration [4]. However, most of present studies mainly focused on alveolar bone regeneration rather than the cementum regeneration. On the other hand, the occurrence and development of periodontitis are closely related to a variety of periodontal pathogens, in which, Porphyromonas gingivalis (P. gingivalis; Pg) is regarded as the keystone pathogen for periodontal destruction [5]. Therefore, improving cementoblast mineralization, the main cells responsible for cementum formation [3], under Pg-induced inflammation, is essential for cementum and further the periodontal regeneration.
Casein kinase 2 interacting protein-1 (Ckip-1) is a scaffold protein with multiple binding domains to regulate different cell functions, biological processes and diseases [6,7]. Our previous work demonstrated that Ckip-1 could negatively regulate cementoblast mineralization and cementum formation [8]. In addition, Ckip-1 was also reported to be a molecular switch during macrophage polarization. Briefly, Ckip-1 silencing restricted M1 macrophage polarization and enlarged M2 macrophage polarization under the induction with certain cytokines [9]. However, the regulatory effects of Ckip-1 on macrophage polarization needs further clarification.
Macrophages are the major cells in the immune system with the plasticity polarizing into pro-inflammatory M1 phenotype and anti-inflammatory M2 phenotype under certain conditions [10]. M2 macrophages were demonstrated to promote a variety of tissue regeneration, including periodontal bone regeneration [11,12]. Further, our previous work also verified the positive effect of M2 macrophages on cementoblast mineralization [13]. However, whether Ckip-1-silenced macrophages share the similarities with M2 macrophages in facilitating cementoblast mineralization remain unclear.
Exosomes are extracellular vesicles secreted by cells with diameters ranging from 30 to 160 nm. They contain biomolecules such as miRNAs, and act as mediators for intercellular communication [14]. Relevant studies have confirmed that M2 macrophage-derived exosomes (M2-EXO) mediated M2 macrophage-promoted tissue regeneration, and could be applied for muscle regeneration [15], vascular regeneration [16], and bone regeneration [17]. Nevertheless, these exosome-producing M2 macrophages were usually induced by exogenous IL-4/IL-13 with transitory and unstable effects [10]. Studies have revealed that the gene modification method by transfection were beneficial to manufacturing effective, stable and sustainable exosomes rich in endogenous functional target molecules [18,19]. Also, exosomes derived from genetically modified macrophages that share characteristics with M2-EXO may be promising for cementoblast mineralization and cementum regeneration.
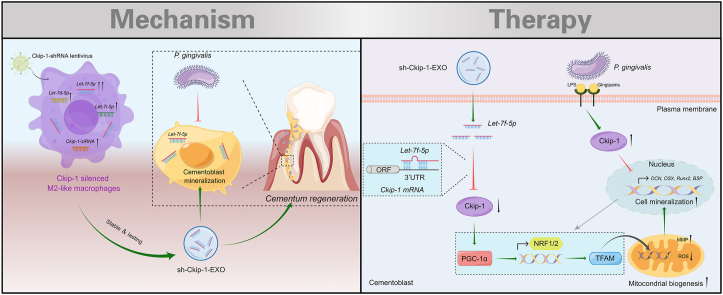
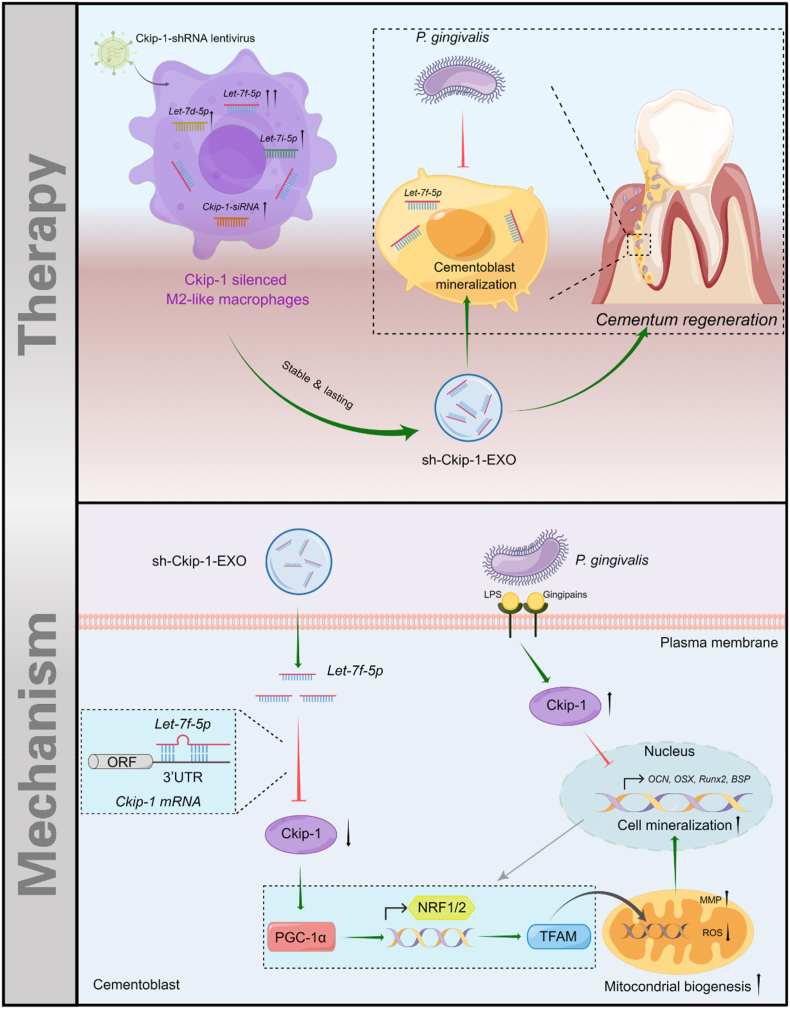
In this study, we found Ckip-1 silencing by lentivirus transfection could directly promote M2 macrophage polarization and induce the transformation of Pg-stimulated macrophages from M1 to M2 phenotype. Based on it, exosomes derived from Ckip-1-silenced M2-like macrophages (sh-Ckip-1-EXO) was applied to rescue Pg-suppressed cementoblast mineralization. In terms of mechanism, this gene-modified M2-like macrophages transferred exosomal Let-7f-5p to cementoblasts, and then accelerated cementoblast mineralization by targeting Ckip-1 to activate the peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α)-dependent mitochondrial biogenesis (Scheme 1). In all, this study may provide a new strategy for Pg-suppressed cementum regeneration.
Scheme 1.
Genetically engineered M2 macrophage-derived exosomes for Pg-suppressed cementum regeneration. Inspired by M2 macrophage-derived exosome (M2-EXO)-based tissue regeneration, exosomes derived from Ckip-1-silenced M2-like macrophages (sh-Ckip-1-EXO) was constructed and demonstrated to show therapeutic effect on Pg-suppressed cementum regeneration. Mechanismly, sh-Ckip-1-EXO delivered Let-7f-5p that accelerated cementoblast mineralization by targeting Ckip-1 to activate the PGC-1α-dependent mitochondrial biogenesis, which may provide a strategy for cementum regeneration, and further the whole periodontal regeneration under Pg-dominated inflammation. P. gingivalis, Pg, Porphyromonas gingivalis; LPS, lipopolysaccharide; ORF, open reading frame; 3′UTR, 3′ untranslated regions; MMP, mitochondrial membrane potential; ROS, reactive oxygen species.
2. Materials and methods
2.1. Cell culture
OCCM‐30 (osteocalcin-T-antigen transgenic mice first mandibular molar-derived immortalized cementoblasts-30; gifted and authorized by Dr. M.J. Somerman; cells from passage 15–20 were used) and RAW264.7 cell lines were maintained in high-glucose Dulbecco's modified Eagle medium (DMEM; Hyclone) supplemented with 10 % fetal bovine serum (FBS; Every Green) in a humidified atmosphere with 5 % CO2 at 37 °C. Induction medium for OCCM-30 mineralization was prepared with DMEM, 10 % FBS, 50 μg/mL vitamin C, 10 mM sodium β-glycerophosphate and 10 nM dexamethasone (Sigma).
2.2. Bacteria culture
ATCC 33277 P. gingivalis (Pg) was maintained in trypticase soy broth medium containing 1 μg/mL vitamin K1, 5 μg/mL hemin and 0.1 % yeast extract (Sigma) in an anaerobic atmosphere with 80 % N2, 10 % H2 and 10 % CO2 at 37 °C. Pg concentration was determined in line with the rule that one optical density (OD) at 600 nm is equal to 109 Pg/mL. RAW264.7 or OCCM‐30 was stimulated with different multiplicity of infection (MOIs) of Pg by the direct coculture method.
2.3. Lentiviral transfection
RAW264.7 cells (4 × 105) and OCCM-30 cells (2 × 105) were seeded in 6-well plates and cultured for 24 h. Lentiviruses (GenePharma) containing a Ckip-1-specific short hairpin RNA (sh-Ckip-1; sense, 5′-GTGACTATGAGAAGTGCGA-3′) and the control (sh-NC; sense, 5′-TTCTCCGAACGTGTCACGT-3′) were constructed to silence Ckip-1 in RAW264.7 cells and OCCM-30 cells at a MOI of 50:1 with 5 ng/mL polybrene (GenePharma) for 24 h. Then, the transfection medium was discarded. Transfected cells were normally cultured with complete medium with puromycin (2 μg/mL) for about 5 d. The successfully transfected green fluorescent protein (GFP)-expressing cells were observed by a fluorescence microscope. Finally, the transfected cells were identified by detecting the mRNA and protein expression of Ckip-1 in cells.
2.4. Macrophage induction
6 × 105 RAW264.7 cells were seeded in 6-well plates and cultured for 24 h. The resting RAW264.7 cells were regarded as the M0 macrophages. M1 macrophage polarization was induced by LPS (E. coli; 100 ng/mL; PeproTech) plus IFN-γ (20 ng/mL; PeproTech) for 24 h. M2 macrophage polarization was induced by IL-4 (20 ng/mL; PeproTech) for 24 h. Different macrophages were also induced with Pg at a MOI of 100 or Pg-LPS (1 μg/mL; InvivoGen) for 24 h.
2.5. Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR)
500 ng RNA extracted from cells or exosomes by TRIzol (TaKaRa) was reverse transcribed into cDNA by PrimeScript RT Reagent (TaKaRa) or miRNA 1st Strand cDNA Synthesis Kit (Vazyme) with stem-loop primers (RiboBio) for Let-7d-5p, Let-7f-5p, Let-7i-5p, Ckip-1-siRNA (si-Ckip-1), U6 following the manufacturers' instructions. SYBR qPCR Master Mix (Vazyme) or miRNA Universal SYBR qPCR Master Mix (Vazyme), specific primers for mRNA (Sangon) or miRNA (RiboBio), cDNA were applied to perform qPCR by the Applied Biosystems QuantStudio 6 with the following thermocycling conditions (mRNA: 95 °C for 30 s; 40 cycles at 95 °C for 10 s, 62 °C for 34 s; 72 °C for 30 s miRNA: 95 °C for 300 s; 40 cycles at 95 °C for 10 s, 60 °C for 30 s; 95 °C for 15 s, 60 °C for 60 s, 95 °C for 15 s). Values were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6, and quantified by the 2−ΔΔCt method. Primers for Ckip-1, IL-1β, IL-6, inducible nitric oxide synthase (iNOS), prostaglandin E Synthase (Ptges), prostaglandin endoperoxide Synthase 2 (Ptgs2), CD206, transforming growth factor-β1 (TGF-β1), IL-10, osteocalcin (OCN), Osterix (OSX), runt-related transcription factor 2 (Runx2), bone sialoprotein (BSP), PGC-1α, nuclear respiratory factor 1 (NRF1), nuclear respiratory factor 2 (NRF2), mitochondrial transcription factor A (TFAM) and GADPH were listed in Table S1. Sequences for Let-7d-5p, Let-7f-5p, Let-7i-5p, si-Ckip-1 were listed in Table S2.
2.6. Western blotting
Proteins were extracted by the M-PER protein lysis buffer (Thermo Scientific), collected by centrifugation and quantified by a BCA kit (Beyotime) according to the manufacturers' instructions. 20 μg protein from each group was added to 10 % or 12 % SDS-PAGE for electrophoresis, and transferred onto methanol-activated membranes (Millipore). The membranes were then blocked with 5 % nonfat milk for 1.5 h at room temperature (RT), and incubated with the following primary antibodies at 4 °C overnight: anti-IL-1β (1:1000; ABclonal), anti-ARG-1 (1:500; ABclonal), anti-IL-6 (1:1000; Proteintech), anti-CD206 (1:1000; CST), anti-Ckip-1 (1:1000; Proteintech), anti-OCN (1:500; Santa Cruz), anti-BSP (1:1000; Affinity), anti-OSX (1:1000; Abcam), anti-Runx2 (1:1000; Abcam), anti-TSG-101 (1:1000; Abcam), anti-CD63 (1:1000; ABclonal), anti-HSP90 (1:1000; Proteintech), anti-Calnexin (1:1000; Abcam), anti-F4/80 (1:1000; CST), anti-PGC-1α (1:800; NOVUS), anti-NRF1 (1:1000; Abcam); anti-NRF2 (1:1000; Proteintech); anti-TFAM (1:1000; Proteintech); anti-β-actin (1:15000; Proteintech). After incubating with horseradish peroxidase (HRP)-conjugated goat anti-mouse (1:10000; Proteintech) or goat anti-rabbit (1:8000; Proteintech) secondary antibodies for 1 h at RT, bands were visualized with an Enhanced Chemiluminescence Detection Reagent (Advansta) by the Odyssey LI-COR scanner.
2.7. Alkaline phosphatase (ALP) staining
Normal or transfected OCCM-30 cells were seeded in 6-well plates at a density of 3 × 105/well, and induced with macrophages, exosomes, Pg or SR (SR18292, 10 μM; MCE) for 4 d. After PBS washing, 4 % paraformaldehyde (PFA; Servicebio) fixation at RT for 15 min. ALP staining was carried out by the BCIP/NBT ALP Color Development Kit (Beyotime) following the manufacturer's protocol. Images were taken, and quantified by Image J.
2.8. Conditional medium (CM)-based coculture and transwell coculture
1 × 106 macrophages (sh-NC or sh-Ckip-1) were seeded in 10 cm dishes and cultured for 24 h. The supernatants from the two macrophages were collected and filtered through 0.22 μm filters. To prepare CM-sh-NC and CM-sh-Ckip-1, the supernatants were mixed with an equal volume of induction medium. OCCM-30 cells were seeded in 6-well plates at a density of 3 × 105/well, and induced with CM-sh-NC or CM-sh-Ckip-1 for 2 d.
The effects of the two macrophages on OCCM-30 mineralization were also studied by coculturing sh-NC macrophages (Trans-sh-NC) or sh-Ckip-1 macrophages (Trans-sh-Ckip-1) with OCCM-30 cells in a 6-well-transwell coculture system (0.4 μm; Corning). Briefly, macrophages were seeded in the upper chamber at a density of 6 × 105/insert. OCCM-30 cells were seeded in the lower chamber at a density of 3 × 105/well, and induced with the macrophages for 2 d.
2.9. Exosome extraction, identification and quantification
6 × 105 normal macrophages were seeded in 10 cm dishes and cultured for 24 h. For the collection of M0 and M2 (20 ng/mL IL-4 for 1 d; with PBS washing) macrophage-derived supernatants, serum-free medium was added for another 24 h, and supernatants were collected. sh-NC and sh-Ckip-1 macrophage-derived supernatants were also collected after culturing with serum-free medium for 1 d. To extract M0 and M2 macrophage-derived exosomes (M0-EXO; M2-EXO), sh-NC and sh-Ckip-1 macrophage-derived exosomes (sh-NC-EXO; sh-Ckip-1-EXO), the above supernatants were subjected to centrifugation at 300g for 10 min; 2000 g for 10 min; 10000 g for 30 min. After 0.22 μm filtration, the supernatants were centrifugated at 120000 g for 90 min (XE-100; Beckman). The precipitation was resuspended with PBS, and again centrifugated at 120000 g for 90 min. The obtained exosomes were dissolved with 50–100 μL PBS according to the amount of precipitation.
For exosome identification, 20 μg exosomes resuspended with loading buffer were denatured at 95 °C for 10 min. Positive protein markers and negative protein marker of exosomes were detected by Western blotting. Also, the morphology of exosomes was determined by a transmission electron microscope (TEM; JEM-2100; JEOL). The sizes of exosomes were examined by dynamic light scattering (DLS) method with a nanometer particle potentiometer (Zetasizer Nano ZSP; Malvern). The concentration of exosomes dissolved in PBS was determined by the BCA kit (Beyotime) according to the manufacturer's instruction.
2.10. Exosome internalization
Dil-labeled exosomes were obtained by incubating 5 μg exosomes with Dil dye at 37 °C for 30 min, followed by ultracentrifugation as mentioned above to remove the excess dye. OCCM-30 cells were seeded in 24-well plates at a density of 5 × 104/well and cultured for 24 h. OCCM-30 cells were then incubated with the resuspended Dil-labeled exosomes at 37 °C for 12 h. After PBS washing, 4 % PFA (Servicebio) fixation at RT for 15 min and 0.5 % Triton X-100 (Biofroxx) permeabilization at RT for 15 min, cells were stained with Actin-Tracker Green-phalloidine (Beyotime) at RT for 30 min and incubated with Antifade Mounting Medium with 4,6-diamino-2-phenyl indole (DAPI) (Beyotime) at RT for 5 min. The internalization of M0-EXO, M2-EXO, sh-NC-EXO and sh-Ckip-1-EXO by OCCM-30 cells were observed by a fluorescence microscope.
2.11. Exosome stimulation
OCCM-30 cells were seeded in 6-well plates at a density of 3 × 105/well and cultured for 24 h. 2.5 μg/mL M0-EXO and M2-EXO, 500 ng/mL sh-NC-EXO and sh-Ckip-1-EXO were applied to induce cementoblast mineralization for 2, 4 d. OCCM-30 cells were also induced with 500 ng/mL sh-NC-EXO and sh-Ckip-1-EXO in the existence of Pg (MOI = 100). The effect of different exosomes on cementoblast mineralization, the rescue effect of sh-Ckip-1-EXO on Pg-suppressed cementoblast mineralization, and the effect of sh-Ckip-1-EXO on the expression of Ckip-1 in OCCM-30 cells were detected. In addition, the effect of sh-Ckip-1-EXO on Pg-induced OCCM-30 cell inflammation was detected.
OCCM-30 cells were seeded in 6-well plates at a density of 3 × 105/well and cultured for 24 h. 25 μg sh-NC-EXO and sh-Ckip-1-EXO were added to stimulate OCCM-30 cells for 1 d. The expression of different miRNAs in OCCM-30 cells after sh-NC-EXO and sh-Ckip-1-EXO stimulation was examined.
2.12. Animal experiments
1 × 107 OCCM-30 cells were loaded in 6 × 6 × 6 mm sterilized gelatin sponges (Ethicon) and induced for 4 days. Then, 7-week-old BALB/c nude male mice (22 ± 1 g) provided by the laboratory were anesthetized with pentobarbital sodium at a dose of 20 mg/kg. Pg (5 × 108) and exosomes (200 μg) absorbed by gelatin sponges were implanted subcutaneously into the dorsa of mice (CON, Pg, Pg+sh-NC-EXO, Pg+sh-Ckip-1-EXO groups were set symmetrically in each mouse, n = 6). Samples were harvested after 4 weeks, and then fixed with 4 % paraformaldehyde, decalcified, paraffin embedded, and sectioned at a 5 μm thickness. The sections were applied for hematoxylin and eosin (HE) and immunohistochemistry (IHC) staining. IHC was conducted with UltraSensitive SP IHC Kit (MAIXIN Biotech) following the manufacturer's instructions (anti-BSP, 1:200, Affinity; anti-OSX, 1:200, Abcam). Images were taken, and quantified by Image J. Animal experiments were approved by the Ethics Committee of the School of Medicine, Wuhan University (WP20230091).
2.13. Oligo transfer experiment
To determine whether the oligos including miRNAs in macrophages could be transferred to OCCM-30 cells, the oligo transfer experiment was performed. Briefly, macrophages were seeded in the upper chamber at a density of 6 × 105/insert. The adherent macrophages were transfected with FAM-oligo (5′-UUCUCCGAACGUGUCACGUTT-3’; 50 nM; GenePharma) by GP-transfect-Mate (GenePharma) for 24 h following the manufacturer's instruction. OCCM-30 cells were seeded in the lower chamber at a density of 2.8 × 105/well and cultured for 24 h. Then, OCCM-30 cells were cocultured with transfected macrophages in a transwell system (0.4 μm; Corning) for 24 h. The transfection effect and the transfer of FAM-oligo from macrophages to OCCM-30 cells were detected by a fluorescence microscope.
2.14. MiRNA sequencing
OCCM-30 cells induced for 0, 3, 14, 21 d were treated with TRIzol (TaKaRa), and sent to RiboBio for RNA extraction, purification, quality control, library construction and sequencing. The data were analyzed according to the time point. log2 |fold change| > 1 and FDR (false discovery rate) < 0.05 were the screening criteria both need to be satisfied for miRNAs with significance. The heatmap for the miRNA expression of Let-7 family was produced by TBtools (https://github.com/CJ-Chen/TBtools/releases).
2.15. Mimic/inhibitor transfection
OCCM-30 cells were seeded in 6-well plates at a density of 2.8 × 105/well. The next day, cells were transfected with mimic-NC/mimic (50 nM), inhibitor-NC/inhibitor (100 nM) of Let-7f-5p (RiboBio) by GP-transfect-Mate (GenePharma) for 24 h following the manufacturer's instruction, and induced for 2, 4 d. Then, the regulatory effect of Let-7f-5p on cementoblast mineralization, and also the expression of Ckip-1 in OCCM-30 cells were determined.
2.16. Immunofluorescence
OCCM-30 cells were seeded in 24-well plates at a density of 5 × 104/well and cultured for 24 h. Cells were transfected with mimic/inhibitor of Let-7f-5p for 24 h as mentioned above, or stimulated with 5 μg/mL sh-NC-EXO and sh-Ckip-1-EXO for 24 h. After PBS washing, 4 % PFA (Servicebio) fixation and 1 % Triton X-100 (Biofroxx) permeabilization at RT for 15 min each, cells were blocked with goat serum at RT for 1 h and incubated with anti-Ckip-1 antibody (1:200; Proteintech) at 4 °C overnight. The next day, cells were incubated with secondary antibody conjugated with Cy3 (1:200; ABclonal) for 1 h, and Antifade Mounting Medium with DAPI (Beyotime) for 5 min at RT. A fluorescence microscope was applied to observe the expression of Ckip-1 in OCCM-30 cells.
2.17. Dual-luciferase reporter assay
The targeting relationship between Let-7 family and Ckip-1 was predicted by TargetScan (https://www.targetscan.org/mamm_31/) and miRBase (https://www.mirbase.org/). To construct the luciferase reporter, a fragment located at position 76 to 82 of the Ckip-1 mRNA 3′-UTR containing Let-7f-5p targeting sites or mutant sites was cloned into pmirGLO plasmids (Miaoling). OCCM-30 cells were seeded in 12-well plates at a density of 1 × 105/well and cultured for 24 h. The plasmids and Mimic-NC/Mimic of Let-7f-5p were transfected into OCCM-30 cells with TurboFect (Thermo Scientific) for 24 h. After culturing for 2 d, activities of firefly and renilla luciferase were detected by a dual luciferase reporter assay kit (Vazyme Biotech Co., Ltd). Renilla luciferase activity was normalized to firefly luciferase activity.
2.18. mRNA sequencing
OCCM-30 cells induced for 0 d (CON; control) or 7 d (osteogenic induction medium; OIM) were treated with TRIzol (TaKaRa) for RNA extraction. Sample quality inspection, NGS, data processing and analysis were performed by ANOROAD (Beijing, China). A series of mineralization-related genes, Ckip-1 and PGC-1α were selected. The heatmap was produced by TBtools (https://github.com/CJ-Chen/TBtools/releases).
2.19. Mitochondrial membrane potential (MMP) detection
MitoTracker™ Red CMXRos (Invitrogen) was used to detect MMP according to the manufacturer's instruction. Briefly, Ckip-1-silenced OCCM-30 cells and control cells were seeded in 48-well plates at a density of 1.5 × 104/well and cultured for 24 h. After inducing with or without SR (10 μM; MCE) for 1 d, the cells were incubated with 200 nM working solution for 30 min in the dark. After PBS washing, 4 % PFA fixation and DAPI staining (Beyotime), MMP was observed by a fluorescent microscope.
2.20. Mitochondrial reactive oxygen species (ROS) detection
MitoSOX™ Red (YEASEN) was used to detect mitochondrial ROS according to the manufacturer's instruction. Ckip-1-silenced OCCM-30 cells and control cells were seeded in 48-well plates at a density of 1.5 × 104/well and cultured for 24 h. After inducing with or without SR (10 μM; MCE) for 1 d, the cells were incubated with 2.5 μM working solution for 15 min in the dark. After HBSS washing, 4 % PFA fixation and DAPI staining (Beyotime), ROS was observed by a fluorescent microscope.
2.21. Statistical analysis
Data was presented as the mean ± SD (standard deviation) of at least 3 independent experiments. Statistical analyses were performed by GraphPad Prism 8. Shapiro-Wilk test was used to determine the normal distribution. The Student's t-test was applied to evaluate the differences between two groups. One-way ANOVA was applied to evaluate the differences between multiple groups, followed by Bonferroni correction. Significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001. P > 0.05 was considered not significant (ns).
3. Results
3.1. M2 macrophage-derived exosomes (M2-EXO) facilitate cementoblast mineralization
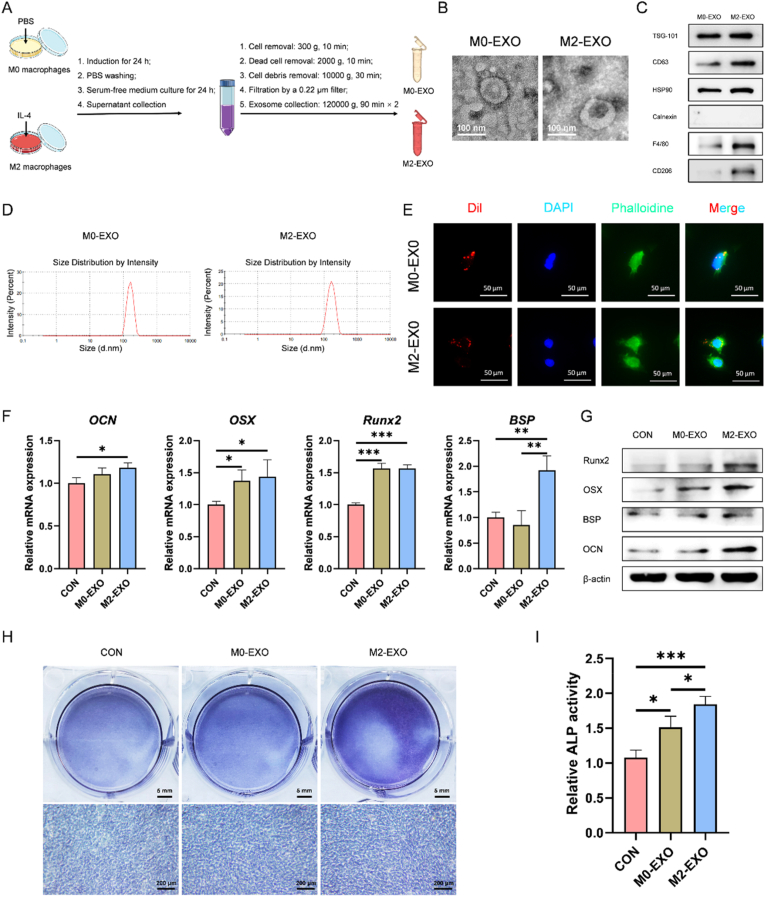
The effect of M2-EXO on cementoblast mineralization was first evaluated. The supernatants from M0 macrophages and M2 macrophages induced with IL-4 were collected, and subjected for gradient centrifugation to extract exosomes (Fig. 1A). The cup-like morphology, positive or negative protein markers and the nano-size about 140 nm of M0-EXO and M2-EXO were identified successfully by TEM (Fig. 1B), western blotting (Fig. 1C) and DLS (Fig. 1D), respectively. The internalization of the two Dil-labeled exosomes by cementoblasts was also verified (Fig. 1E). Cementoblasts were then induced with the M0-EXO and M2-EXO for different days. Cementoblasts with no exosome stimulation was set as the control (CON) group. The results of RT-qPCR, western blotting and ALP staining showed that the expression of mineralization-related markers on day 2 (Fig. 1F–G) and ALP activity of cementoblasts on day 4 (Fig. 1H–I) both increased significantly after exosome stimulation, in which, M2-EXO showed better mineralization-promoting effect. In all, M2-EXO facilitated cementoblast mineralization.
Fig. 1.
M2 macrophage-derived exosomes (M2-EXO) facilitate cementoblast mineralization. (A) Flow diagram for the extraction of M0 macrophage-derived exosomes (M0-EXO) and M2-EXO. Identification of M0-EXO and M2-EXO by transmission electron microscope (TEM) for cup-like morphology (B), western blotting for protein markers (C), and dynamic light scattering (DLS) for nano-size (D). (E) Internalization of Dil-labeled M0-EXO and Dil-labeled M2-EXO by cementoblasts after culturing for 12 h. Upregulated expression of OCN, OSX, Runx2 and BSP in cementoblasts induced with M2-EXO (2.5 μg/mL) for 2 d is demonstrated by RT-qPCR (F) and western blotting (G). (H–I) Increased alkaline phosphatase (ALP) activity of cementoblasts induced with M2-EXO (2.5 μg/mL) for 4 d is detected by ALP staining. *P < 0.05, **P < 0.01, ***P < 0.001.
3.2. Ckip-1 regulates M2 macrophage polarization negatively
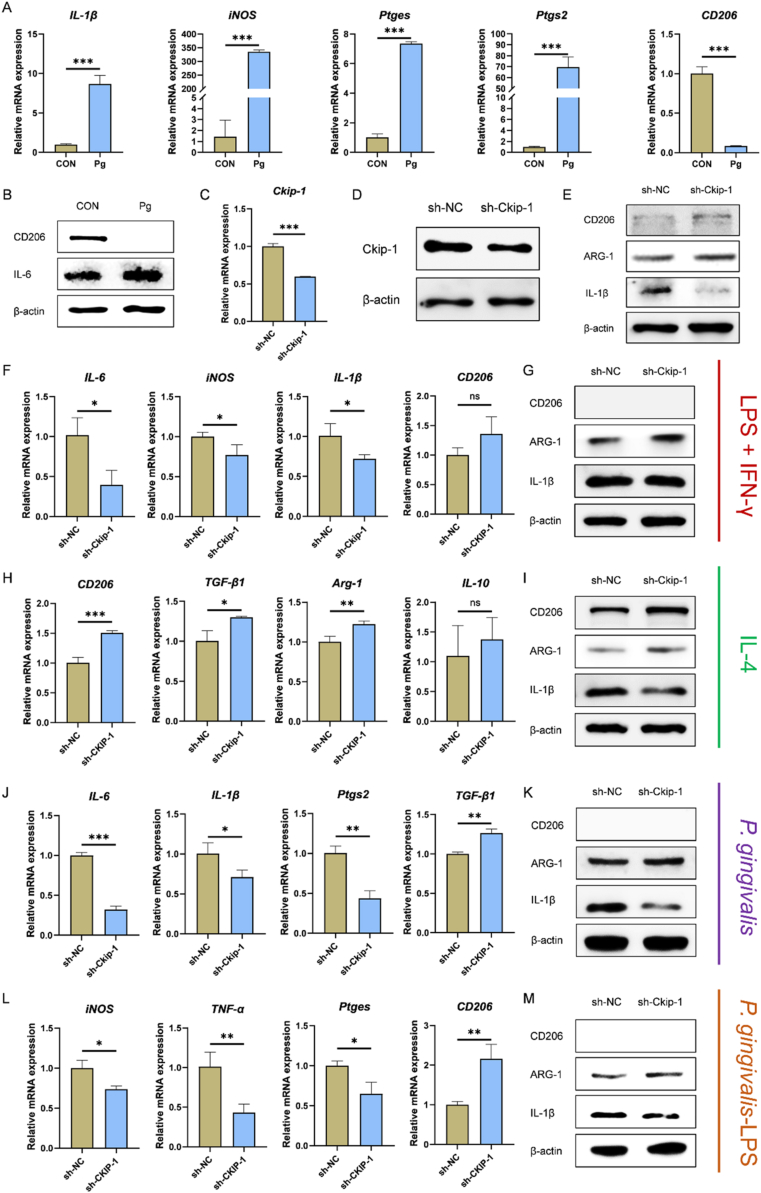
To study the effect of Pg on macrophage-represented immune microenvironment, macrophages were stimulated with Pg (MOI = 100) by the direct coculture method. The results of RT-qPCR and western blotting showed higher expression of IL-1β, iNOS, Ptges, Ptgs2, IL-6 and lower expression of CD206 in Pg group compared with CON group, indicating Pg induced M1 macrophage polarization (Fig. 2A–B). Then, Ckip-1-silenced macrophages were constructed by lentiviral transfection (Fig. S1), and identified successfully (Fig. 2C–D). By western blotting, it was demonstrated that Ckip-1 silencing could promote M2 macrophage polarization directly, with upregulated expression of CD206, ARG-1 and downregulated expression of IL-1β (Fig. 2E). We then determined the M2 macrophage polarization potential in sh-NC group and sh-Ckip-1 group under different conditions by RT-qPCR and western blotting. We found Ckip-1 silencing restricted M1 macrophage polarization induced by LPS (E. coli) + IFN-γ (Fig. 2F–G), and enlarged M2 macrophage polarization induced by IL-4 (Fig. 2H–I). Though the protein expression of CD206 was not detected under Pg (MOI = 100) stimulation, it was confirmed that Pg-induced macrophages were transferred from M1 phenotype to M2 phenotype when Ckip-1 was silenced, characterized by the decreased expression of IL-1β, Ptgs2, IL-6 and increased expression of ARG-1 (Fig. 2J–K). Similar trend was also demonstrated in Pg-LPS-treated macrophages (Fig. 2L-M). Taken together, Ckip-1 silencing regulated M2 macrophage polarization, and might be applied to reshape Pg-dominated immune microenvironment for cementum regeneration.
Fig. 2.
Ckip-1 silencing promotes M2 macrophage polarization. Increased expression of M1 macrophage polarization-related markers and decreased expression of M2 macrophage polarization-related marker CD206 in Pg (MOI = 100, 1 d)-stimulated macrophages are measured by RT-qPCR (A) and western blotting (B). Identification of Ckip-1-silenced macrophages by RT-qPCR (C) and western blotting (D). (E) Increased expression of CD206 and ARG-1 in Ckip-1-silenced macrophages is detected by western blotting. Decreased expression of M1 macrophage polarization-related markers and increased expression of ARG-1 in Ckip-1-silenced macrophages stimulated with LPS (E. coli-LPS; 100 ng/mL) plus IFN-γ (20 ng/mL) for 1 d is detected by RT-qPCR (F) and western blotting (G), respectively. Increased expression of M2 macrophage polarization-related markers in Ckip-1-silenced macrophages stimulated with IL-4 (20 ng/mL) for 1 d is detected by RT-qPCR (H) and western blotting (I). Decreased expression of M1 macrophage polarization-related markers and increased expression of M2 macrophage polarization-related markers in Ckip-1-silenced macrophages stimulated with Pg (MOI = 100) for 1 d is detected by RT-qPCR (J) and western blotting (K). Decreased expression of M1 macrophage polarization-related markers and increased expression of M2 macrophage polarization-related markers in Ckip-1-silenced macrophages stimulated with Pg-LPS (1 μg/mL) for 1 d is detected by RT-qPCR (L) and western blotting (M). *P < 0.05, **P < 0.01, ***P < 0.001. P > 0.05 was considered not significant (ns).
3.3. Ckip-1 silencing-induced M2-like macrophages promote cementoblast mineralization
Ckip-1 silencing-induced permanent M2-like macrophages were then obtained. The CM-based culture method was first used to study the effect of Ckip-1-silenced macrophages on cementoblast mineralization. The supernatants from Ckip-1-silenced macrophages and the control macrophages were collected for CM preparation (Fig. S2A). By RT-qPCR and western blotting, it was demonstrated that the expression of mineralization-related markers Runnx2, OSX, OCN, BSP were all increased after inducing with CM from Ckip-1-silenced macrophages (CM-sh-Ckip-1) for 2 d, compared with CM from the control macrophages (CM-sh-NC) (Figs. S2B–C). Also, ALP staining on day 4 verified the superior mineralization-promoting effect of the supernatant from Ckip-1-silenced macrophages (Figs. S2D–E).
The direct effect of Ckip-1-silenced macrophages on cementoblast mineralization was further studied by coculturing cementoblasts with the two macrophages in a transwell system (Fig. S3A). After induction, the mineralization capacity of cementoblasts in the lower chamber was examined. In consistent with the CM results, increased expression of mineralization-related markers (Figs. S3B–C) on day 2 and increased ALP activity on day 4 (Figs. S3D–E) were found in Trans-sh-Ckip-1 group when compared with the Trans-sh-NC group. Therefore, it was confirmed that Ckip-1 silencing-induced M2-like macrophages could promote cementoblast mineralization.
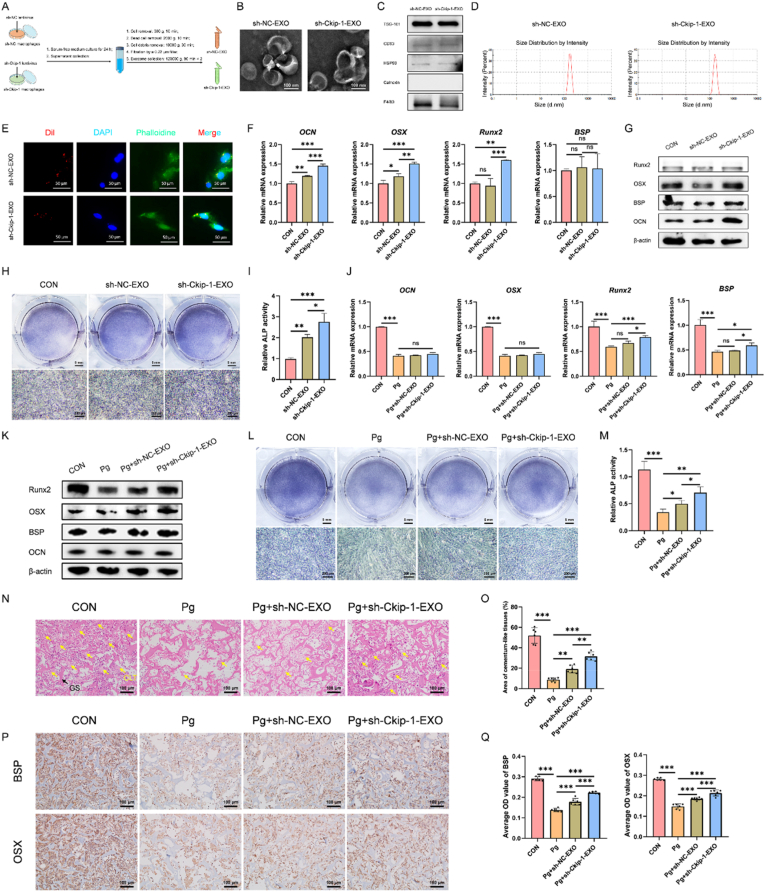
3.4. Ckip-1 silencing-induced M2-like macrophage-derived exosomes (sh-ckip-1-EXO) rescue pg-suppressed cementogenesis
Based on the above findings, it was speculated that sh-Ckip-1-EXO might mediate Ckip-1-silenced macrophage-facilitated cementoblast mineralization. Similarly, sh-Ckip-1-EXO and the control sh-NC-EXO were also extracted by gradient centrifugation (Fig. 3A), and identified successfully by TEM (Fig. 3B), western blotting (Fig. 3C) and DLS (Fig. 3D). The two Dil-labeled exosomes could also be uptaken by cementoblasts after culturing for 12 h (Fig. 3E). The effect of sh-Ckip-1-EXO on cementoblast mineralization was determined by RT-qPCR (Fig. 3F), western blotting (Fig. 3G) on day 2, and ALP staining (Fig. 3H–I) on day 4. All the results demonstrated the mineralization-accelerating effect of the two exosomes, in which, sh-Ckip-1-EXO exhibited a superior effect.
Fig. 3.
Ckip-1-silenced M2-like macrophage-derived exosomes (sh-Ckip-1-EXO) rescue Pg-suppressed cementogenesis. (A) Flow diagram for the extraction of sh-NC macrophage-derived exosomes (sh-NC-EXO) and sh-Ckip-1-EXO. Identification of sh-NC-EXO and sh-Ckip-1-EXO by TEM for cup-like morphology (B), western blotting for protein markers (C), and DLS for nano-size (D). (E) Internalization of Dil-labeled sh-NC-EXO and Dil-labeled sh-Ckip-1-EXO by cementoblasts after culturing for 12 h. Upregulated expression of OCN, OSX, Runx2 and BSP in cementoblasts induced with sh-Ckip-1-EXO (500 ng/mL) for 2 d is demonstrated by RT-qPCR (F) and western blotting (G). (H–I) Increased ALP activity of cementoblasts induced with sh-Ckip-1-EXO (500 ng/mL) for 4 d is detected by ALP staining. Suppressed expression of OCN, OSX, Runx2 and BSP in Pg (MOI = 100)-stimulated cementoblasts induced for 2 d is rescued by sh-Ckip-1-EXO (500 ng/mL), detected by RT-qPCR (J) and western blotting (K). (L–M) Suppressed ALP activity of Pg (MOI = 100)-stimulated cementoblasts induced for 4 d is rescued by sh-Ckip-1-EXO (500 ng/mL), detected by ALP staining. Cementoblast-loaded gelatin sponges (GS) with different stimulations are implanted subcutaneously into the dorsa of nude mice for 4 weeks. Suppressed cementum-like tissue (CLT) formation in Pg-infected (5 × 108) group is rescued by sh-Ckip-1-EXO (200 μg), detected by HE staining (N–O). Suppressed cementogenesis in Pg-infected (5 × 108) group is rescued by sh-Ckip-1-EXO (200 μg), detected by immunohistochemistry (P–Q). Yellow arrow, CLT; *P < 0.05, **P < 0.01, ***P < 0.001. P > 0.05 was considered not significant (ns).
Cementoblasts were also induced with sh-NC-EXO and sh-Ckip-1-EXO in the existence of Pg (MOI = 100), and cementoblasts with no exosome stimulation was set as the CON group. The results of RT-qPCR (Fig. 3J), western blotting (Fig. 3K) on day 2 and ALP staining (Fig. 3L-M) on day 4 showed that the expression of mineralization-related markers and ALP activity were decreased after Pg stimulation. However, when the two exosomes were added, the suppressed mineralization was rescued, in which, the rescuing effect of sh-Ckip-1-EXO was more powerful. Moreover, sh-Ckip-1-EXO was found to alleviate Pg-induced cementoblast inflammation (Fig. S4).
Further, ectopic cementogenesis model in nude mice was also carried out to evaluate the therapeutic effect of sh-Ckip-1-EXO on Pg-suppressed cementogenesis. Pre-induced-cementoblast-loaded gelatin sponges (GS) with Pg (5 × 108) or Pg plus exosomes (200 μg) were implanted subcutaneously into the dorsa of nude mice for 4 weeks. HE staining results showed that the suppressed cementum-like tissue (CLT) formation in Pg-infected group could be rescued by sh-NC-EXO and sh-Ckip-1-EXO, in which, the rescuing effect of sh-Ckip-1-EXO was better (Fig. 3N-O). Immunohistochemistry results also confirmed the better rescuing effect of sh-Ckip-1-EXO on Pg-suppressed cementogenesis (Fig. 3P-Q). Taken together, sh-Ckip-1-EXO might be implicated in Ckip-1-silenced macrophage-facilitated cementum regeneration, which could be applied to rescue Pg-suppressed cementum regeneration.
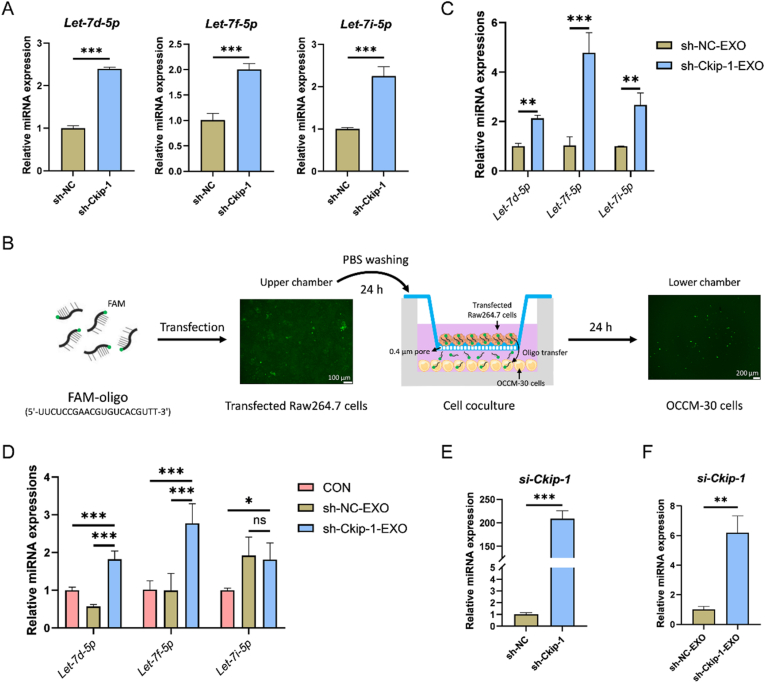
3.5. sh-ckip-1-EXO facilitates cementoblast mineralization by delivering Let-7f-5p
To study the mechanism underlying sh-Ckip-1-EXO-promoted cementoblast mineralization, the inside Let-7 miRNAs were studied. It was found that Let-7d-5p, Let-7f-5p and Let-7i-5p all significantly increased in sh-Ckip-1 macrophages when compared with sh-NC macrophages by RT-qPCR (Fig. 4A). Further, the FAM-oligo transfer experiment showed that the green fluorescence was transferred from the FAM-oligo-transfected macrophages to cementoblasts in a transwell coculture system (Fig. 4B), indicating that miRNAs in macrophages might also be packaged into exosomes to be transferred to the target cells. Then, by RT-qPCR, the 3 miRNAs increased in macrophages were also found to be abundant in sh-Ckip-1-EXO compared with sh-NC-EXO, in which, Let-7f-5p was the most packaged (Fig. 4C). As expected, RT-qPCR showed that the expression of Let-7f-5p in cementoblasts was upregulated mostly after sh-Ckip-1-EXO stimulation (Fig. 4D). Interestingly, higher expression of si-Ckip-1 in Ckip-1-silenced macrophages (Fig. 4E) and the exosomes derived from them (Fig. 4F) was also found. However, the content of si-Ckip-1 in cementoblasts was not significantly increased after exosome stimulation (Fig. S5).
Fig. 4.
sh-Ckip-1-EXO delivers Let-7f-5p to cementoblasts. (A) Increased expression of Let-7d-5p, Let-7f-5p and Let-7i-5p in Ckip-1-silenced macrophages is demonstrated by RT-qPCR. (B) Green fluorescence is observed to be transferred from FAM-oligo-transfected macrophages to cementoblasts in a transwell coculture system for 1 d. (C) The expression of Let-7f-5p increases mostly in sh-Ckip-1-EXO, detected by RT-qPCR. (D) The expression of Let-7f-5p increases mostly in cementoblasts stimulated with sh-Ckip-1-EXO (25 μg) for 1 d, detected by RT-qPCR. (E) Increased expression of si-Ckip-1 in Ckip-1-silenced macrophages is demonstrated by RT-qPCR. (F) Increased expression of si-Ckip-1 in sh-Ckip-1-EXO is confirmed by RT-qPCR. *P < 0.05, **P < 0.01, ***P < 0.001. P > 0.05 was considered not significant (ns).
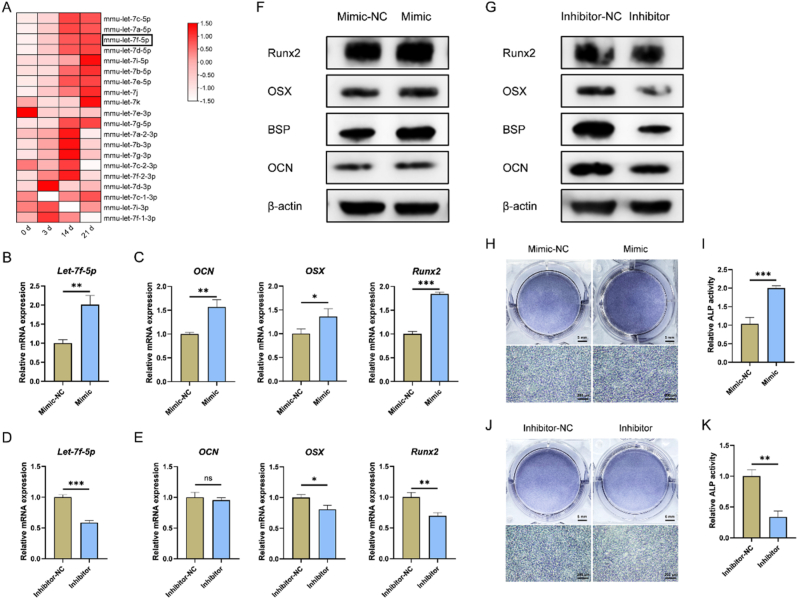
Next, the regulatory effect of Let-7f-5p on cementoblast mineralization was studied. MiRNA sequencing results showed that most of the miRNAs of Let-7 family increased gradually during cementoblast mineralization, including Let-7f-5p (Fig. 5A). Then, cementoblasts were transfected with the mimic or inhibitor of Let-7f-5p, and induced for 2 and 4 d. By RT-qPCR, western blotting and ALP staining, increased expression of mineralization-related markers (Fig. 5B–C, F) and ALP activity (Fig. 5H–I) of cementoblasts were found in Mimic group compared with Mimic-NC group. A contrary trend was demonstrated when Let-7f-5p was knocked down by the inhibitor (Fig. 5D–E, G, J-K). In all, sh-Ckip-1-EXO could deliver Let-7f-5p to promote cementoblast mineralization.
Fig. 5.
Let-7f-5p accelerates cementoblast mineralization. (A) Let-7 family miRNAs increase gradually during cementoblast mineralization, detected by miRNA sequencing of cementoblast induced for 0, 3, 14 and 21 d. Cementoblasts are transfected with Let-7f-5p mimic for 1 d, and induced for 2 d. Increased expression of Let-7f-5p (B), OCN, OSX and Runx2 (C) in the mimic group is detected by RT-qPCR. Cementoblasts are transfected with Let-7f-5p inhibitor for 1 d, and induced for 2 d. Decreased expression of Let-7f-5p (D), OSX, Runx2 (E) in the inhibitor group is detected by RT-qPCR. (F) Increased protein expression of OCN, BSP, OSX and Runx2 in mimic-transfected cementoblasts induced for 2 d by western blotting. (G) Decreased protein expression of OCN, BSP, OSX and Runx2 in inhibitor-transfected cementoblasts induced for 2 d by western blotting. (H–I) Enhanced ALP activity in mimic-transfected cementoblasts induced for 4 d by ALP staining. (J–K) Suppressed ALP activity in inhibitor-transfected cementoblasts induced for 4 d by ALP staining. *P < 0.05, **P < 0.01, ***P < 0.001. P > 0.05 was considered not significant (ns).
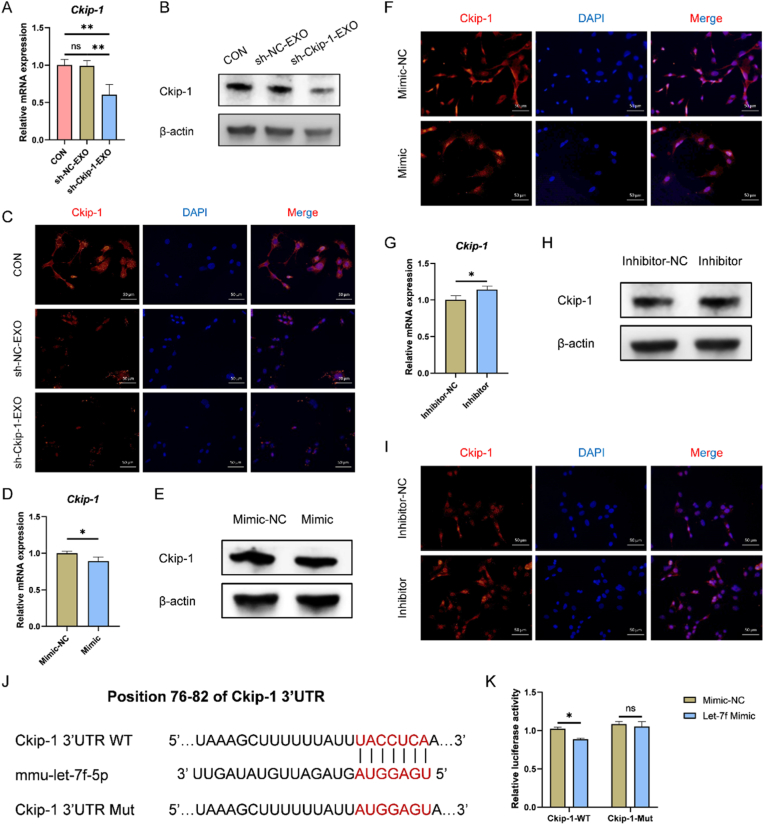
3.6. Let-7f-5p enriched in sh-ckip-1-EXO facilitates cementoblast mineralization by targeting Ckip-1
Inspired by the fact that sh-Ckip-1-EXO could also deliver si-Ckip-1 to cementoblasts, it was then studied that whether Ckip-1, a negative regulator for cementum formation demonstrated by our previous work [8], was downregulated after exosome stimulation. Cementoblasts were induced with sh-NC-EXO and sh-Ckip-1-EXO, or without exosome stimulation, for 1 and 2 d. The 2 d-induction results from RT-qPCR (Fig. 6A), western blotting (Fig. 6B), and the 1 d-incubation results from immunofluorescence (Fig. 6C) all confirmed our supposition that sh-Ckip-1-EXO, whether at a high (5 μg/mL) or low (500 ng/mL) concentration, could both markedly silence the expression of Ckip-1 in cementoblasts. On the other hand, the targeting relationship between Let-7f-5p and Ckip-1 was predicted by TargetScan (Fig. S6A) and miRBase (Fig. S6B). Therefore, whether Let-7f-5p enriched in sh-Ckip-1-EXO could function the Ckip-1 silencing effect was then focused on. It was proved that the expression of Ckip-1 in cementoblasts was downregulated when Let-7f-5p was overexpressed (Fig. 6D–F), whereas upregulated when Let-7f-5p was knocked down (Fig. 6G–I). In addition, Let-7f-5p was demonstrated to target the 3′UTR of Ckip-1 mRNA by a dual-luciferase reporter assay (Fig. 6J–K). Taken together, sh-Ckip-1-EXO delivered Let-7f-5p which targets Ckip-1 to promote cementoblast mineralization.
Fig. 6.
Let-7f-5p targets and silences Ckip-1, a negative regulator for cementum formation and cementoblast mineralization, in cementoblasts. Decreased expression of Ckip-1 in cementoblasts induced with sh-Ckip-1-EXO (500 ng/mL) for 2 d is detected by RT-qPCR (A) and western blotting (B). (C) Decreased expression of Ckip-1 in cementoblasts stimulated with sh-Ckip-1-EXO (5 μg/mL) for 1 d is detected by immunofluorescence. Decreased expression of Ckip-1 in Let-7f-5p mimic-transfected (1 d) cementoblasts induced for 2 d is detected by RT-qPCR (D) and western blotting (E). (F) Decreased expression of Ckip-1 in Let-7f-5p mimic-transfected (1 d) cementoblasts is detected by immunofluorescence. Increased expression of Ckip-1 in Let-7f-5p inhibitor-transfected (1 d) cementoblasts induced for 2 d is detected by RT-qPCR (G) and western blotting (H). (I) Increased expression of Ckip-1 in Let-7f-5p inhibitor-transfected (1 d) cementoblasts is detected by immunofluorescence. The targeting potential of Let-7f-5p to the 3′UTR (J) of Ckip-1 is confirmed by dual-luciferase reporter assay (K). *P < 0.05, **P < 0.01. P > 0.05 was considered not significant (ns).
3.7. Downregulation of Ckip-1 by Let-7f-5p promotes cementoblast mineralization by activating PGC-1α-dependent mitochondrial biogenesis
Then, PGC-1α-dependent mitochondrial biogenesis was taken into consideration. By RT-qPCR and western blotting, it was found that the expression of mitochondrial biogenesis-related markers PGC-1α, NRF1, NRF2 and TFAM was upregulated when Let-7f-5p was overexpressed in Mimic group (Figs. S7A–B), while downregulated when Let-7f-5p was knocked down in Inhibitor group (Figs. S7C–D). Further, PGC-1α suppression by the well-documented inhibitor SR blocked the mineralization-promoting effect of Let-7f-5p (Fig. S7E), suggesting that Let-7f-5p might promote cementoblast mineralization via PGC-1α-dependent mitochondrial biogenesis.
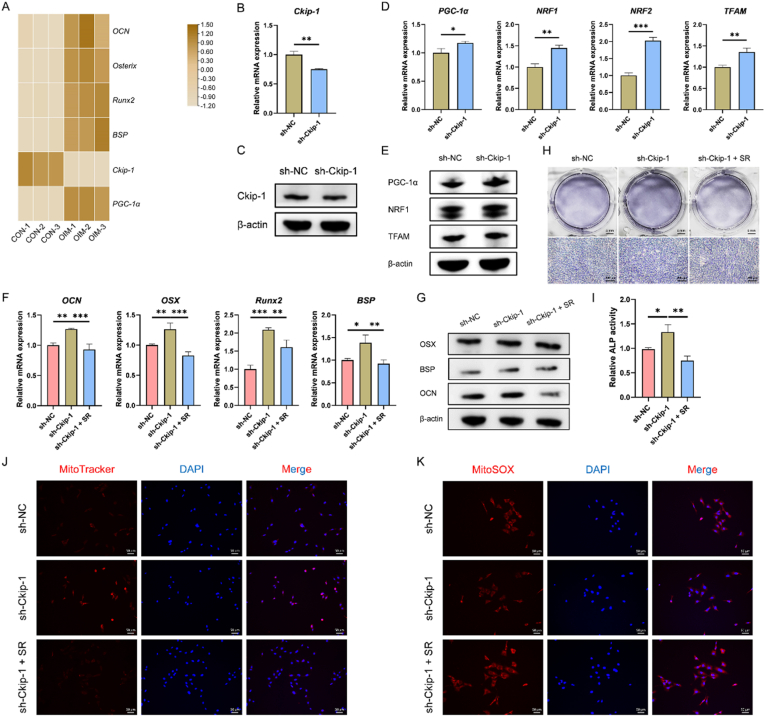
The involvement of mitochondrial biogenesis in Ckip-1 silencing-facilitated cementoblast mineralization was further studied. By mRNA sequencing, the expression of PGC-1α was found to be increased in cementoblasts cultured with OIM, which was positively correlated with the mineralization process. However, the expression of Ckip-1 decreased after induction (Fig. 7A). Then, Ckip-1-silenced cementoblasts were constructed by lentivirus transfection (Fig. S8), and identified successfully (Fig. 7B–C). Further, it was demonstrated that Ckip-1 silencing could activate the PGC-1α/NRF1/NRF2/TFAM axis in cementoblasts (Fig. 7D–E), with increased MMP (Fig. 7J) and decreased ROS production (Fig. 7K). However, inhibition of PGC-1α obstructed Ckip-1 silencing-promoted cementoblast mineralization as detected by RT-qPCR, western blotting and ALP staining (Fig. 7F–I), accompanied with decreased MMP (Fig. 7J) and increased ROS production (Fig. 7K). Taken together, downregulation of Ckip-1 by sh-Ckip-1-EXO-delivered Let-7f-5p might facilitate cementoblast mineralization through activating PGC-1α-dependent mitochondrial biogenesis.
Fig. 7.
Downregulated Ckip-1 by Let-7f-5p activates PGC-1α-dependent mitochondrial biogenesis to facilitate cementoblast mineralization. (A) The expression of mineralization-related markers, Ckip-1 and PGC-1α in control (CON) and osteogenic induction medium (OIM)-induced (7 d) cementoblasts is detected by mRNA sequencing. Identification of Ckip-1-silenced cementoblasts by RT-qPCR (B) and western blotting (C). Increased expression of PGC-1α, NRF1, NRF2 and TFAM in Ckip-1-slienced cementoblasts induced for 2 d is detected by RT-qPCR (D) and western blotting (E). Ckip-1 silencing-promoted expression of OCN, OSX, Runx2 and BSP in cementoblasts induced for 2 d is blocked by SR18292 (SR), a well-documented inhibitor of PGC-1α, detected by RT-qPCR (F) and western blotting (G). (H–I) Ckip-1 silencing-promoted ALP activity of cementoblasts induced for 4 d is blocked by SR, demonstrated by ALP staining. Ckip-1 silencing-enhanced MMP and ckip-1 silencing-suppressed ROS production in cementoblasts induced for 1 d is reversed by SR, demonstrated by MitoTracker (J) and MitoSOX (K) staining, respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
4. Discussion
In recent years, though the research on cementum regeneration has increased, there is still a long way to go to achieve a well-recognized cementum regeneration. More and more studies have focused on cementoblast mineralization under Pg-associated inflammation, while the problem how to effectively achieve cementum regeneration under Pg infection has not been well studied and solved. In this study, sh-Ckip-1-EXO derived from macrophages rich in endogenous Let-7 miRNAs and si-Ckip-1 was constructed genetically by a simple lentivirus transfection method to compensate for the transitory and unstable effect of M2-EXO for Pg-suppressed cementoblast mineralization and cementum regeneration. It was demonstrated that Ckip-1 silencing could directly promote M2 macrophage polarization and remodel the inflammatory microenvironment induced by Pg. Based on it, sh-Ckip-1-EXO was proved to rescue Pg-suppressed cementoblast mineralization through exosomal Let-7f-5p that targets Ckip-1 to activate the PGC-1α-dependent mitochondrial biogenesis, which may provide a novel strategy for cementum, and further the whole periodontal regeneration under inflammation.
Macrophage polarization plays important roles in various diseases or physiological and pathological processes, such as infectious, autoimmune, metabolic diseases, tumors, tissue repair and regeneration [20,21]. Manipulating macrophage polarization is an effective way for cementum regeneration as our and other's previous works have demonstrated the positive effect of M2 macrophages on cementoblast mineralization and the cementoblastic differentiation of periodontal ligament stem cells (PDLSCs) [13,22]. So far, many strategies have been confirmed to be involved in regulating macrophage polarization, including classical cytokines, extracellular vesicles, drug components, nanomaterials and genes [20,[23], [24], [25], [26]]. Different from other strategies such as IL-4/IL-13 stimulation, gene modification seems to be more effective, stable and sustainable without continued addition of exogenous inducers to maintain the M2 phenotype of macrophages. Ckip-1 was reported to act as an adjuster during macrophage polarization [9]. However, we proved that Ckip-1 silencing could not only restrict M1 macrophage polarization and enlarge M2 macrophage polarization under cytokine stimulation, but also directly regulate M2 macrophage polarization. Moreover, Ckip-1 silencing transformed the phenotype of Pg-stimulated macrophages from M1 to M2, remodeling an anti-inflammatory environment for cementum regeneration. As expected, Ckip-1 silenced M2 macrophages showed a promoting-effect on cementoblast mineralization.
Due to their high permeability, low clearance, non-toxicity, therapeutic functions and the application of drug carriers, exosomes were widely studied for tissue regeneration [14]. Similar to M2 macrophages, M2-EXO was well-documented in inflammation targeting and elimination, angiogenesis, tumor growth, and bone formation [17,[27], [28], [29]]. However, its role in periodontal regeneration was rarely reported. In this work, we first uncovered that M2-EXO could facilitate cementoblast mineralization. Further, sh-Ckip-1-EXO inspired from M2-EXO was also demonstrated to mediate Ckip-1-silenced macrophage-promoted cementoblast mineralization and rescue the cementoblast mineralization suppressed by Pg, with a lower effective concentration. These findings encouraged us to figure out what's in sh-Ckip-1-EXO functioned this effect.
The membrane structure of exosomes prevents the RNA contained in exosomes from being degraded, thus playing a more stable role. Let-7 miRNAs were the first discovered human miRNA with highly conserved sequence and function between species [30], and were revealed to be involved in different biological processes, including osteogenesis [[31], [32], [33], [34]]. Related studies have found that Let-7 miRNAs were closely related to M2 macrophage polarization [35,36]. For example, the expression of Let-7b-5p in tumor-associated M2 macrophages of prostate cancer was significantly upregulated [37]. In consistent with the studies, we also observed increased expression of Let-7d-5p, Let-7f-5p, Let-7i-5p in Ckip-1 silencing-induced M2 macrophages. Moreover, relevant studies have also shown that several subtypes, including Let-7f-5p, Let-7i-5p could promote osteoblast mineralization, the osteogenic differentiation of stem cells, and bone formation [34,[38], [39], [40]]. Further, the enhanced osteogenic differentiation was partially attributed to the delivered Let-7 miRNAs in exosomes [41], suggesting the possibility of exosomal transfer of Let-7 miRNAs from Ckip-1-silenced macrophages to cementoblasts. As excepted, we found that Let-7d-5p, Let-7f-5p, Let-7i-5p in Ckip-1-silenced macrophages could be packaged into exosomes, in which, Let-7f-5p was the mostly packaged and transferred. In addition, the transfer process of miRNA was simulated successfully. Based on the similarities between bone and cementum [42], and also the finding that most of the Let-7 family miRNAs increased during cementoblast mineralization by miRNA sequencing, we confirmed that Let-7f-5p also facilitated cementoblast mineralization. However, how the transferred Let-7f-5p promoted cementoblast mineralization remains unknown.
To our knowledge, only two studies reported that exosomes derived from lentivirus-transfected sh-RNA-expressing cells were rich in this type of sh-RNA or cleavaged si-RNA, and capable of silencing the corresponding mRNA expression in target cells [18,19]. It seemed like that these cells worked as an active machine to produce si-RNA-loaded exosomes to reshape the microenvironment. Consistent with the previous studies, we also detected higher expression of si-Ckip-1 in Ckip-1-silenced macrophages and the exosomes derived from them. Actually, endogenous loading is a method based on parental cells. First, the source cells are modified (direct transfection, co-incubation) to introduce target molecules (Let-7 miRNAs, si-Ckip-1) into the source cells. In the process of producing exosomes, cargos are internally sorted and loaded into the intracellular multivesicular bodies (MVBs), and then followed by the secretion of intraluminal vesicles (ILVs) as exosomes [14]. Though increased trend of si-Ckip-1 expression in cementoblasts was found in sh-Ckip-1-EXO-stimulated group when compared with sh-NC-EXO-stimulated group, the differences were not significant. Strangely, the si-Ckip-1 expression in cementoblasts showed a decreased trend in the two exosome-stimulated groups. We speculated that this negative result might be caused by insufficient exosome application, improper stimulation time or the preferential degradation characteristic of si-Ckip-1 rather than Let-7 miRNAs in cementoblasts.
Ckip-1, also known as PLEKHO1 (pleckstrin homology domain-containing family O, member 1), was a scaffold protein interacting with various signals to be involved in tumorigenesis, bone formation and macrophage polarization as mentioned above [6,7]. Besides, we also demonstrated that Ckip-1 could negatively regulate cementum formation and cementoblast mineralization with or without Pg stimulation [8]. Based on the above findings, we then confirmed that sh-Ckip-1-EXO and the enriched Let-7f-5p also silenced Ckip-1 like si-Ckip-1. Moreover, the targeting role of Let-7f-5p to the 3′UTR of Ckip-1 mRNA was detected. Therefore, sh-Ckip-1-EXO might contain various biomolecules that shared similarities with Let-7 miRNAs and si-Ckip-1 to facilitate cementoblast mineralization.
PGC-1α was an essential regulator for mitochondrial biogenesis through activating NRF1 and NRF2 to accelerate TFAM synthesis [43]. It has been demonstrated that PGC-1α-dependent mitochondrial biogenesis was positively correlated with the osteogenesis in many cell and animal models [44,45]. Our group also confirmed the impaired PGC-1α-dependent mitochondrial biogenesis in cementoblast under hypoxic condition, and PGC-1α activation could protect CoCl2-suppresed cementoblast mineralization [46]. Further, one recent literature uncovered its therapeutic potential in the treatment of periodontitis [47]. In consistent with the above works, we further confirmed that Ckip-1 silencing could activate PGC-1α-dependent mitochondrial biogenesis with enhanced PGC-1α/NRF1/NRF2/TFAM expression, increased MMP and decreased ROS production, thus facilitating cementoblast mineralization.
There also exist some shortcomings or matters could be further explored in our study. For example, the mineralization-facilitating capacity of M2-EXO and sh-Ckip-1-EXO was not compared. Besides Let-7f-5p, whether there are other more effective biomolecules in sh-Ckip-1-EXO. Also, whether sh-Ckip-1-EXO has the same inflammation tropism as M2-EXO, and whether there is a possibility that sh-Ckip-1-EXO could rescue cementoblast mineralization by influencing bacteria behaviors remain unclear. Finally, more efforts are needed to build a stable, reliable and typical animal model for cementum regeneration in our future work.
5. Conclusion
In conclusion, a novel type of exosome derived from Ckip-1 silencing-induced M2-like macrophages was designed for Pg-inhibited cementoblast mineralization and cementum regeneration. Mechanismly, sh-Ckip-1-EXO were similar to M2-EXO induced by IL-4 in promoting cementoblast mineralization, and rescued Pg-inhibited cementoblast mineralization by delivering Let-7f-5p and many other molecules that targeted Ckip-1 to facilitate cementoblast mineralization through activating the PGC-1α-dependent mitochondrial biogenesis. Our study may provide a new strategy for cementum, and further the whole periodontal regeneration under Pg-dominated inflammation.
Ethics approval and constent to participate
Animal experiments in this study were approved by the Ethics Committee of the School of Medicine, Wuhan University (WP20230091). We also confirm our compliance with all relevant ethical regulations.
Ethics approval and constent to participate
Animal experiments in this study were approved by the Ethics Committee of the School of Medicine, Wuhan University (WP20230091). We also confirm our compliance with all relevant ethical regulations.
CRediT authorship contribution statement
Xin Huang: Conceptualization, Methodology, Validation, Data curation, Writing – original draft. Yifei Deng: Validation, Formal analysis. Junhong Xiao: Validation, Formal analysis. Huiyi Wang: Investigation, Formal analysis. Qiudong Yang: Investigation, Formal analysis. Zhengguo Cao: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
None.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82370967, No. 82170963). We also thank Figdraw (www.figdraw.com) for the assistance in creating schematic diagram.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2023.10.009.
Contributor Information
Xin Huang, Email: huangxin1994@whu.edu.cn.
Zhengguo Cao, Email: caozhengguo@whu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kinane D.F., Stathopoulou P.G., Papapanou P.N. Periodontal diseases. Nat. Rev. Dis. Prim. 2017;3 doi: 10.1038/nrdp.2017.38. [DOI] [PubMed] [Google Scholar]
- 2.Tonetti M.S., Jepsen S., Jin L., Otomo-Corgel J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: a call for global action. J. Clin. Periodontol. 2017;44(5):456–462. doi: 10.1111/jcpe.12732. [DOI] [PubMed] [Google Scholar]
- 3.Arzate H., Zeichner-David M., Mercado-Celis G. Cementum proteins: role in cementogenesis, biomineralization, periodontium formation and regeneration. Periodontol. 2000;67(1):211–233. doi: 10.1111/prd.12062. 2015. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J., Faure L., Adameyko I., Sharpe P.T. Stem cell contributions to cementoblast differentiation in healthy periodontal ligament and periodontitis. Stem Cell. 2021;39(1):92–102. doi: 10.1002/stem.3288. [DOI] [PubMed] [Google Scholar]
- 5.Hajishengallis G., Darveau R.P., Curtis M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012;10(10):717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie J., Liu L., He F., Fu X., Han W., Zhang L. CKIP-1: a scaffold protein and potential therapeutic target integrating multiple signaling pathways and physiological functions. Ageing Res. Rev. 2013;12(1):276–281. doi: 10.1016/j.arr.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Fu L., Zhang L. Physiological functions of CKIP-1: from molecular mechanisms to therapy implications. Ageing Res. Rev. 2019;53 doi: 10.1016/j.arr.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Huang X., Ma L., Wang X., Wang H., Peng Y., Gao X., Huang H., Chen Y., Zhang Y., Cao Z. Ckip-1 mediates P. Gingivalis-suppressed cementoblast mineralization. J. Dent. Res. 2022;101(5):599–608. doi: 10.1177/00220345211054744. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y., Liu W., Wang Y., Zhang L., Wei J., Zhang X., He F., Zhang L. Casein Kinase 2 Interacting Protein-1 regulates M1 and M2 inflammatory macrophage polarization. Cell. Signal. 2017;33:107–121. doi: 10.1016/j.cellsig.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Locati M., Curtale G., Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu. Rev. Pathol. 2020;15:123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A., Sinha M., Datta S., Abas M., Chaffee S., Sen C.K., Roy S. Monocyte and macrophage plasticity in tissue repair and regeneration. Am. J. Pathol. 2015;185(10):2596–2606. doi: 10.1016/j.ajpath.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang Z., Yoshizawa-Smith S., Glowacki A., Maltos K., Pacheco C., Shehabeldin M., Mulkeen M., Myers N., Chong R., Verdelis K., Garlet G.P., Little S., Sfeir C. Induction of M2 macrophages prevents bone loss in murine periodontitis models. J. Dent. Res. 2019;98(2):200–208. doi: 10.1177/0022034518805984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang X., Wang X., Ma L., Wang H., Peng Y., Liu H., Xiao J., Cao Z. M2 macrophages with inflammation tropism facilitate cementoblast mineralization. J. Periodontol. 2023;94(2):290–300. doi: 10.1002/JPER.22-0048. [DOI] [PubMed] [Google Scholar]
- 14.Kalluri R., LeBleu V.S. The biology function and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou M., Li B., Liu C., Hu M., Tang J., Min J., Cheng J., Hong L. M2 Macrophage-derived exosomal miR-501 contributes to pubococcygeal muscle regeneration. Int. Immunopharm. 2021;101(Pt B) doi: 10.1016/j.intimp.2021.108223. [DOI] [PubMed] [Google Scholar]
- 16.Luo Z., Peng W., Xu Y., Xie Y., Liu Y., Lu H., Cao Y., Hu J. Exosomal OTULIN from M2 macrophages promotes the recovery of spinal cord injuries via stimulating Wnt/β-catenin pathway-mediated vascular regeneration. Acta Biomater. 2021;136:519–532. doi: 10.1016/j.actbio.2021.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Kang M., Huang C.-C., Lu Y., Shirazi S., Gajendrareddy P., Ravindran S., Cooper L.F. Bone regeneration is mediated by macrophage extracellular vesicles. Bone. 2020;141 doi: 10.1016/j.bone.2020.115627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hareendran S., Albraidy B., Yang X., Liu A., Breggia A., Chen C.C., Loh Y.P. Exosomal carboxypeptidase E (CPE) and CPE-shRNA-loaded exosomes regulate metastatic phenotype of tumor cells. Int. J. Mol. Sci. 2022;23(6):3113. doi: 10.3390/ijms23063113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang C.-H., Qin L., Gao Y.-C., Chen T.-Y., Xu K., Liu T., Ren T. APE1 shRNA-loaded cancer stem cell-derived extracellular vesicles reverse Erlotinib resistance in non-small cell lung cancer via the IL-6/STAT3 signalling. Clin. Transl. Med. 2022;12(5):e876. doi: 10.1002/ctm2.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sica A., Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L.-X., Zhang S.-X., Wu H.-J., Rong X.-L., Guo J. M2b macrophage polarization and its roles in diseases. J. Leukoc. Biol. 2019;106(2):345–358. doi: 10.1002/JLB.3RU1018-378RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., He X.-T., Kong D.-Q., Xu X.-Y., Wu R.-X., Sun L.-J., Tian B.-M., Chen F.-M. M2 macrophages enhance the cementoblastic differentiation of periodontal ligament stem cells via the akt and JNK pathways. Stem Cell. 2019;37(12):1567–1580. doi: 10.1002/stem.3076. [DOI] [PubMed] [Google Scholar]
- 23.Kim W., Lee E.J., Bae I.-H., Myoung K., Kim S.T., Park P.J., Lee K.-H., Pham A.V.Q., Ko J., Oh S.H., Cho E.-G. Lactobacillus plantarum-derived extracellular vesicles induce anti-inflammatory M2 macrophage polarization in vitro. J. Extracell. Vesicles. 2020;9(1) doi: 10.1080/20013078.2020.1793514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu H., Wu L., Liu L., Ruan Q., Zhang X., Hong W., Wu S., Jin G., Bai Y. Quercetin ameliorates kidney injury and fibrosis by modulating M1/M2 macrophage polarization. Biochem. Pharmacol. 2018;154:203–212. doi: 10.1016/j.bcp.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S., Xie F., Li K., Zhang H., Yin Y., Yu Y., Lu G., Zhang S., Wei Y., Xu K., Wu Y., Jin H., Xiao L., Bao L., Xu C., Li Y., Lu Y., Gao J. Gold nanoparticle-directed autophagy intervention for antitumor immunotherapy via inhibiting tumor-associated macrophage M2 polarization. Acta Pharm. Sin. B. 2022;12(7):3124–3138. doi: 10.1016/j.apsb.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao X., Sharma N., Kapadia F., Zhou G., Lu Y., Hong H., Paruchuri K., Mahabeleshwar G.H., Dalmas E., Venteclef N., Flask C.A., Kim J., Doreian B.W., Lu K.Q., Kaestner K.H., Hamik A., Clément K., Jain M.K. Krüppel-like factor 4 regulates macrophage polarization. J. Clin. Invest. 2011;121(7):2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G., Zhang J., Zhao Q., Zhuang W., Ding J., Zhang C., Gao H., Pang D.-W., Pu K., Xie H.-Y. Molecularly engineered macrophage-derived exosomes with inflammation tropism and intrinsic heme biosynthesis for atherosclerosis treatment. Angew Chem. Int. Ed. Engl. 2020;59(10):4068–4074. doi: 10.1002/anie.201913700. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y., Guo Z., Chen W., Wang X., Cao M., Han X., Zhang K., Teng B., Cao J., Wu W., Cao P., Huang C., Qiu Z. M2 macrophage-derived exosomes promote angiogenesis and growth of pancreatic ductal adenocarcinoma by targeting E2F2. Mol. Ther. 2021;29(3):1226–1238. doi: 10.1016/j.ymthe.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao X.-M., Guan Z., Yang Z.-J., Ma L.-Y., Dai Y.-J., Liang C., Hu J.-T. Comprehensive analysis of M2 macrophage-derived exosomes facilitating osteogenic differentiation of human periodontal ligament stem cells. BMC Oral Health. 2022;22(1):647. doi: 10.1186/s12903-022-02682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roush S., Slack F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Letafati A., Najafi S., Mottahedi M., Karimzadeh M., Shahini A., Garousi S., Abbasi-Kolli M., Sadri Nahand J., Tamehri Zadeh S.S., Hamblin M.R., Rahimian N., Taghizadieh M., Mirzaei H. MicroRNA let-7 and viral infections: focus on mechanisms of action. Cell. Mol. Biol. Lett. 2022;27(1):14. doi: 10.1186/s11658-022-00317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y., Shen N., Wicha M.S., Luo M. The roles of the let-7 family of MicroRNAs in the regulation of cancer stemness. Cells. 2021;10(9):2415. doi: 10.3390/cells10092415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Zhao J., Chen S., Li D., Yang J., Zhao X., Qin M., Guo M., Chen C., He Z., Zhou Y., Xu L. Let-7 as a promising target in aging and aging-related diseases: a promise or a pledge. Biomolecules. 2022;12(8):1070. doi: 10.3390/biom12081070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei J., Li H., Wang S., Li T., Fan J., Liang X., Li J., Han Q., Zhu L., Fan L., Zhao R.C. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cell. Dev. 2014;23(13):1452–1463. doi: 10.1089/scd.2013.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rong J., Xu L., Hu Y., Liu F., Yu Y., Guo H., Ni X., Huang Y., Zhao L., Wang Z. Inhibition of let-7b-5p contributes to an anti-tumorigenic macrophage phenotype through the SOCS1/STAT pathway in prostate cancer. Cancer Cell Int. 2020;20:470. doi: 10.1186/s12935-020-01563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L., Liu T., Chen G., Li Y., Zhang S., Mao L., Liang P., Fasihi Harandi M., Li T., Luo X. Exosomal microRNA let-7-5p from cysticercus prompted macrophage to M2 polarization through inhibiting the expression of C/EBP δ. Microorganisms. 2021;9(7):1403. doi: 10.3390/microorganisms9071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Xu L., Hu Y., Huang Y., Zhang Y., Zheng X., Wang S., Wang Y., Yu Y., Zhang M., Yuan K., Min W. miRNA let-7b modulates macrophage polarization and enhances tumor-associated macrophages to promote angiogenesis and mobility in prostate cancer. Sci. Rep. 2016;6 doi: 10.1038/srep25602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen G.-Y., Ren H., Shang Q., Zhao W.-H., Zhang Z.-D., Yu X., Huang J.-J., Tang J.-J., Yang Z.-D., Liang D., Jiang X.-B. Let-7f-5p regulates TGFBR1 in glucocorticoid-inhibited osteoblast differentiation and ameliorates glucocorticoid-induced bone loss. Int. J. Biol. Sci. 2019;15(10):2182–2197. doi: 10.7150/ijbs.33490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C., Liu S., Li J., Cheng Y., Wang Z., Feng T., Lu G., Wang S., Song J., Xia P., Hao L. Biological functions of let-7e-5p in promoting the differentiation of MC3T3-E1 cells. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.671170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Cheng W., Han B., Guo Y., Wei S., Yu L., Zhang X. Let-7i-5p functions as a putative osteogenic differentiation promoter by targeting CKIP-1. Cytotechnology. 2021;73(1):79–90. doi: 10.1007/s10616-020-00444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi S.-Y., Han E.-C., Hong S.-H., Kwon T.-G., Lee Y., Lee H.-J. Regulating osteogenic differentiation by suppression of exosomal MicroRNAs. Tissue Eng. 2019;25(15–16):1146–1154. doi: 10.1089/ten.TEA.2018.0257. [DOI] [PubMed] [Google Scholar]
- 42.Salmon C.R., Tomazela D.M., Ruiz K.G.S., Foster B.L., Paes Leme A.F., Sallum E.A., Somerman M.J., Nociti F.H. Proteomic analysis of human dental cementum and alveolar bone. J. Proteonomics. 2013;91:544–555. doi: 10.1016/j.jprot.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88(2):611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 44.Liu L., Cheng Y., Wang J., Ding Z., Halim A., Luo Q., Song G. Simulated microgravity suppresses osteogenic differentiation of mesenchymal stem cells by inhibiting oxidative phosphorylation. Int. J. Mol. Sci. 2020;21(24):9747. doi: 10.3390/ijms21249747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal S., Porwal K., Rajak S., Sinha R.A., Chattopadhyay N. Selective dietary polyphenols induce differentiation of human osteoblasts by adiponectin receptor 1-mediated reprogramming of mitochondrial energy metabolism. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110207. [DOI] [PubMed] [Google Scholar]
- 46.Wang H., Wang X., Ma L., Huang X., Peng Y., Huang H., Gao X., Chen Y., Cao Z. PGC-1 alpha regulates mitochondrial biogenesis to ameliorate hypoxia-inhibited cementoblast mineralization. Ann. N. Y. Acad. Sci. 2022;1516(1):300–311. doi: 10.1111/nyas.14872. [DOI] [PubMed] [Google Scholar]
- 47.Sun X., Ping Y., Li X., Mao Y., Chen Y., Shi L., Hong X., Chen L., Chen S., Cao Z., Chen P., Song Z., Wismeijer D., Wu G., Ji Y., Huang S. Activation of PGC-1α-dependent mitochondrial biogenesis supports therapeutic effects of silibinin against type I diabetic periodontitis. J. Clin. Periodontol. 2023;50(7):964–979. doi: 10.1111/jcpe.13811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.