Abstract
Summary
The interpretation of pathway enrichment analysis results is frequently complicated by an overwhelming and redundant list of significantly affected pathways. Here, we present an R package aPEAR (Advanced Pathway Enrichment Analysis Representation) which leverages similarities between the pathway gene sets and represents them as a network of interconnected clusters. Each cluster is assigned a meaningful name that highlights the main biological themes in the experiment. Our approach enables an automated and objective overview of the data without manual and time-consuming parameter tweaking.
Availability and implementation
The package aPEAR is implemented in R, published under the MIT open-source licence. The source code, documentation, and usage instructions are available on https://gitlab.com/vugene/aPEAR as well as on CRAN (https://CRAN.R-project.org/package=aPEAR).
1 Introduction
Pathway enrichment analysis (PEA) is indispensable when interpreting high-throughput omics data and identifying the underlying biological processes that are dysregulated in a particular condition or disease (Khatri et al. 2012). Despite the comprehensive insights provided by the vast number of available pathway gene set annotations in various databases, analysing large amounts of pathways introduces the redundancy problem: a single gene can be involved in multiple biological processes, resulting in pathways being highly correlated and containing overlapping sets of genes (Merico et al. 2010, Reimand et al. 2019). This causes a profusion of significantly affected pathways and impedes the interpretation of the PEA results. Ultimately, there is a need to aggregate similar pathways and analyse their interactions.
Here, we present an R package aPEAR (Advanced Pathway Enrichment Analysis Representation), which aids in the interpretation of the PEA results. The aPEAR package implements multiple metrics to calculate similarities between pathway gene sets, detects pathway clusters, and assigns biologically relevant names to them. Finally, aPEAR builds a visual representation of an enrichment network that can be explored interactively to elucidate the biological processes affected by the experimental conditions.
2 Methods and implementation
The R package aPEAR exports a single main function enrichmentNetwork() which visualizes the PEA results as a network where nodes and edges represent the pathways and similarity between them, respectively. While it was created with the clusterProfiler (Wu et al. 2021) and gprofiler2 (Kolberg et al. 2020) output in mind, any enrichment result is accepted as long as it is formatted correctly (Supplementary Text S1). The network is constructed in several steps:
The pairwise similarity between all pathway gene sets is evaluated using the Jaccard index (default), cosine similarity, or correlation similarity metrics.
The similarity matrix is then used to detect clusters of redundant pathways using Markov (default) (Van Dongen 2008), hierarchical, or spectral (John et al. 2020) clustering algorithms.
Each cluster is assigned a biologically meaningful name. Network analysis is used to determine the pathway with the most connections, using either PageRank (Page, 1998) (default) or HITS (Kleinberg 1999) algorithm. Alternatively, the highest absolute NES value or the lowest P-value can be used to select the most important pathway in the cluster. The description of this pathway is used as the cluster label.
A ggplot2 (Wickham 2016) graph is constructed using the similarity matrix and the annotated clusters. An interactive graph is visualized using plotly (Sievert 2020). Pathways and their assigned clusters are returned as output as well (Supplementary Text 2).
3 Results and discussion
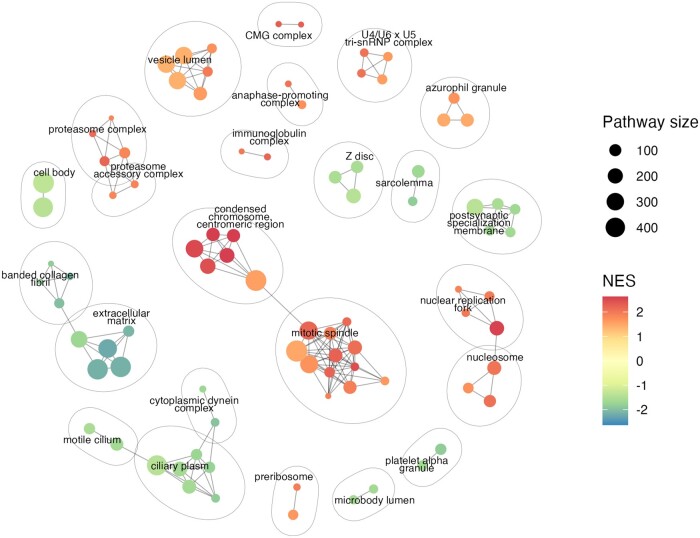
Currently, the most frequently used tools for gene set visualization include the emapplot function from the R package enrichplot (Yu, 2022) and the Cytoscape plugin Enrichment Map (Merico et al. 2010). These methods use a word cloud-like algorithm to assign a cluster name which results in labels that are not semantically meaningful. The emapplot has some limitations when handling large datasets and produces numerous small clusters with nonintuitive labels (Supplementary Methods, Supplementary Fig. S1). Cytoscape, while a powerful software tool that is able to work with large amounts of data, requires many time-consuming manual adjustments that can introduce bias into the network (Supplementary Methods, Supplementary Figs S2–S4). In contrast, aPEAR makes it easy to work with numerous pathways, highlights the biological context of the clusters and does not require additional manipulation of the graph, making it ideal for automated data visualization as well as interactive investigations (Fig. 1).
Figure 1.
Example enrichment network generated by aPEAR. The nodes represent the significant pathways, and the edges represent similarity between them. Coloured by normalized enrichment score (NES).
To determine which similarity metric and clustering algorithm is best suitable for pathway cluster analysis, 180 tests were performed using PEA results from 10 real-world datasets (Supplementary Methods, Supplementary Tables S1 and S2). Based on cluster quality evaluation using the Dunn index (Dunn 1974), the Silhouette index (Rousseeuw 1987) and the Davies-Bouldin index (Davies and Bouldin 1979), the Jaccard similarity metric and the Markov clustering algorithm were found to be best suited for such analysis and, thus, were set as the default parameters in the aPEAR package (Supplementary Text S3, Supplementary Fig. S5). Note that the Jaccard coefficient can be affected by the pathway size and may under-connect the smaller pathways contained within the larger ones (Salvatore et al. 2020).
4 Conclusion
We developed an R package, aPEAR, that visualizes clusters of similar pathways as an enrichment network and, consequently, enables better interpretation of the PEA results.
Supplementary Material
Acknowledgements
We would like to express our sincere gratitude to Migle Gabrielaite for her invaluable manuscript reviews and to Milda Milčiūtė for her excellent work testing the aPEAR package.
Contributor Information
Ieva Kerseviciute, VUGENE, K. Donelaitis street, Kaunas, LT-44248, Lithuania.
Juozas Gordevicius, VUGENE, K. Donelaitis street, Kaunas, LT-44248, Lithuania.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
This work was supported by VUGENE (www.vugene.com).
Data availability
The complete analysis used to evaluate the package can be found at https://github.com/ievaKer/aPEAR-publication.
References
- Davies DL, Bouldin DW.. A cluster separation measure. IEEE Trans Pattern Anal Mach Intell 1979;1:224–7. [PubMed] [Google Scholar]
- Dunn JC. Well-separated clusters and optimal fuzzy partitions. J Cybern 1974;4:95–104. [Google Scholar]
- John CR, Watson D, Barnes MR. et al. Spectrum: fast density-aware spectral clustering for single and multi-omic data. Bioinformatics 2020;36:1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Sirota M, Butte AJ.. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 2012;8:e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberg JM. Authoritative sources in a hyperlinked environment. J ACM 1999;46:604–32. [Google Scholar]
- Kolberg L, Raudvere U, Kuzmin I. et al. gprofiler2—an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res 2020;9:709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O. et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010;5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page L. The pagerank citation ranking: Bringing order to the web. Technical report, Stanford Digital Library Technologies Project, 1998. [Google Scholar]
- Reimand J, Isserlin R, Voisin V. et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, cytoscape and enrichmentmap. Nat Protoc 2019;14:482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 1987;20:53–65. [Google Scholar]
- Salvatore S, Dagestad Rand K, Grytten I. et al. Beware the Jaccard: the choice of similarity measure is important and non-trivial in genomic colocalisation analysis. Brief Bioinform 2020;21:1523–30. [DOI] [PubMed] [Google Scholar]
- Sievert C. Interactive Web-Based Data Visualization with R, Plotly, and Shiny (Chapman & Hall/CRC the R Series). 1st ed. Boca Raton, FL: Chapman and Hall/CRC, p. 470, 2020. [Google Scholar]
- Van Dongen S. Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal Appl 2008;30:121–41. [Google Scholar]
- Wickham H. Ggplot2 – Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag New York, 2016. [Google Scholar]
- Wu T, Hu E, Xu S. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. enrichplot: Visualization of Functional Enrichment Result, Bioconductor, 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete analysis used to evaluate the package can be found at https://github.com/ievaKer/aPEAR-publication.