Abstract
The presence of a hemolysin-encoding gene, elyA or hlyA, from Shiga toxin-producing Escherichia coli (STEC) was detected by PCR in each of 95 strains tested. PCR products of elyA from human STEC isolates of serovars frequently detected in Germany, such as O157:H−, O103:H2, O103:H−, O26:H11, and O26:H−, showed nucleotide sequences identical to previously reported ones for O157:H7 and O111:H− strains. Compared to them, four elyA amplicons derived from human isolates of rare STEC serovars showed identity of about 98% but lacked an AluI restriction site. However, the nucleotide sequence of an amplicon derived from a porcine O138:K81:H− STEC strain was identical to the corresponding region of hlyA, encoding alpha-hemolysin, from E. coli. This hlyA amplicon showed 68% identity with the nucleotide sequence of the corresponding elyA fragment. It differed from the elyA PCR product in restriction fragments generated by AluI, EcoRI, and MluI. Of the 95 representative STEC strains, 88 produced hemolysin on blood agar supplemented with vancomycin (30 mg/liter), cefixime (20 μg/liter), and cefsulodin (3 mg/liter) (BVCC). The lowest added numbers of two to six STEC CFU per g of stool or per ml of raw milk were detectable on BVCC plates after seeding of the preenrichment broth, modified tryptic soy broth (mTSB) supplemented with novobiocin (10 mg/liter), with 16 STEC strains. These strains represented the seven prevailing serovars diagnosed from German patients. However, with ground-beef samples, PCR was essential to identify the lowest added numbers of two to six STEC CFU among colonies of hemolyzing Enterobacteriaceae, such as Serratia spp. and alpha-hemolysin-producing E. coli. We conclude that preenrichment of stool and food samples in mTSB for 6 h followed by overnight culturing on BVCC is a simple method for the isolation and presumptive identification of STEC.
Shiga toxin-producing Escherichia coli (STEC) is increasingly recognized as the cause of severe diseases, such as hemorrhagic colitis and hemolytic-uremic syndrome in humans (15), edema disease in piglets (14), and diarrhea in calves (21). In addition to the major virulence factor, Shiga toxin (Stx), and its variants (30), STEC frequently produces intimin (19), which is involved in attaching of the organisms and effacing of gut mucosal cells. Furthermore, STEC secretes pore-forming hemolysins. Iron acquisition by lysis of erythrocytes and impairment of the immune response due to cytotoxicity for leukocytes are assumed to be the main pathogenic functions of the hemolysins (9).
Two different plasmid-encoded hemolysins, both members of the RTX toxin family (9, 28), have been described for STEC. Alpha-hemolysin is formed by porcine edema disease-causing STEC strains of serovars O138:K81, O139:K82, and O141:K85, which produce Stx variant 2e (14), and by E. coli causing urinary tract infections and septicemia (17, 22). It generates a clear, broad zone of hemolysis surrounding the colony and is visible after only 4 h on blood agar containing washed sheep erythrocytes and CaCl2 (enterohemolysin agar) (4). The second hemolysin, secreted exclusively by human STEC strains, produces a narrow, turbid, hemolytic halo after overnight incubation on enterohemolysin agar (4).
The elyA genes in human STEC isolates of serovars O157:H7 and O111:H− are 62 to 64% identical to hlyA, encoding alpha-hemolysin, from E. coli (18, 29). Furthermore, the RTX hemolysin- or leucotoxin-encoding genes apxIA and apxIIIA of Actinobacillus pleuropneumoniae and aaltA of A. actinomycetemcomitans show similarities in the range of 56 to 60% with elyA and hlyA, respectively (18).
Humans are infected either by contaminated food, especially of bovine origin, such as ground beef and raw milk, or by person-to-person transmission, by the fecal-oral route (24). Detection of strains of the prominent STEC serovar O157:H7 from food and stool samples can be conducted on special media because these strains are unable to ferment sorbitol within 24 h and lack β-glucuronidase activity. However, STEC isolates from humans now comprise at least 160 different serovars with variable distributions in different countries (1, 7, 31). In Germany, STEC serovars O157:H7, O157:H−, O111:H−, O103:H2, O103:H−, O26:H11, and O26:H− prevail (7). Because of the biochemical and serological diversity, detection of major virulence factor Stx by cytotoxicity assays with Vero cells, Stx enzyme-linked immunosorbent assay (ELISA), or stx PCR is the method of choice for identifying STEC. For simple detection of STEC by culturing, Beutin et al. (4) described a blood agar medium called enterohemolysin agar. Unfortunately, only 74% of 54 STEC strains tested showed hemolysis on this agar (6). Additionally, the nonselective enterohemolysin agar allows concomitant flora of fecal and food specimens to overgrow STEC as well as competing hemolysis of Enterobacteriaceae and gram-positive bacteria.
In the present study, the occurrence of elyA and its variants in different STEC serovars was evaluated by restriction fragment length polymorphisms of PCR products (PCR-RFLP). Additional sequence information for elyA and its variants in strains of emerging or rare STEC serovars is provided. After short-term preenrichment, blood agar supplemented with vancomycin, cefixime, and cefsulodin (BVCC) was tested for efficient recognition of hemolysing STEC among the concomitant flora of food and stool specimens. Suspected hemolytic colonies from the modified blood agar were confirmed by PCR detection of elyA and stx genes.
MATERIALS AND METHODS
Bacterial strains and serotyping.
A total of 92 STEC isolates from different patients were collected from 1993 to 1996. Stool specimens or suspected isolates were received from laboratories in different parts of Germany. The determination of O and H antigens from E. coli was performed as described previously (5). The STEC collection comprised 51 strains of serogroup O157 and 41 strains of other serovars (Table 1). Furthermore, the reference strain EDL 933, a human STEC isolate of serovar O157:H7 (Centers for Disease Control and Prevention, Atlanta, Ga.), porcine O138:K81:H− strain E57 (16), and an Orough:H4 isolate from raw milk were added to the strain collection.
TABLE 1.
STEC strains tested in this study
| Serovar | No. of strains | Source | stx/eaeAa |
|---|---|---|---|
| O157:H7 | 41 | Human | 2× stx1/eaeA; 36× stx2/eaeA; 5× stx1+2/eaeA |
| O157:H− | 11 | Human | 8× stx2/eaeA; 3× stx1+2/eaeA |
| O156:H27 | 1 | Human | 1× stx1 |
| O146:H− | 1 | Human | 1× stx1+2 |
| O138:K81:H− | 1 | Pig (strain E57) | 1× stx2e |
| O117:H7 | 1 | Human | 1× stx1 |
| O113:H53 | 1 | Human | 1× stx2 |
| O111:H− | 5 | Human | 3× stx1/eaeA; 1× stx2/eaeA; 1× stx1+2/eaeA |
| O103:H2 | 5 | Human | 4× stx1; 1× stx2 |
| O103:H− | 2 | Human | 2× stx1 |
| O95:H− | 1 | Human | 1× stx1 |
| O89:H− | 1 | Human | 1× stx1 |
| O69:H− | 1 | Human | 1× stx1+2 |
| O28:H35 | 1 | Human | 1× stx2 |
| O26:H12 | 1 | Human | 1× stx1 |
| O26:H11 | 5 | Human | 3× stx1; 1× stx2; 1× stx1+2 |
| O26:H− | 4 | Human | 2× stx1; 2× stx2 |
| O25:H14 | 1 | Human | 1× stx1 |
| O12:H− | 1 | Human | 1× stx1 |
| O8:H− | 1 | Human | 1× stx2 |
| O1:H− | 1 | Human | 1× stx1+2 |
| Ont:H19 | 1 | Human | 1× stx1 |
| Ont:H1 | 1 | Human | 1× stx2 |
| Ont:H− | 2 | Human | 1× stx1; 1× stx2 |
| Orough:H11 | 2 | Human | 1× stx1; 1× stx2 |
| Orough:H4 | 1 | Raw milk | 1× stx2 |
| Orough:H− | 1 | Human | 1× stx1 |
Numbers of strains of the corresponding serovar which harbor virulence genes encoding Shiga toxin (stx) and intimin (eaeA). For specification of stx genes, the nomenclature proposed by Calderwood et al. (8a) was used.
Media.
The supplementation of modified tryptic soy broth (mTSB) (23) with bile salts no. 3 (Difco, Detroit, Mich.) and novobiocin (Sigma, St. Louis, Mo.) was reduced to 1.12 g and 10 mg/liter, respectively. Buffered peptone water (BPW) supplemented with vancomycin (8 mg/liter), cefixime (50 μg/liter), and cefsulodin (10 mg/liter) (BPW-VCC) was prepared as described by Wallace and Jones (32).
BVCC was made from 33 g of tryptose blood agar base (Difco), 5 g of tryptose (Difco), 5 g of soluble starch (E. Merck AG, Darmstadt, Germany), 441 mg of CaCl2 · 2H2O, 3 g of agar (Difco), and 970 ml of distilled water. The agar suspension was adjusted to pH 7.0 with 1 N HCl and heated at 100°C for 1 h. After the agar suspension was cooled to 50°C, 30 ml of defibrinated, sterile sheep blood (Oxoid) and sterile solutions of 30 mg of vancomycin hydrochloride (Lilly, Giessen, Germany), 20 μg of cefixime (a gift from Merck), and 3 mg of cefsulodin sodium salt (Sigma) were added. Before use, the sheep blood was washed three times with 50 ml of sterile saline. After the mixture was stirred, 25-ml quantities of BVCC were poured into petri dishes. Enterohemolysin agar was prepared as described by Beutin et al. (4).
Bacteriological examination of specimens.
Fecal samples were collected from 50 healthy persons. Twenty-seven ground-beef samples were obtained from six butcher shops. Raw-milk samples originated from 53 dairy farms and two health food shops in northern Germany.
Counts of aerobic mesophilic bacteria, Enterobacteriaceae, and E. coli were determined in accordance with Section 35 of the German Federal Foods Act (10–13).
The efficiencies of preenrichment broths for the propagation of STEC strains were examined with BPW-VCC and mTSB shaken at 120 rpm and 37°C. For preenrichment of STEC strains from food and fecal specimens, 1 g or 1 ml of sample was added to 9 ml of mTSB and shaken at 120 rpm and 37°C for 6 h. Subsequently, 0.1 ml of broth culture was streaked on a BVCC plate and incubated at 37°C. Hemolytic growth was evaluated after 4 and 20 h. Colonies of hemolyzing Enterobacteriaceae were identified with the API 20E system (Biomerieux, Nürtingen, Germany).
With the described enrichment procedure, the detection limit for STEC was determined by seeding the food and fecal specimens with different numbers of each of 16 STEC strains. These human strains belonged to the seven predominant STEC serovars in Germany. Four of the strains were O157:H7; the remaining serovars, O157:H−, O111:H−, O103:H2, O103:H−, O26:H11, and O26:H−, were represented by two strains each.
PCR and analysis of amplicons.
For release of DNA, suspensions in 50 μl of sterile bidistilled water of single hemolyzing colonies or bacterial growth from a BVCC plate were boiled for 10 min. Cell debris was centrifuged for 5 min at 8,240 × g. Supernatant (0.5 μl) was mixed with 40 pmol of primer (synthesized by GIBCO BRL, Eggenstein, Germany), 200 μM each deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, 1 U of cloned Thermus brockianus DNA polymerase, and assay buffer (Biometra, Göttingen, Germany). The stx1B gene was detected by PCR as described by Rüssmann et al. (26). A 691- or 692-bp fragment of each of the stx2AB genes and their variants was amplified with the primers stx2-start (5′-TTT CCA TGA CRA CGG ACA GCA GTT AT-3′) and stx2-end (5′-CTC ATT ATA CTT RGA RAA CTC AAT TTT SCC T-3′). PCR of the stx2 amplicon was conducted in 35 cycles with denaturation for 20 s at 94°C, annealing for 60 s at 50°C, and polymerization for 60 s at 72°C.
STEC eaeA was detected by PCR as described by Schmidt et al. (27). For detection of the estA and astA genes, encoding heat-stable enterotoxins, we amplified a 155-bp fragment of estA by using the primers estA-start (5′-CCT TTC SCT CAG GAT GCT AAA CC-3′) and estA-end (5′-CAA GCA GGA TTA CAA CAC AAT TCA CAG-3′) as well as a 112-bp fragment of astA by using the oligonucleotides astA-start (5′-GCC ATC AAC ACA GTA TAT CCG RAG GC-3′) and astA-end (5′-GGT CGC GAG TGA CGG CTT TGT-3′). Amplification was conducted as described for stx2 PCR.
Primers hlyA-start (5′-AGG AAG TYG TKA AGG ARC AGG AGG-3′) and hlyA-end (5′-CCA TCY GCG CCA TGG AAK ATA TCA-3′) were used to amplify nucleotides 2033 to 2234 of hlyA and nucleotides 1988 to 2186 of elyA and its variants, respectively. Amplification of the hlyA and elyA fragments was performed as described for stx2 PCR, but the temperature of annealing was raised to 60°C. For typing, the hlyA and elyA amplicons were digested separately with AluI, EcoRI, and MluI as recommended by the manufacturer (Amersham, Braunschweig, Germany).
Nucleotide sequence determination of the hlyA and elyA amplicons was performed with an automated DNA sequencer (LI-COR 4200) as recommended by the manufacturer (MWG-Biotech, Ebersberg, Germany).
RESULTS
Detection of hemolysin in STEC strains.
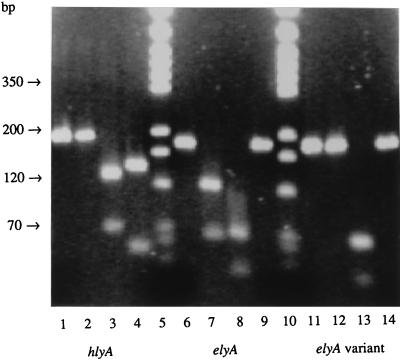
Using primers for conserved C-terminal nucleotide sequences from hlyA and elyA, we obtained PCR products from lysates of the 95 representative STEC strains. Five amplicons revealed restriction patterns that were generated by AluI, EcoRI, and MluI and that deviated from that of O157:H7 reference strain EDL 933. Four of the strains yielded elyA amplicons that were not digested by AluI (Fig. 1). These strains were characterized by the presence of stx1 genes, the absence of eaeA, and an association with unusual serovars, such as Ont:H−, Ont:H19, Orough:H4, and O89:H−. The hlyA amplification product derived from the porcine isolate of serovar O138:K81:H− was digested by MluI but not by AluI and showed larger EcoRI subfragments corresponding to hlyA, encoding alpha-hemolysin (Fig. 1), than elyA amplicons. This strain harbored stx2e, estA, and astA genes but not the eaeA gene.
FIG. 1.
PCR of hlyA and elyA as well as their subsequent typing by digestion with AluI, EcoRI, and MluI. The corresponding amplicon of each hemolysin gene (lanes 1, 6, and 11) as well as its AluI (lanes 2, 7, and 12)-, EcoRI (lanes 3, 8, and 13)-, and MluI (lanes 4, 9, and 14)-generated restriction fragments are shown from the left to the right. Lanes 1 to 4, products from hlyA; lanes 6 to 9, products from elyA; lanes 11 to 14, products from elyA variants; lanes 5 and 10, markers (pGEM-3 DNA digested separately with HinfI, RsaI, and SinI; Promega, Mannheim, Germany). The agarose gel was documented by the Gel Doc 1000 video gel documentation system from Bio-Rad (Munich, Germany).
Nucleotide sequence variations of elyA indicated by PCR-RFLP were confirmed by sequence analysis of the amplicons (Fig. 2). The four elyA amplification products which were not digested by AluI showed a sequence identity of 98 to 98.7% with the elyA fragment derived from O157:H7 and O111:H− STEC strains. Nucleotide sequences of amplicons derived from STEC strains of the prevailing serovars O157:H−, O103:H2, O103:H−, O26:H11, and O26:H− corresponded to that for the O157:H7 reference strain. The PCR product of the O138:K81:H− porcine isolate, which contained MluI subfragments, showed the same nucleotide sequence as hlyA, encoding alpha-hemolysin. Derived amino acid sequences of each sequenced elyA amplicon were identical. However, elyA and hlyA amplicons showed only 68% identities for nucleotide sequences and 72.7% identities for amino acid sequences.
FIG. 2.
Nucleotide sequences of elyA and hlyA amplicons. elyA represents strains of serovars O157:H7 (28), O111:H− (29), and O157:H−, O103:H2, O103:H−, O26:H11, and O26:H−; strain 1114/93 was serovar Ont:H−; strain 18-6/95 was serovar Orough:H−; strain 849/95 was serovar O89:H−; strain 6780/96 was serovar Ont:H19; and hlyA represents strains used by Kuhnert et al. (18) and strain E57 (serovar O138:K81:H−). Dashes represent nucleotides identical to elyA. Asterisks symbolize gaps in the aligned nucleotide sequence.
Eighty-eight (92.6%) of the 95 STEC strains produced hemolysin on BVCC after 20 h. After serial propagation of these strains, they showed hemolysis on BVCC within just 4 to 6 h. Fifty (96.2%) of 52 O157 strains and 38 (88.4%) of 43 non-O157 strains lysed erythrocytes on this agar. In comparison, only 44 (84.6%) of the O157 STEC strains and 32 (74.4%) of the non-O157 STEC strains formed a hemolysis zone on enterohemolysin agar, resulting in a rate of 80% hemolyzing STEC strains on this agar. In contrast to other STEC strains, O103 strains hemolyzed on BVCC as strongly as did most alpha-hemolysin-producing E. coli strains. On the other hand, 2 of 13 alpha-hemolysin-producing E. coli strains isolated from stool specimens produced narrow and turbid zones of hemolysis overnight, like most STEC strains.
Identification of STEC strains in stool and food specimens.
BPW-VCC and mTSB were tested for their efficiency as preenrichment broths for STEC isolation. In mTSB, strains grew to the stationary phase within 12 h. However, in BPW-VCC, strains grew slower and four (4.2%) of the isolates did not grow within 48 h. In BPW supplemented only with 20 μg of cefixime per liter, these four isolates grew within 48 h. However, their growth remained poor with 50 μg of cefixime per liter of BPW. These isolates belonged to serovars Orough:H−, O8:H−, O26:H−, and O138:K81:H−.
After preenrichment in mTSB, a limited number of hemolyzing Enterobacteriaceae lacking stx genes were isolated from food and fecal samples on BVCC. In 13 (26%) of 50 stool specimens, hlyA-containing E. coli colonies were confirmed by PCR-RFLP. While 11 of them secreted alpha-hemolysin on BVCC plates after 4 h of incubation, the remaining 2 produced narrow and turbid zones of hemolysis only after overnight incubation. Eleven (20%) of 55 raw-milk samples contained alpha-hemolysin-producing E. coli colonies characterized by a clear and broad zone of beta-hemolysis on BVCC. Each ground-beef sample contained colonies of hemolyzing Enterobacteriaceae that did not harbor stx genes. Amplicons of hlyA were obtained from hemolyzing E. coli colonies isolated from 15 ground-beef samples (55.6%). Three of these samples showed beta-hemolysis after 4 h on BVCC. Mixtures of hemolyzing Serratia spp. and E. coli colonies were detected in seven samples. Serratia spp. as the only hemolyzing colonies were isolated from 11 (40.7%) of 27 ground-beef samples. They were easily distinguished from E. coli colonies by their white color and their musty smell after 24 h of incubation. Hemolyzing Citrobacter freundii was isolated from one ground-beef specimen.
To determine the limit of detection of STEC by preenrichment with mTSB and consecutive plating on BVCC, 16 STEC strains of the seven predominant serovars in Germany were added separately to preenrichment broth with five specimens each of ground beef, raw milk, and stool. Even the smallest amounts of 2.3 CFU of STEC added per g of stool and 6.2 CFU of STEC added per g of ground beef and per ml of raw milk were detected. However, detection of STEC by culturing on BVCC was hampered in ground-beef samples and one stool specimen by hemolysis of Serratia spp. and non-STEC E. coli, respectively. In these, the inoculated STEC colonies were recognized by PCR of genes encoding Stx and hemolysin from single hemolytic colonies or total growth.
Determination of the concomitant flora resulted in total counts of 1.2 × 106 to 4.8 × 108 aerobic mesophilic bacteria per g of stool, counts of 5.3 × 106 to 4.3 × 107 per g of ground beef, and counts of 1.63 × 103 to 4.4 × 107 per ml of raw milk. E. coli counts varied from 2.1 × 105 to 4.3 × 107 per g of stool, from 3 to 750 per g of ground beef, and from <0.3 to 9.3 per ml of raw milk. The numbers of Enterobacteriaceae in ground-beef samples ranged from 2.4 × 104 to 5 × 105 per g.
DISCUSSION
The described PCR method proved efficient for detecting hemolysin genes from E. coli: elyA from STEC strains and hlyA from E. coli producing alpha-hemolysin. PCR-RFLP showed that all human STEC isolates harbored elyA. In comparison to the results for the O157:H7 reference strain, PCR-RFLP and subsequent nucleotide sequencing of elyA amplicons revealed only four STEC strains with minor sequence variations. These strains were of rare non-O157 serovars associated with stx1 genes and lacked eaeA. However, among the STEC strains, hlyA was restricted to an O138:K81:H− strain. Alpha-hemolysin of this porcine strain was associated with stx2e and estA genes, as shown by Meyer and Karch (20). In the present study, astA, encoding a second heat-stable enterotoxin, was detected in this strain. The close association of elyA, located on the 94- to 103-kb STEC virulence plasmid (28), and stx genes, harbored by a lysogenic lambdoid phage (25), was remarkable. This situation could also be true for hlyA from porcine strains containing stx2e.
The different rates of detection of hemolysin from STEC by PCR (100%) and by culturing on BVCC (92.6%) might have been due to the repression of gene expression under growth conditions in the laboratory, faulty transport of hemolysin to the cell surface, or mutations of elyA not targeted by the PCR method described here. In comparison to STEC strains of rare serovars, a slightly higher proportion of strains of serogroup O157 showed hemolysis on BVCC and enterohemolysin agar. Essential ingredients of blood agar are required to detect hemolysis of STEC. These include calcium (2) and washed sheep blood (4). An increase in the vancomycin concentration to 250 mg per liter of BVCC allowed us to recognize the hemolysis of two additional strains in our STEC collection. Presumably, vancomycin facilitated the secretion of hemolysin through an increase in the permeability of the cell wall. In previous studies, the rates of detection of hemolyzing STEC on enterohemolysin agar varied from 97.6% (3) to 75.2% (7). In the present study, BVCC was superior to enterohemolysin agar for the detection of hemolysis by STEC. After serial propagation of STEC on BVCC, hemolysin production was observed during the logarithmic growth phase, as has been reported for other hemolysins of the RTX type (9).
A vancomycin (8 mg/liter), cefixime (50 μg/liter), and cefsulodin (10 mg/liter) supplementation which differed from that in BVCC was used in BPW-VCC for preenrichment of STEC (32). The higher quantities of cefixime in BPW-VCC caused 4.2% growth inhibition of the strains in our culture collection. This inhibition was avoided by use of mTSB instead of BPW-VCC for preenrichment of STEC. After preenrichment with mTSB, the frequent association of Stx production with the formation of hemolysin allowed us to isolate most STEC strains from special blood agar, such as BVCC. The antibiotic supplements of BVCC allowed us to detect resistant, hemolyzing Enterobacteriaceae as E. coli, C. freundii, and Serratia spp. after 16 h of incubation but suppressed the growth of gram-positive bacteria, Proteus spp., and Pseudomonas spp. However, after 24 h of incubation, colonies of Serratia spp. differed in color, shape, and odor from other species of Enterobacteriaceae in ground-beef samples. As members of the Proteae, Serratia spp. produce a type of hemolysin different from the RTX cytolysins (8).
Examining 180 fecal non-STEC E. coli isolates from healthy children and patients, Bettelheim (3) identified 3 (1.7%) weakly hemolyzing and 52 (28.9%) alpha-hemolysin-producing strains. Beutin et al. (4) detected no weakly hemolyzing strains but 40 (15%) alpha-hemolysin-producing strains among 267 fecal non-STEC E. coli isolates from 200 healthy infants. In good accordance with these studies of fecal non-STEC E. coli isolates, we detected such strains in 4% of fecal samples from healthy patients by the production of a narrow, turbid, hemolytic halo and in 22% of such fecal samples by the production of strong hemolysis.
In conclusion, BVCC considerably facilitated the isolation, presumptive identification, and enumeration of most STEC strains in stool and raw-milk specimens. In ground-beef specimens and a few stool specimens, STEC strains were sensitively detected by PCR of stx and elyA genes from single, weakly hemolyzing colonies grown on BVCC. Thus, preenrichment of stool and food samples in mTSB for 6 h followed by subculturing on BVCC may be recommended for the isolation and presumptive identification of STEC strains. The identities of such strains should be confirmed by proof of stx genes and Stx itself.
ACKNOWLEDGMENT
This study was financially supported by the German Federal Ministry of Health as part of the National Reference Centre for Salmonella and Other Bacterial Enteropathogens.
REFERENCES
- 1.Acheson D W K, Keusch G T. Which Shiga toxin-producing types of E. coli are important? ASM News. 1996;62:302–306. [Google Scholar]
- 2.Bauer M E, Welch R A. Characterization of an RTX toxin from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1996;64:167–175. doi: 10.1128/iai.64.1.167-175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettelheim K A. Identification of enterohaemorrhagic Escherichia coli by means of their production of enterohaemolysin. J Appl Bacteriol. 1995;79:178–180. doi: 10.1111/j.1365-2672.1995.tb00932.x. [DOI] [PubMed] [Google Scholar]
- 4.Beutin L, Montenegro M A, Ørskov I, Ørskov F, Prada J, Zimmermann S, Stephan R. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J Clin Microbiol. 1989;27:2559–2564. doi: 10.1128/jcm.27.11.2559-2564.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bockemühl J, Aleksic S, Karch H. Serological and biochemical properties of Shiga-like toxin (verocytotoxin)-producing strains of Escherichia coli, other than O-group 157, from patients in Germany. Zentbl Bakteriol. 1992;276:189–195. doi: 10.1016/s0934-8840(11)80005-8. [DOI] [PubMed] [Google Scholar]
- 6.Bockemühl J, Karch H. Zur aktuellen Bedeutung der enterohämorrhagischen Escherichia coli (EHEC) in Deutschland (1994–1995) Bundesgesundheitsblatt. 1996;39:290–296. [Google Scholar]
- 7.Bockemühl J, Karch H, Tschäpe H. Infektionen des Menschen durch enterohämorrhagische Escherichia coli (EHEC) in Deutschland, 1996. Bundesgesundheitsblatt. 1997;40:194–197. doi: 10.1007/s00103-002-0458-4. [DOI] [PubMed] [Google Scholar]
- 8.Braun V, Focareta T. Pore-forming bacterial protein hemolysins (cytolysins) Crit Rev Microbiol. 1991;18:115–158. doi: 10.3109/10408419109113511. [DOI] [PubMed] [Google Scholar]
- 8a.Calderwood S B, Acheson D W K, Keusch G T, Barrett T J, Griffin P M, Strockbine N A, Swaminathan B, Kaper J P, Levine M M, Kaplan B S, Karch H, O’Brien A D, Obrig T O, Takeda Y, Tarr P I, Wachsmuth I K. Proposed new nomenclature of SLT (VT) family. ASM News. 1996;62:118–119. [Google Scholar]
- 9.Coote J G. Structural and functional relationships among the RTX toxin determinants of Gram-negative bacteria. FEMS Microbiol Rev. 1992;88:137–162. doi: 10.1111/j.1574-6968.1992.tb04961.x. [DOI] [PubMed] [Google Scholar]
- 10.German Federal Foods Act. Bestimmung der coliformen Keime in Milch, Milchprodukten, Butter und Käse, L 01.00-2. Berlin, Germany: Beuth Verlag; 1980. [Google Scholar]
- 11.German Federal Foods Act. Bestimmung der Escherichia coli in Milch, Milchprodukten, Butter, Käse und Speiseeis, L 01.00-25. Berlin, Germany: Beuth Verlag; 1987. [Google Scholar]
- 12.German Federal Foods Act. Bestätigung von Escherichia coli durch zusätzliche Identifizierungsverfahren, L 00.00-21. Berlin, Germany: Beuth Verlag; 1990. [Google Scholar]
- 13.German Federal Foods Act. Bestimmung der Keimzahl: Gußverfahren. Verfahren zur Qualitätssicherung im Laboratorium, L 01.00-00. Berlin, Germany: Beuth Verlag; 1991. [Google Scholar]
- 14.Imberechts H, De Greve H, Lintermans P. The pathogenesis of edema disease in pigs. A review. Vet Microbiol. 1992;31:221–233. doi: 10.1016/0378-1135(92)90080-d. [DOI] [PubMed] [Google Scholar]
- 15.Karmali M A. Infections by verotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konowalchuk J, Speirs J I, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korhonen T K, Valtonen M W, Parkkinen J, Väisänen-Rhen V, Finne J, Ørskov F, Ørskov I, Svenson S B, Mäkelä P H. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect Immun. 1985;48:486–491. doi: 10.1128/iai.48.2.486-491.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhnert P, Heyberger-Meyer B, Burnens A P, Nicolet J, Frey J. Detection of RTX toxin genes in gram-negative bacteria with a set of specific probes. Appl Environ Microbiol. 1997;63:2258–2265. doi: 10.1128/aem.63.6.2258-2265.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louie M, De Azavedo J C S, Handelsman M Y C, Clark C G, Ally B, Dytoc M, Sherman P, Brunton J. Expression and characterization of the eaeA gene product of Escherichia coli O157:H7. Infect Immun. 1993;61:4085–4092. doi: 10.1128/iai.61.10.4085-4092.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer T, Karch H. Genes coding for Shiga-like toxin and heat stable enterotoxin in porcine strains of Escherichia coli. FEMS Microbiol Lett. 1989;57:115–120. doi: 10.1016/0378-1097(89)90353-4. [DOI] [PubMed] [Google Scholar]
- 21.Mohammad A, Peiris J S M, Wijetwanta E A. Serotypes of verocytotoxigenic Escherichia coli from cattle and buffalo calf diarrhea. FEMS Microbiol Lett. 1986;35:261–265. [Google Scholar]
- 22.Ørskov I, Ørskov F. Escherichia coli in extra-intestinal infections. J Hyg. 1985;95:551–575. doi: 10.1017/s0022172400060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padhye N V, Doyle M P. Rapid procedure for detecting enterohemorrhagic Escherichia coli O157:H7 in food. Appl Environ Microbiol. 1991;57:2693–2698. doi: 10.1128/aem.57.9.2693-2698.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padhye N V, Doyle M P. Escherichia coli O157:H7: epidemiology, pathogenesis, and methods for detection in food. J Food Prot. 1992;55:555–565. doi: 10.4315/0362-028X-55.7.555. [DOI] [PubMed] [Google Scholar]
- 25.Rietra P J, Willshaw G A, Smith H R, Field A M, Scotland S M, Rowe B. Comparison of Vero-cytotoxin-encoding phages from Escherichia coli of human and bovine origin. J Gen Microbiol. 1989;135:2307–2318. doi: 10.1099/00221287-135-8-2307. [DOI] [PubMed] [Google Scholar]
- 26.Rüssmann H, Schmidt H, Heesemann J, Caprioli A, Karch H. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with a haemolytic uraemic syndrome. J Med Microbiol. 1994;40:338–343. doi: 10.1099/00222615-40-5-338. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt H, Rüssmann H, Karch H. Virulence determinants in nontoxigenic Escherichia coli O157 strains that cause infantile diarrhea. Infect Immun. 1993;61:4894–4898. doi: 10.1128/iai.61.11.4894-4898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt H, Karch H. Enterohemolytic phenotypes and genotypes of Shiga toxin-producing Escherichia coli O111 strains from patients with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol. 1996;34:2364–2367. doi: 10.1128/jcm.34.10.2364-2367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarr P I. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infections. Clin Infect Dis. 1995;20:1–10. doi: 10.1093/clinids/20.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Thomas A, Chesty T, Frost J A, Chart H, Smith H R, Rowe B. Vero cytotoxin-producing Escherichia coli, particularly serogroup O157, associated with human infections in England and Wales: 1992–4. Epidemiol Infect. 1996;117:1–10. doi: 10.1017/s0950268800001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace J S, Jones K. The use of selective and differential agars in the isolation of Escherichia coli O157 from dairy herds. J Appl Bacteriol. 1996;81:663–668. doi: 10.1111/j.1365-2672.1996.tb03562.x. [DOI] [PubMed] [Google Scholar]