Abstract
Efficient, nonselective methods to obtain DNA from the environment are needed for rapid and thorough analysis of introduced microorganisms in environmental samples and for analysis of microbial community diversity in soil. A small-scale procedure to rapidly extract and purify DNA from soils was developed for in-the-field use. Amounts of DNA released from bacterial vegetative cells, bacterial endospores, and fungal conidia were compared by using hot-detergent treatment, freeze-thaw cycles, and bead mill homogenization. Combining a hot-detergent treatment with bead mill homogenization gave the highest DNA yields from all three microbial cell types and provided DNA from the broadest range of microbial groups in a natural soil community. Only the bead mill homogenization step was effective for DNA extraction from Bacillus globigii (B. subtilis subsp. niger) endospores or Fusarium moniliforme conidia. The hot-detergent–bead mill procedure was simplified and miniaturized. By using this procedure and small-scale, field-adapted purification and quantification procedures, DNA was prepared from four different soils seeded with Pseudomonas putida cells or B. globigii spores. In a New Mexico soil, seeded bacterial targets were detected with the same sensitivity as when assaying pure bacterial DNA (2 to 20 target gene copies in a PCR mixture). The detection limit of P. putida cells and B. globigii spores in different soils was affected by the amount of background DNA in the soil samples, the physical condition of the DNA, and the amount of DNA template used in the PCR.
PCR analysis provides a sensitive and specific means to detect and monitor microorganisms in complex environmental samples. Successful detection and characterization of microbial DNA in the environment require efficient extraction of the DNA from environmental samples and adequate purification from the coextracted contaminants that inhibit PCR. Soils and sediments vary greatly in chemical and organic composition. They also contain abundant humic and fulvic acids that are inhibitory to Taq DNA polymerase and other enzymes (24, 26, 28; for a recent review, see reference 29). Soils are therefore one of the most challenging environmental matrices from which to obtain microbial DNA that will support PCR.
Two applications in environmental microbial assessment require simultaneous extraction of the DNA from a wide range of microorganisms in a single sample. For analysis of the diversity and dynamics of natural microbial communities, a broad-based, nonselective DNA extraction procedure is desirable to obtain unbiased representation of community members. For forensic and other investigative analyses, a simple, small-scale procedure is needed to provide rapid, sensitive detection of a wide variety of potentially released organisms, including several medically important bacterial and fungal pathogens, for in-the-field analysis of environmental samples.
Direct comparisons of the relative effectiveness of different extraction and purification procedures for simultaneous preparation of both bacterial and fungal propagules have not been made. Most studies describing recovery of microbial DNA from soils or sediments have focused on extraction of DNA from a single introduced microorganism, usually vegetative cells of a gram-negative organism, or have examined only a single environmental sample. Sometimes native DNA was removed from the sample prior to introducing the target microorganism (4, 24). DNA extraction from gram-positive and spore-forming bacteria in the soil has been described elsewhere (14, 24, 33), but the methods used in these studies resulted in severely sheared DNA that does not provide for the highest possible PCR detection sensitivity. Comparisons of methods for lysis of indigenous soil bacteria indicate that the portion of bacteria lysed by a particular method depends greatly on the method employed and the types and sizes of cells in the sample (11, 37). The relative ability of different extraction techniques, either singly or in combination, to simultaneously obtain high-molecular-weight DNA from multiple cell types of bacteria and fungi has not been established. Such studies are required to provide unbiased representation of all the DNA in an environmental sample for simultaneous detection of a wide variety of introduced microorganisms and for analysis of microbial communities.
To date, all reported procedures have been developed for laboratory implementation and are not directly adaptable to rapid field use. Although numerous methods have been reported for direct DNA isolation and purification from microorganisms in soil (4, 6, 9, 11, 13–16, 18, 20, 23–25, 27, 33, 36), the sample preparation procedures and experimental conditions used in different studies vary widely. Published procedures vary tremendously in the time (a few hours to several days), equipment, and laboratory space necessary to prepare DNA from environmental samples. Many of the reported procedures use specialized laboratory equipment, such as high-speed centrifuges, gel electrophoresis units, and ultracentrifuges, and most require chemicals or enzymes that are labile or that require special handling, storage, and disposal.
The objective of this work was to develop and test a nonselective, small-scale procedure for DNA sample preparation to support rapid in-the-field PCR analysis (12, 34) for sensitive detection of microbial spores and cells in environmental samples. The efficacy with which three extraction methods, alone and in combination, released DNA from bacterial vegetative cells (Pseudomonas putida), bacterial endospores (Bacillus globigii), and fungal conidia (Fusarium moniliforme) is presented. The most-effective extraction and purification methods were scaled down and miniaturized for field use, and several steps that were found to be unnecessary for PCR were eliminated. These methods were tested with both pure microbial cultures and complex soil samples. A method to quantify DNA in crude, humic acid-contaminated extracts from environmental samples was developed and used to determine the amounts of DNA released from different soils. Usefulness of the small-scale procedure for specific detection of target microorganisms in the environment was tested by determining the detection sensitivity for introduced bacterial cells and spores in different soils, and several factors that impact the detection limit of target microorganisms in soils were investigated. Effectiveness of the procedure for microbial community analysis was tested by identifying the native microbial groups that could be detected in a soil by using rRNA gene-targeted PCR primers.
MATERIALS AND METHODS
Microbial species used to compare DNA extraction methods.
Three microbial species that represent a wide range of microbial cell types were used in the following experiments designed to compare the DNA extraction efficiencies of various methods. These microbes were selected for our studies because they are closely related to animal and human pathogens that are of interest in forensics-related detection applications.
The gram-negative bacterium Pseudomonas putida mt-2 (ATCC 33015) was used for analysis of bacterial vegetative cells. This strain possesses a 117-kb plasmid (pWW0, TOL plasmid [1 to 10 copies per cell] [reviewed in reference 1; see also reference 35]) that carries genes for toluene degradation. P. putida cells were grown in nutrient broth (Difco Laboratories, Detroit, Mich.) at 30°C overnight in an orbital shaker at 150 rpm. One milliliter of the culture grown overnight was used to start a 100-ml culture that was grown to mid-log phase (absorbance at 600 nm of 0.6). Vegetative cells were collected, and dilution series were generated in water. A portion of each dilution was plated on nutrient agar to determine CFU titer. Mid-log-phase cells were selected, because at this growth stage, the maximum proportion of cells are viable and a minimum of dead cells and free DNA are present. Since extracellular DNA present in the cell preparation can bias the observed DNA extraction results, the amount of extracellular DNA present in the cultures was examined in preliminary experiments by testing our ability to detect the PCR target gene in cell-free medium from mid-log-phase cultures. The target DNA was not detected in the culture medium, suggesting that there was not a measurable amount of extracellular DNA in the P. putida cell preparations used for DNA extraction and soil seeding studies.
The gram-positive bacterium Bacillus globigii (B. subtilis subsp. niger, ATCC 9372) was used for analysis of bacterial spores. This species produces endospores typical of the Bacillus genus. Two spore preparations were provided by the United States Army (Dugway Proving Ground, Utah). Spore preparation 93 contained 1 × 1011 CFU/g, and spore preparation 95 contained 4 × 1011 CFU/g. Spore preparation 93 contained little extracellular DNA that could bias the extracted DNA yield or the achieved detection limit. Free DNA was not detected in water washes of this spore preparation by agarose gel electrophoresis and ethidium bromide staining or by PCR. In addition, PCR detection limits were identical for spores treated with DNase and untreated spores. In contrast, spore preparation 95 contained considerable amounts of extracellular DNA. Both preparations were used in the studies presented here to compare the amounts of extracellular DNA present in different preparations and because only limited amounts of each spore preparation were available. Spore preparation 93 (low extracellular DNA) was used in all soil seeding experiments described below.
Spores of the fungus Fusarium moniliforme (ATCC 24088) were also used to compare effectiveness of different DNA extraction methods. Conidia were prepared by growing fungal plate cultures on potato dextrose agar (Difco Laboratories) at room temperature under ambient light to induce sporulation. A mixture of macroconidia and microconidia was harvested from the plates by rubbing the surface mycelium gently with a rubber swab and collecting the spores in 0.05% Tween 20 solution. Hyphal debris was removed from the spores by centrifuging the crude spore preparation through a 40% sucrose pad, through which the spores settled to the bottom of the tube, leaving the remainder of the cellular debris on the surface of the sucrose pad. Spores were quantified by counting in a hemocytometer.
Extraction methods tested for maximum DNA yield from microbial cells and spores.
Three extraction techniques were used for comparisons of DNA yield. The three techniques were a hot-detergent treatment (13), freeze-thaw cycles (25), and bead mill homogenization (13). The bead mill homogenization method was modified substantially from the original published report (13), and all of the methods were miniaturized as much as possible. To identify critical steps for efficient DNA recovery from a combination of microbial cell and spore types, suspensions of P. putida vegetative cells (109 CFU/ml), B. globigii endospores (preparations 93 and 95) (108 spores/ml), and F. moniliforme conidia (107 conidia/ml) were subjected to permutations of the three-step procedure described below, in which one, two, or all steps were omitted. Since the overall goal was to develop a procedure for eventual field use, the liquid nitrogen (liquid N) freezing used in the freeze-thaw cycles was raised to −20 or 4°C to determine if the very low temperature freezing step was essential or if it could be replaced with a temperature more easily achieved outside the laboratory. This experimental design gave 16 possible procedure combinations (see Fig. 1).
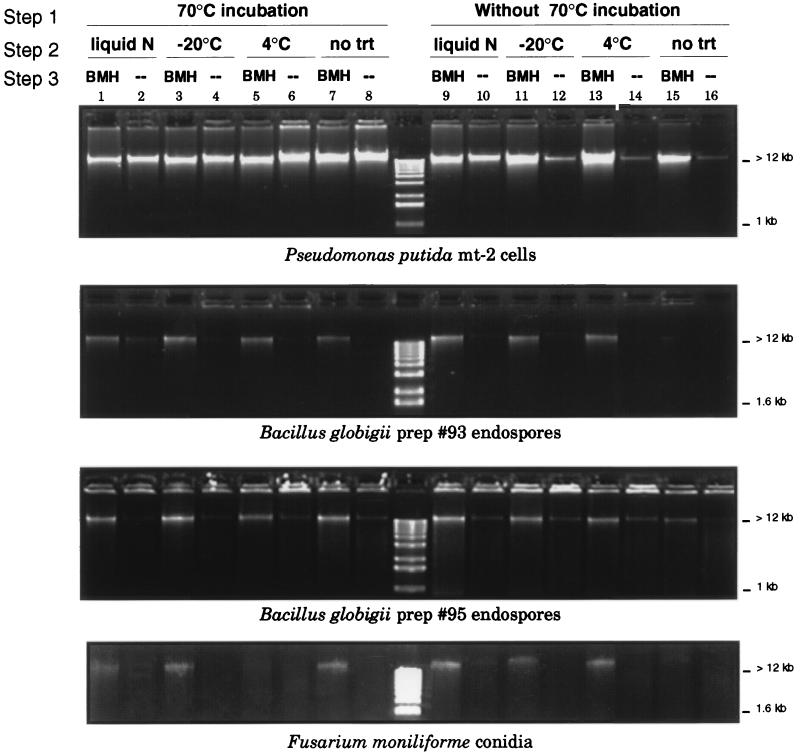
FIG. 1.
Relative amounts of DNA extracted from P. putida cells, B. globigii endospores, and F. moniliforme conidia processed by variations of the three-step procedure described in Materials and Methods. (Step 1) Cells or spores were suspended in TENS buffer and either incubated at 70°C for 20 min, or this step was omitted and samples were immediately carried to step 2. (Step 2). Samples were frozen in liquid nitrogen to −20°C or chilled to 4°C and then rapidly heated in a boiling water bath. This step was repeated three times or was not done (no trt). (Step 3) Samples were either homogenized for 3 min at 5,000 rpm in a mini-bead beater (BMH), or this step was omitted (--). Equal volumes of DNA extract were separated by agarose gel electrophoresis on a 3% agarose gel, and DNA was visualized under UV light after staining with ethidium bromide. Comparison of the DNA band intensities in a gel illustrates the different amounts of DNA released from equal amounts of starting material by each procedure or procedure combination. Lane 16 in each panel illustrates the combined amount of DNA present as extracellular background DNA and released by room temperature incubation in TENS buffer containing the detergent SDS. prep, preparation.
Each 1-ml cell or spore suspension was divided into two 0.5-ml samples, and DNA was extracted independently from the duplicate sets. A 0.5-ml portion of 2× TENS buffer (1× TENS is 50 mM Tris HCl [pH 8.0], 20 mM EDTA, 100 mM NaCl, 1% [wt/vol] sodium dodecyl sulfate [SDS]) was added to a 0.5-ml cell or spore suspension in a 2-ml bead-beater tube containing 1,800 mg of a mixture of glass beads (600 mg each of beads with a diameter of 710 to 1,180 μm, 425 to 600 μm, and 106 μm [Sigma Chemical Company, St. Louis, Mo., or BioSpec Products, Inc., Bartlesville, Okla.]). This bead combination disrupts soil colloids and plant tissue (710- to 1,180-μm-diameter beads), fungal, plant, and other eukaryotic cells (425- to 600-μm-diameter beads), and bacterial cells (106-μm-diameter beads) (6a and cell disruption guidelines in the mini-bead beater instruction manual, BioSpec Products, Inc.). The following treatments were conducted sequentially, with the least harsh treatment first, followed by increasingly harsh extraction treatments.
(i) Step 1. Hot-detergent treatment.
Samples were vortexed briefly and incubated at 70°C for 20 min. During this time, samples were suspended and mixed by vortexing for 5 s every 10 min.
(ii) Step 2. Freeze-thaw cycles.
Samples were frozen for 2 min in liquid N for 5 min at −20°C or placed in the refrigerator until they reached 4°C and then immediately placed in a boiling water bath to rapidly thaw the sample. This process was repeated three times.
(iii) Step 3. Bead mill homogenization.
Samples were homogenized for 3 min at 5,000 rpm in a mini-bead beater cell disruptor (type BX-4, catalog no. 311OBX; Bio-Spec Products). They were then centrifuged at 12,000 × g for 10 min at room temperature to pellet the bead mix. The supernatant containing DNA was collected and stored on ice. Nucleic acids in fungal extractions were precipitated by using 1/10 volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethyl alcohol. The pellet containing DNA was suspended in 20 μl of TE buffer (10 mM Tris HCl [pH 8.0], 1 mM EDTA). Bacterial DNA extractions were not precipitated prior to gel electrophoresis to determine yield.
For each test organism, equal volumes (10 μl per sample) of DNA extracts were applied to agarose gels containing 3% (wt/vol) SeaKem agarose (FMC Bioproducts, Rockland, Maine) gels prepared and run in 1× TAE buffer [1× TAE is 40 mM Tris-acetate [pH 8.0], 1 mM EDTA]) and separated by electrophoresis. DNA in the gel was stained with ethidium bromide (5 μg/ml) and visualized under UV light, and the DNA concentration was determined by using densiometry measurements of digitized gel images using Kodak Digital Science ID software (Eastman Kodak Co., Rochester, N.Y.). Lambda DNA standards (10 to 200 ng) were interspersed between sample lanes on each quantification gel to provide a standard curve for quantification. Gels containing 3% agarose were used to concentrate the extracted DNA into a band width comparable to that of the lambda DNA standards to improve quantification accuracy. Each experiment was repeated two to four times.
To further compare the DNA extracted from Bacillus spores by using the freeze-thaw cycles and bead mill homogenization, a spore suspension (109 CFU/ml) of B. globigii preparation 95 was prepared in TE buffer and divided into 12 replicate samples. Three replicate samples were exposed to either 3 min of bead mill homogenization, 3 cycles of liquid N freeze-thawing, 40 cycles of liquid N freeze-thawing, or no treatment. The number of cycles was increased to 40 to maximize the chances of extracting DNA from the spores. Samples were centrifuged at 12,000 × g for 10 min. Equal volumes of DNA extract were applied to a 1% agarose gel, separated by electrophoresis, and stained with ethidium bromide. For this experiment, the DNA concentration in each preparation was quantified by the PicoGreen assay described below.
Data analysis.
DNA yields quantified from gel images or PicoGreen assays were analyzed by using indicator variables in a linear regression analysis using the general linear model procedure of SAS (SAS Institute, Inc., Cary, N.C.). Treatment means were compared by using Tukey’s studentized range test (honestly significant difference [HSD]) with a 0.95 experimental error level.
Small-scale, in-the-field soil DNA extraction procedure.
Based on the results of the above experiments, a two-step method (hot-detergent and bead mill homogenization treatments) that provided the highest DNA yield from the combination of test organisms was selected to extract DNA from natural and seeded soil samples. This procedure was also miniaturized, and incubation and sample treatment times were calibrated to determine the minimum times needed.
Soil samples were sieved through a 2-mm-pore-size screen and mixed well prior to use. DNA extractions of soil samples (0.5 g [wet wt]) were conducted on three to four replicate samples for each soil. One milliliter of TENS buffer was added to each 0.5-g soil sample in a 2-ml mini-bead beater vial containing 900 mg of a mixture of glass beads (300 mg each of beads with a diameter of 710 to 1,180 μm, 425 to 600 μm, and 106 μm). The hot-detergent and bead mill homogenization procedures were conducted as described above for microbial cells and spores. The soil-bead pellet was washed once with 1 ml of TENS buffer and centrifuged as described above, and the wash supernatant was pooled with the original supernatant. Nucleic acids were precipitated by using 1/10 volume of 3 M sodium acetate (pH 5.2) and 2.5 volumes of ethyl alcohol. The pellet containing DNA was suspended in 100 to 500 μl of TE buffer or sterile water.
PicoGreen quantification of DNA extracted from soils.
Soil extracts are often contaminated with high humic acid concentrations that absorb UV and interfere with accurate quantification by UV absorbance at 260 nm. A fluorescence-based assay was developed to quantify the double-stranded DNA present in highly pigmented crude extracts (22). Samples were diluted 1/500 in 0.1× TAE, and UV absorbance in the 200- to 300-nm range was determined. Samples were further diluted to an absorbance maximum in this range of less than 0.05. An equal volume of a 200-fold dilution of PicoGreen dye (Molecular Probes, Eugene, Oreg.) was added, and samples were incubated at room temperature in the dark for 20 min. Fluorescence of the intercalated PicoGreen dye was determined in a Turner Fluorometer, using band pass filters with an excitation wavelength of 486 nm and an emission wavelength of 510 to 700 nm. DNA concentrations were determined relative to a lambda DNA standard curve. Humic acids in the samples were measured by their fluorescence (excitation wavelength of 471 nm and emission wavelength of 529 nm), using humic acid (Sigma Chemical Co.) as a reference standard.
DNA purification through G200 microcolumns.
Chromatography through a Sephadex G200 microcolumn was used to remove humic acids from soil DNA (4, 8, 26). The procedure was miniaturized to work in a microcentrifuge, to reduce processing time, and to allow simultaneous processing of several samples. It was also modified to provide better cleanup of soils containing very large amounts of humic acid. Sephadex G200 resin was equilibrated for several hours at 4°C in TE buffer (pH 7.6), and the fines were removed. For samples containing very large amounts of extracted humic acids (over 2,500 μg/g of soil), Sephadex G200 resin containing 20 mg of polyvinylpolypyrrolidone (PVPP) per ml was prepared by adding granular PVPP (Sigma Chemical) to a 2:1 (vol/vol) resin-buffer suspension. Sephadex microcolumns were prepared in 0.22-μm-pore-size Micropure Separators (catalog no. 42512; Amicon Inc., Beverly, Mass.) or 96-well coarse polypropylene filter plates (catalog no. 58812; Advanced Genetic Technology Corp., Gaithersburg, Md.). Five hundred microliters of resin-buffer suspension was applied to each microcolumn and packed to about 400 μl by centrifugation in a swinging bucket rotor at 750 × g. Up to 200 ng of crude DNA (in 50-μl maximum total volume) was pipetted slowly onto the top center of the packed column, and the column was centrifuged at 750 × g for 15 min at room temperature. An equal volume of eluate containing DNA was collected and precipitated by adding 2/3 volume of 5 M ammonium acetate with 2 volumes of 100% ethyl alcohol. The amount of DNA that could be applied to each column for maximum recovery and effective removal of humic acids was tested. DNA yields were determined from digitized images following electrophoresis through agarose gels as described above.
DNA extraction from different soils.
The extraction, quantification, and purification methods described above were used in PCR experiments to detect microbial populations in a natural soil and to determine the detection sensitivity of P. putida cells and B. globigii spores in four different soils. All soils were of neutral pH (about 7.6) but varied considerably in texture, chemical composition, and organic matter content. An Anthony fine sandy loam from New Mexico (N.Mex. soil) (78% sand, less than 1% organic matter) was collected from an agricultural field at the New Mexico State University plant research facility, near Las Cruces, N.Mex. (10). An Ohio sandy loam (Ohio soil) (1.8% organic matter; total N about 1,140 mg/kg of soil) was collected from an agricultural field in north-central Ohio. An Arizona sandy loam soil (Ariz. soil) (2.75% organic matter, about 10 mg of total N per kg of soil) was collected from a mature pinyon rhizosphere, within 1 cm of active mycorrhizal roots, in a native woodland in Coconino National Forest near Flagstaff, Ariz. An Arizona cinders soil (Ariz. cinders) was collected from a pinyon rhizosphere at Sunset Crater National Monument, Ariz. This soil is comprised of coarse, black cinders (3) but had organic matter and nitrogen concentrations similar to those of the Ariz. soil. All soils were stored at 4°C after collection.
Detection of different microbial groups in soil.
DNA was extracted from the N.Mex. soil by the above soil DNA extraction procedure or by only the hot-detergent lysis treatment to determine the effects of bead mill treatment on soil DNA yield and detection of different phylogenic groups of the soil microbial community. The extracted DNA was quantified by gel electrophoresis and purified as described above. Usefulness of the soil DNA extraction procedure or the partial procedure for community analysis was tested by PCR amplification of rRNA gene fragments of several phylogenetic groups representing different components of the soil microbial community. These groups include members with a broad range of microbial cell sizes, composition, and growth habits. The PCR primers used in the analysis and the groups they represent are listed in Table 1.
TABLE 1.
Primer pairs used in PCR to detect phylogenetic groups of native microorganisms and plant material in a N.Mex. sandy loam soil
| Organism group | Primer pair [reference(s)] | DNA sequence | Product size (bp) |
|---|---|---|---|
| Bacteria | P3MOD (31, 32) | ATTAGATACCCTDGTAGTCC | 723 |
| PC5B | TACCTTGTTACGACTT | ||
| Bacillus species and relatives | BacF (30) | AGGGTCATTGGAAACTGGG | 600 |
| BacR | CGTGTTGTAGCCCAGGTCATA | ||
| High-G+C gram-positive bacteria | ActinoF (30) | GGCCTTCGGGTTGTAAACC | 542 |
| ActinoR | CTTTGAGTTTTAGCCTTGCGGC | ||
| Streptomyces species and related taxaa | StreF (21) | GAGTTTGATCCTGGCTCAG | 1,243 |
| StrepR | GCCATTGTAGCACGTGTGCA | ||
| Fungi, protists, and green algae | NS1 (28) | GTAGTCATATGCTTGTCTC | 555 |
| NS2 | GGCTGCTGGCACCAGACTTGC | ||
| Fungi | ML7 (28) | GACCCTATGCAGCTTCTACTG | 735 |
| ML8 | TTATCCCTAGCGTAACTTTTATC | ||
| Plants | NS3 (5) | GCAAGTCTGGTGCCAGCAGCC | 597 |
| NS4 | CTTCCGTCAATTCCTTTAAG |
Also amplifies members of the firmicutes, Proteobacteria, planctomycetes, chlamydia, and other relatives (21).
Soil seeding experiments.
Each of the four soils was seeded with either P. putida cells or B. globigii spores. While Pseudomonas species are common in many soils, those carrying the TOL plasmid are not. Therefore, we were able to detect this specific Pseudomonas isolate seeded into soil samples using PCR primers that amplify sequences from the toluene degradation genes. B. globigii is also not commonly found in soils. The N.Mex. soil was seeded with either P. putida cells or B. globigii spores to determine the detection limit in a soil sample having low background DNA and humic acid content. P. putida cells were seeded at 4 × 107, 4 × 105, 4 × 103, 4 × 102, and 0 cells/g of soil in a volume of 100 μl/g of soil. Dry B. globigii spores (preparation 93; low extracellular DNA) were suspended in solutions of 0.5% Tween 20 and seeded into the N.Mex. soil at concentrations of 2.5 × 107, 2.5 × 106, 2.5 × 105, 2.5 × 104, 2.5 × 103 and 0 spores/g of soil in 100-μl delivery volume. The Tween 20 did not affect extraction efficiency but facilitated accurate microscopic counting and more-uniform distribution of spores in the suspensions. The same concentrations of the two bacterial target organisms were seeded into separate samples of the Ariz. and Ohio soils and into Ariz. cinders to compare the effects of soil background DNA and humic acids on PCR detection limits. Seeded soils were mixed well and allowed to equilibrate for 20 min before the DNA was extracted, quantified, and purified as described above. DNA was extracted as two 0.5-g samples and then pooled.
PCR primers for the detection of P. putida cells and B. globigii spores seeded into soils.
The P. putida target was detected by using the PCR primer pair XylR-F1 (TCGCTAAACCAACTGTCA) and XylR-R1 (GCACCATAAGGAATACGG). This primer pair was designed to amplify a 259-bp fragment of the xylR gene on the pWW0 plasmid. This primer pair detects as little as 10 to 100 fg (about 2 to 20 cells) of P. putida DNA in a standard PCR assay (where 1/10 of the reaction mixture is detected on ethidium bromide-stained gels [8]). The B. globigii target was detected by using the PCR primer pair BG007F (GGGCACAACCTTTATCCA) and BG007R (ACCTTCTTTTCTGTGGGC). This primer pair amplifies a 301-bp fragment of the chromosome that contains no known open reading frames. Detection using this primer pair is specific for B. globigii, and this primer pair does not amplify DNA from the closely related organisms, Bacillus subtilis, Bacillus anthracis, Bacillus cereus, Bacillus thuringiensis, Bacillus mycoides, or species outside the Bacillus genus (8). In standard PCR assays, this primer pair detects B. globigii DNA with a sensitivity of 10 to 100 fg (2 to 20 cells) (8).
PCR assays to detect specific microbial targets.
PCR mixtures contained 50 mM KCl, 10 mM Tris (pH 8.3), 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, 2 mM each of the four deoxynucleoside triphosphates, 1 μM (each) oligonucleotide primer, 2.5 U of Taq DNA polymerase (Perkin-Elmer Inc., Branchburg, N.J.), and 1 pg to 100 ng of template DNA in a total reaction volume of 100 μl. Control PCRs included DNA of the target organism (10 pg), DNA of the target organism added to soil template DNA (to control for negative results due to contaminants in the soil DNA sample), and a negative control containing no template DNA. PCR assays were conducted in a Perkin-Elmer 480 or Perkin-Elmer 9600 thermal cycler, using the following cycling conditions: 2 min at 94°C; 35 cycles, with 1 cycle consisting of 1 min at 55°C, 1 min at 72°C, and 0.5 min at 94°C; 1 min at 55°C; and 5 min at 72°C.
Amplified PCR products (10 μl of each 100-μl reaction mixture) were analyzed by agarose gel electrophoresis and staining with ethidium bromide. Analysis of large amplicons (500 to 1,000 bp) was through agarose gels containing 1.5% (wt/vol) SeaKem agarose (FMC Bioproducts). Analysis of smaller products was through gels containing 1% (wt/vol) SeaKem agarose plus 2% (wt/vol) NuSieve agarose (FMC Bioproducts). Agarose gels were prepared and run in 1× TAE buffer. PCR assay samples on the gels were bracketed by a 1-kb DNA molecular size ladder (New England Biolabs, Beverly, Mass.) to estimate the size of the amplified product. Results were scored as positive or negative by ability to visually detect a product when 1/10 of the PCR product was applied to the gel.
Effects of template concentration on PCR success.
To determine the best soil DNA template concentration to use for optimal detection sensitivity, several template concentrations were generated from quantified soil DNA and tested in PCR analysis using the P3MOD-PC5B primer pair (Table 1) to amplify any bacterial genes coding for 16S rRNA (rDNA) present in the sample. The two template concentrations that produced the largest amount of amplified 16S rDNA product were then used in PCR with species-specific primer pairs to determine the detection limit for the sample.
DNA extraction and PCR detection of dilute and degraded samples.
Samples collected for forensic analysis are sometimes degraded and may be very dilute. Effectiveness of the soil DNA extraction procedure in recovering DNA from dilute bacterial soil suspensions and from spores and DNA damaged by autoclaving was tested with B. globigii spores. Tenfold dilutions from 5 × 109 to 2.5 × 102, and 0 B. globigii spores/ml were divided into two 0.5-ml sets. One set of dilutions was autoclaved for 30 min using slow exhaust prior to extracting the DNA. Extracted DNA was precipitated using ethyl alcohol, with 10 μg of yeast tRNA per ml added as a carrier to aid precipitation. The DNA was suspended in 100 μl of TE buffer, and 10 μl of each sample was used as a template in PCR assays.
RESULTS AND DISCUSSION
DNA extraction methods for maximum yield from microbial cells and spores.
Hot-detergent treatment, freeze-thaw cycles, and bead mill homogenization were selected as cell and spore disruption techniques that were potentially amenable to in-the-field use for microbial DNA preparation. These three methods, alone or in all possible combinations, were tested for their ability to release genomic DNA from three microorganisms that represent very different cell and spore types to identify a single procedure for efficient DNA extraction from multiple organisms. Figure 1 illustrates the relative amounts of DNA that were extracted from P. putida vegetative cells, two preparations of B. globigii endospores, and F. moniliforme conidia by using different method combinations. All extraction schemes comprised of these three methods resulted in large DNA, generally between 12 and 24 kb. The different DNA extraction methods and combinations resulted in vastly different amounts of released DNA from the three target microorganisms. This can be seen in Fig. 1 as the different DNA band intensities in the lanes of a specific panel.
P. putida cells could be lysed by most of the methods and method combinations that were tested. The hot-detergent treatment, the liquid N freeze-thaw treatment, and bead mill homogenization all released DNA from P. putida cells. Analysis of variance of the effects of the three treatments on DNA yield indicated that each treatment had a significant effect (Table 2). However, the three methods released different amounts of DNA from equal amounts of starting material. A DNA yield of 1.0 (±0.3) μg/ml was obtained from P. putida cells simply by suspending them in the TENS buffer at room temperature (probably by action of the SDS [Fig. 1, lane 16]). Bead mill homogenization and liquid N freeze-thaw treatments had similar effects and increased the DNA yield to an average of 9.4 (±5.1) μg/ml. The hot (70°C)-detergent treatment was the most effective treatment for P. putida cells, and DNA yields from this treatment averaged 20.8 (±6.8) μg/ml. The liquid N freezing temperature appeared essential to success of the freeze-thaw cycle treatment. The −20°C freeze-thaw treatment was less effective than the liquid N freeze-thaw treatment (e.g., compare lanes 10 and 12 of Fig. 1), and the 4°C cycling with heating had little effect (e.g., compare lanes 14 and 16).
TABLE 2.
Results of testing the effects of three DNA extraction methods on DNA yields from P. putida cells, B. globigii endospores, and F. moniliforme conidiaa
| Organism | Probabilityb
|
||
|---|---|---|---|
| 70°C incubation in TEN buffer | Three freeze-thaw cycles | Bead mill homogenization | |
| P. putida | 0.002 | 0.029 | 0.047 |
| B. globigii prepn 93 | NSD | NSD | 0.0001 |
| B. globigii prepn 95 | NSD | NSD | 0.0001 |
| F. moniliforme | NSD | NSD | 0.0002 |
The three DNA extraction methods were described in detail in Materials and Methods. Data were analyzed by using indicator variables in the general linear model as described in the text.
Probability of a greater F value (P values) when testing whether the extraction procedure had an effect on the DNA yields as described by SAS type III sums of squares. P values greater than 0.05 were considered significant. NSD, values that were not significantly different (P values of 0.18 to 0.95).
DNA extraction comparisons were conducted on two dry spore preparations of B. globigii. The hot-detergent treatment and the freeze-thaw treatments had no effect on B. globigii spores, and the bead mill homogenization treatment was the only effective method of extracting DNA from the two spore preparations. This result can be seen visually in the two B. globigii panels of Fig. 1, by comparing the relative amounts of DNA in alternate lanes (odd-numbered lanes included a bead mill homogenization step; even-numbered lanes did not), and by comparing the P values for the different treatments in Table 2. DNA extraction schemes that included the bead mill homogenization step significantly increased DNA yields (Table 3). Yields achieved with the bead mill averaged 2.53 μg/ml spore suspension for preparation 93 and 1.30 μg/ml spore suspension for preparation 95 (Table 3).
TABLE 3.
Effect of bead mill homogenization on B. globigii and F. moniliforme sporesa
| Treatment | DNA yield (μg/ml)b
|
||
|---|---|---|---|
| B. globigii prepn 93 | B. globigii prepn 95 | F. moniliforme | |
| Bead mill | 2.53 (0.94) A | 1.30 (0.37) A | 0.74 (0.35) A |
| No bead mill | 0.18 (0.36) B | 0.36 (0.09) B | 0.06 (0.09) B |
DNA yields were quantified by densiometry measurements of DNA bands on calibrated agarose gels as described in the text.
Values in the table are means of eight values, with the standard deviations in parentheses. Means followed by different letters are significantly different at the P = 0.05 level by using Tukey’s studentized range (HSD) test.
The results with F. moniliforme conidia were similar to those obtained for the B. globigii spores. The hot-detergent and freeze-thaw treatments had little effect on F. moniliforme conidia. An example of this result can be seen in Fig. 1. Analysis of variance of the quantified DNA yields from F. moniliforme conidia indicates that only the bead mill provided significantly increased yields (Table 2). Comparison of the quantified DNA yields (Table 3) illustrates that DNA yields were about 12 times higher when the bead mill homogenization treatment was included.
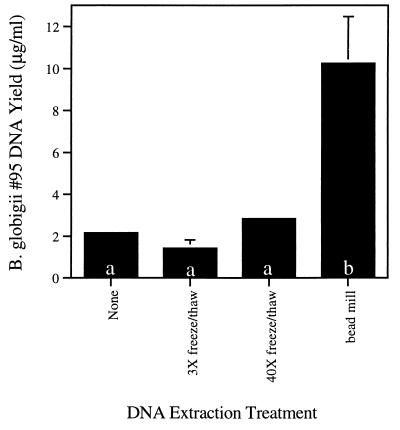
Freeze-thaw cycles have been used in previous studies as a component of procedures for DNA preparation from spore-forming organisms and soils (2, 6, 14, 25), and the ineffectiveness of this technique in rupturing Bacillus or Fusarium spores was surprising. To further compare the effectiveness of liquid N freeze-thaw cycles with bead mill homogenization, we increased the number of freeze-thaw cycles to 40 in attempts to improve the extraction potential of freeze-thaw cycles. Results from this experiment are presented in Fig. 2. Even forty cycles of freeze-thaw were ineffective on B. globigii spores. It is probable that previous reports of success with freeze-thaw cycles, determined by successful PCR or hybridization detection, were due to the presence of extracellular DNA in the spore preparation (7), not spore lysis. Our B. globigii spore preparation 95 contained measurable amounts of extracellular DNA (2.15 ± 0.04 μg/109 spores determined by the PicoGreen assay), and presumably other B. globigii spore preparations could contain extracellular DNA as well. Bacillus spores are impervious to freeze-drying and 85°C heat (19). Using microscopic counts, Moré et al. (11) have shown that 94% of Bacillus subtilis endospores survived a freeze-thaw treatment. Our results confirm these findings and illustrate that only the bead mill disruption effectively releases large amounts of DNA from B. globigii and F. moniliforme spores.
FIG. 2.
Amounts of DNA extracted by three DNA extraction methods. DNA was extracted from the B. globigii 95 endospore suspension (109/ml) by either three cycles of liquid nitrogen freeze-thawing (3× freeze/thaw), 40 cycles of liquid nitrogen freeze-thawing (40× freeze/thaw), or bead mill homogenization and quantified by PicoGreen assay as described in the text. Values are averages of three replicate extractions, and error bars illustrate the standard deviations of the mean values. Standard deviations for the no treatment and 40× freeze-thaw treatment are too small to be visualized graphically (0.04 and 0.02, respectively). Bars with different letters (a or b) indicate treatments that were significantly different by Tukey’s studentized range test (HSD) as described in the text. The B. globigii spore preparation 95 contained about 2 μg of extracellular DNA per ml that was primarily low-molecular-weight, sheared DNA. In this comparison, only the bead mill homogenization extracted significant amounts of large-molecular-size DNA (≥24 kb) from the spores.
On the basis of the DNA extraction comparisons presented here for three target microorganisms, we selected a procedure for field use consisting of the hot-detergent lysis treatment, followed by bead mill homogenization (see “Small-scale, in-the-field soil DNA extraction procedure” in Materials and Methods). Hot-detergent lysis effectively released DNA from bacterial cells (P. putida) and was easy to conduct in a small-scale manner in the field. The bead mill homogenization relied only on a small mini-bead beater (10 in. long by 4.75 in. wide by 7.75 in. tall; BioSpec Products, Inc.) and was the most effective treatment for microbial spore disruption. In addition, conducting the bead mill step after the hot-detergent lysis step did not significantly shear DNA that had been released by the hot-detergent lysis (e.g., compare lanes 1 to 7 to lane 8 in Fig. 1).
Small-scale DNA extraction, purification, and quantification for field use.
Our goal was to develop a small-scale, field, DNA preparation procedure that provided high yields of purified DNA from a wide range of microbial cell and spore types that could be used as template in PCR assays to detect target microorganisms. The procedures needed to be small-scale, rapid, and could not rely on labile or hazardous chemicals that required special storage or on bulky laboratory equipment.
Variations of procedures employing either hot-detergent treatment and bead mill homogenization have been used to extract DNA from cultivated bacteria and indigenous soil bacteria (7, 11, 13, 17, 23, 33). In preliminary comparisons of several DNA sample preparation procedures (8), a combination of these two techniques plus freeze-thawing released more DNA from a soil sample than methods using lysozyme (4, 6, 24, 26) or sonication (14). The soil DNA extraction procedure described here differs substantially from the original (13, 25) and other modified presentations of the hot-detergent lysis and bead mill techniques (11, 23, 33). Buffers and reagents were changed from the originally presented methods, and minimum incubation and optimum (maximum yield with little shearing) homogenization times were determined. Purification steps in the original procedures that we found to substantially reduce the DNA yield (e.g., gel electrophoresis, glass-fines precipitation) or that had no overall effect on PCRs (e.g., phenol-chloroform extraction) were removed. The procedures were miniaturized and optimized to provide maximum DNA yields from several soil, water, and aerosol samples with minimal shearing and to provide maximum DNA yields from multiple microorganisms (8). DNA yields using this procedure were constant between replicate extractions (e.g., see Table 4).
TABLE 4.
PCR detection of P. putida mt-2 cells and B. globigii spores in soils that differ in humic acid and background DNA concentrations
| Soil type | Amt (μg) of soil DNAa | Amt (μg) of humic acidsb | Organism | Visible amplicon with P. putida CFU or B. globigii spores seeded per g of soilc
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 107 | 106 | 105 | 104 | 103 | 0 | ||||
| N. Mex. soil | 0.18 (0.006) | 49 (10) | P. putida | ++ | NT | ++ | ++ | ++ | ---- |
| B. globigii | ++ | ++ | ++ | ++ | ++ | ---- | |||
| Ohio soil | 21.3 (5.6) | 908 (440) | P. putida | ++ | NT | ++ | ++ | ---- | ---- |
| B. globigii | ++ | ++ | ++ | ++ | ---- | ---- | |||
| Ariz. soil | 13.3 (2.6) | 2,200 (762) | P. putida | ++ | NT | ++ | ++ | ---- | ---- |
| B. globigii | ++ | ++ | ++ | ++ | ---- | ---- | |||
| Ariz. cinders | 14.6 (5.4) | 1,933 (614) | P. putida | ++ | NT | ++ | ---- | ---- | ---- |
| B. globigii | ++ | ++ | ++ | ---- | ---- | ---- | |||
Per gram (wet weight) of soil. Values are means of three to four replicate extractions, and numbers in parentheses are standard deviations. DNA was quantified by a PicoGreen spectrophotometric assay as described in Materials and Methods.
Per gram (wet weight) of soil. Values are means of three to four replicate extractions, and numbers in parentheses are standard deviations. Humic acids were quantified by UV spectroscopy as described in Materials and Methods. Values presented are the amounts of humic material present in crude extracts prior to removal by Sephadex G200 spin microcolumns.
P. putida cells were seeded at 4 × 107, 4 × 105, 4 × 104, and 4 × 103 CFU per g of soil, and B. globigii endospores were seeded at 2.5 × 107, 2.5 × 106, 2.5 × 105, 2.5 × 104, and 2.5 × 103 spores per g of soil. Results are the presence (++) or absence (----) of a visible amplicon, detected by ethidium bromide staining 1/10 of the PCR mixture in an agarose gel. All reactions were positive for amplification of general bacterial and fungal rDNA fragments using the P3MOD-PC5B and NS1-NS2 primer pairs, respectively, at template concentrations of 100 pg and 1 ng (data not shown), indicating that they were sufficiently free of inhibitors to support PCR. NT, not tested.
The bead mill homogenization step used in our soil DNA extraction procedure differs in two substantive ways from previously presented uses of this technique. First, three sizes of glass beads were combined into a single reaction mixture to provide more-efficient rupture of a wider range of microbial cell and spore sizes. Second, the homogenization time and intensity were empirically calibrated with multiple soils and other environmental samples to obtain maximum yield without severely shearing the DNA (data not shown). The original bead mill procedure resulted in sheared DNA that was 10 kb to less than 500 bp in size (9, 13). For the soils and the microorganisms presented here, a 3-min homogenization at 5,000 rpm (mini-bead beater, Bio-Spec) provided the highest DNA yields without significantly shearing the DNA. DNA extracted from soil was predominantly large DNA (Fig. 3, lane 3, ≥24-kb fragments). Although target microorganisms can be detected from sheared DNA using PCR assays and closely spaced primers, we required extraction procedures that damaged the extracted DNA as little as possible to maximize the potential detection sensitivity.
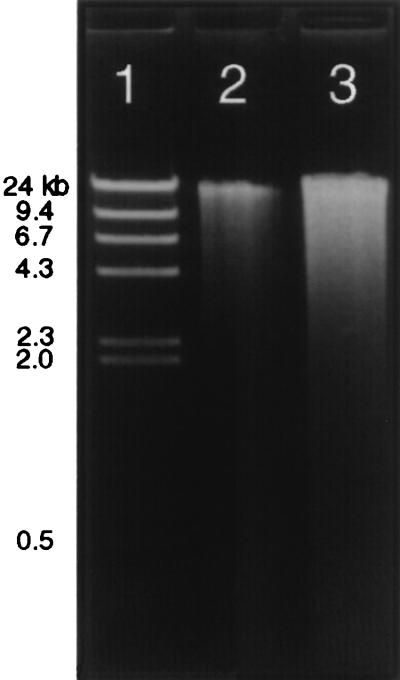
FIG. 3.
DNA extracted from soil. Equal amounts of a N.Mex. soil were extracted by the small-scale soil DNA extraction procedure described in Materials and Methods (lane 3) or a variation of this procedure omitting the bead mill homogenization step (lane 2). Lane 1 contains HindIII-digested lambda DNA molecular size markers. Equal volumes of extract were applied to all the lanes in the gel. DNA fragments were predominantly ≥24 kb in size. DNA yields were determined by the PicoGreen assay as described in the text. Yields obtained when the bead mill step was omitted from the procedure were half that of the combined procedure (0.1 versus 0.2 μg/g of soil).
A simple procedure was needed to purify the extracted DNA from humic acids and other contaminants that coextract with DNA from soils to support in-the-field preparation of DNA samples. Previously reported methods to remove humic acid contaminants from environmental DNA have multiple steps, are laboratory equipment- and labor-intensive, and result in loss of extracted material (see references 6, 13, 18, 20, 23–25, 36, and 37 for examples). They also use chemicals such as phenol, chloroform, guanidinium hydrochloride, cesium chloride, and butanol that require special storage, handling, and disposal. We found that purification of crude extracted DNA through Sephadex G200 spin microcolumns removed all visible humic acids, and over 90% of the applied DNA was consistently recovered from the microcolumns (data not shown). The original syringe column procedure (4, 26) was simplified and miniaturized for operation in prepackaged Micropure column housings (for field application) or 96-well plates for simultaneous processing many samples in the laboratory. Percent DNA recovery and successful humic acid removal were assessed when different amounts of crude DNA (1 μg to 10 pg) were applied to the 500-μl column. Application of 200 ng or less DNA applied to each 500-μl column in a loading volume of 10 to 50 μl provided the best percent DNA recovery and humic acid removal for the four soils presented here and for over a dozen other soils (8). In addition, use of the Micropure or 96-well formats with 0.2-μm-pore-size filters supporting the resin effectively removes any viable bacteria and spores from the sample, producing a sterile DNA solution free of microbial contamination. The Sephadex G200 spin microcolumns provided simple, effective removal of humic acid contaminants in a single step from DNA extracted from soil. PVPP added to the column buffer provided improved removal of humic acids from very highly contaminated samples (8).
We found that to achieve the most effective purification using the Sephadex G200 microcolumns and the highest detection sensitivity using PCR (described below), it was necessary to estimate the concentration of extracted DNA at two points during sample preparation. The first point was to estimate the amount of DNA present in the crude extract so that an appropriate amount of DNA sample could be applied to the Sephadex G200 purification column. The second point was after purification prior to PCR to determine the amount of DNA template to add to the PCR mixtures. In a laboratory setting, DNA quantification of crude extracts is typically accomplished by agarose gel electrophoresis of the DNA along with standards of known concentration and ethidium bromide staining. However, this is not applicable to field use. To quantify DNA in both the crude and purified DNA samples for small-scale, field applications, a sensitive PicoGreen assay was developed and portable equipment was manufactured (22), to be used in conjunction with the DNA extraction and purification methods presented here.
The sample preparation procedures presented here represent a significant simplification of widely used methods for DNA extraction, purification, and quantification from environmental samples. They provide efficient extraction of DNA from both vegetative cells and spores of a variety of microorganisms, simple effective cleanup in a single step using Sephadex G200 microcolumns, and quantification of the amount of extracted DNA for purification and PCR analysis using the PicoGreen assay. By scaling down the sample volume, multiple samples could be easily be processed for PCR in less than 2 h. The entire extraction and purification procedure is conducted in 1.5- to 2-ml plastic tubes. The extraction tubes can be prepared in advance to contain dry buffer ingredients and glass beads and can be stored at room temperature for several months. The extraction and purification can easily be accomplished outside the laboratory using a battery-powered (automobile battery, for example) heat block, mini-bead mill homogenizer, and small microcentrifuge. Portable, battery-powered equipment for DNA quantification has been developed for field use with the above extraction and purification procedures (22). Using these simple DNA preparation methods, coupled with sensitive micro-PCR techniques (12, 34), it is now feasible to use DNA-based detection schemes in a field-deployable kit to detect and monitor release of microorganisms in the environment.
The ability to simultaneously extract DNA from many types of microorganisms by using one simple, small-scale procedure in the field is desirable for many applications in microbial ecology. It is especially useful for unbiased analysis of microbial communities in the environment and for simultaneous detection of a variety of target microorganisms in environmental samples collected during forensic investigations. These methods have broad application to agricultural, industrial, forensic, and investigative needs to monitor microorganisms in the environment.
PCR detection of microbial targets in soils.
The DNA extraction and purification procedures described above were used to prepare template DNA for PCR assays to detect target microbial DNAs in soils. First, using a matrix of 16S rRNA gene primer pairs (Table 1), we determined in a broad sense the microbial groups that were represented in DNA extracted from the N.Mex. soil. Second, we determined the sensitivity with which P. putida cells and B. globigii spores could be detected when seeded into four different soils, and we identified factors that greatly affect the detection sensitivity when using DNA from PCR assays to detect target microogranisms in soil.
Detection of microbial groups in soil using 16S rRNA gene primers.
PCR detection of several microbial groups native to soils was tested with DNA extracted by the soil DNA extraction procedure or by just the hot-detergent treatment part of the procedure. All target microbial groups (Table 1) were easily detected in standard PCR assays using either 1 or 0.1 ng as template DNA when the combined procedure was used to extract DNA from the N.Mex. soil. A product was not detected when the NS3-NS4 primer pair for plant materials was used. Detection of the fungal groups with the NS1-NS2 and ML7-ML8 primers was very faint when the bead mill extraction was omitted from the procedure (data not shown).
The combination of hot-detergent treatment and bead mill homogenization using multiple sizes of beads provided effective extraction of DNA from microbial cells and spores as well as plant material in soil samples. This procedure is therefore useful to reduce sampling biases inherent in DNA-based diversity analysis of soil microbial communities. We and others have found that the total amount of DNA extracted from soil was greater when a grinding or bead mill step was included in the procedure (9, 11, 37) (Fig. 3). Our PCR results indicate that including the bead mill method improved PCR detection of native fungal and plant materials in soil, and the observed increase in total DNA yield may be due in part to better extraction of spore-forming organisms residing in the soil.
Effect of soil DNA template concentration on PCR amplification.
DNA extracted and purified from the four soils used in bacterial seeding experiments was tested as template in PCR amplifications at different concentrations (1 pg to 100 ng per reaction mixture) to determine the effects of different template concentrations on the amount of PCR product obtained. Figure 4 illustrates the effects of different DNA template concentrations of DNA extracted from the N.Mex. soil on the amount of amplified PCR product obtained using the P3MOD-PC5B primer pair (Table 1). The amount of PCR product that could be obtained varied tremendously with the amount of DNA template used to initiate the reaction (Fig. 4). DNA template was titrated in similar tests for all four of the soils examined here, and template concentrations of 100 pg to 1 ng per reaction mixture yielded the most abundant PCR product for all four soils (data shown only for one soil in Fig. 4). This result illustrates that quantification of the extracted, purified DNA and subsequent determination of the optimum amount of soil template DNA used to initiate PCRs are critical to achieving the best PCR detection sensitivity of target microorganisms in soil samples.
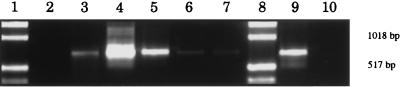
FIG. 4.
Effect of soil DNA template concentration on PCR amplification success. Different amounts (1 pg to 100 ng) of purified DNA from the N.Mex. soil was added to standard PCR mixtures as described in the text, and the P3MOD-PC5B primers were used to amplify native bacterial DNA present in the soil DNA. One-tenth of the PCR mixture was applied to an agarose gel, and the amplified fragment was separated by electrophoresis and stained with ethidium bromide. Lanes 1 and 8 contain molecular size markers. Amplified PCR products resulting from 100 ng, 10 ng, 1 ng, 100 pg, 10 pg, and 1 pg of soil DNA template are shown in lanes 2 through 7, respectively. Lanes 9 and 10 contain the results of positive- and negative-control reactions as described in the text. Addition of more than 1 ng of soil DNA template resulted in considerable inhibition of the PCR, as indicated by substantially reduced product formation. Addition of less than 100 pg of soil DNA template resulted in increasingly less amplified product.
Factors affecting detection of P. putida cells and B. globigii spores in soils.
DNA was extracted, purified, and quantified from four soils each seeded with either P. putida cells or B. globigii spores to compare detection ability of target bacterial species in different environments and to identify environmental factors that affect PCR detection sensitivity in soils. The amounts of background DNA and humic acids present in different seeded soils were measured to determine the effect these characteristics had on detection sensitivity. The four soils differed greatly in their organic matter content, and the amount of DNA that could be extracted from them ranged from 0.18 to 21.3 μg of DNA/g of soil. Extracted humic acids differed dramatically in quantity (Table 4) and in pigmentation. The N.Mex. soil extracts were light tan, the Ohio and Ariz. soil extracts were brown, and the Ariz. cinders extract was black.
The sensitivity with which a seeded bacterial target could be detected in soil was affected by the amount of background DNA present in the soil sample. The N.Mex. soil contained small amounts (0.18 μg of DNA/g of soil) of background DNA, and as few as 4 × 103 P. putida cells/g of soil or 2.5 × 103 B. globigii spores (the lowest concentration tested) were detected in this soil (Table 4). One picogram of the total extracted DNA that was used to initiate a PCR would theoretically contain about 22 copies of the P. putida genome or about 14 copies of the B. globigii genome. Thus, detection sensitivity in this soil was comparable to using pure target bacterial DNA as the template (2 to 20 copies of target DNA in a PCR mixture), indicating that the extraction and purification procedures employed here were highly effective in releasing DNA. This result also indicates that with this soil the amount of background DNA in the sample did not interfere with the PCR assay and that the humic acid and other contaminants had been sufficiently removed from the DNA template. In contrast, the detection limit for both bacterial targets was 10- to 100-fold less sensitive in the other three soils. These soils contained at least 100-fold-more background DNA than the N.Mex. soil (Table 4). The effect of diluting the target DNA into a larger pool of sample DNA is to lower the achievable detection sensitivity.
Although the four soils differed greatly in the amount and types of extracted humic acids, a single passage through a Sephadex G200 microcolumn appeared to sufficiently purify DNA of each soil to support PCR. Column elutes of DNA from all four soils were clear, and PCR amplification of 100 pg and 1 ng of each soil DNA template was positive with the P3MOD-PC5B primer pair (e.g., Fig. 4). The achieved detection sensitivity with primers specific for the two bacterial targets was 10-fold less sensitive in the Ariz. cinders than in the Ariz. soil, even though both soils contained similar extractable DNA and humic acid concentrations (Table 4). This may be attributed to additional unpigmented contaminants present in the unusual cinders sample that were not removed by Sephadex G200 chromatography or ethanol precipitation, to differences in DNA extraction efficiency in the soil samples due to DNA binding to the soil matrix or other factors, or to limitations in our ability to detect faint PCR products using the ethidium bromide-stained gels.
Environmental samples collected during forensic investigations are often difficult to obtain and are therefore valuable. Target microbial DNA in environmental samples collected during forensic investigations may be present in very low concentrations and/or partially degraded or compromised by chemical treatment. It also may be present in very low concentrations. To determine the efficiency with which the soil DNA extraction procedure could release DNA from increasingly dilute solutions, DNA was extracted from suspensions of B. globigii spores over a wide concentration range from 5 × 109 to 250 spores/ml. Standard PCR assays detected as few as 250 spores/ml (about 25 spores in the PCR mixture) using the BG007 primer pair (Fig. 5). This was the lowest dilution tested and is comparable to the detection limit of pure B. globigii DNA using this primer pair. This result indicates that the extraction methods effectively released DNA from a wide range of spore concentrations, including very dilute suspensions. Autoclaving B. globigii spores prior to extraction sheared the DNA into smaller fragments and decreased detection sensitivity by 1 to 2 orders of magnitude (Fig. 5).
FIG. 5.
PCR detection of DNA from B. globigii preparation 93 spores (low extracellular DNA) in dilute suspensions and from autoclaved samples. DNA was extracted from B. globigii spore suspensions as described in the text and used as template DNA in PCRs. The BG007F-BG007R primer pair amplified a 301-bp product, which was separated by electrophoresis in an agarose gel and stained with ethidium bromide as described in the text. Values above each gel lane are the number of spores/milliliter in the original suspension. If extraction were 100% efficient, about 25 copies of spore DNA would be present in the 250-spore/ml PCR mixture. Autoclaving the spore suspension prior to extraction decreased detection sensitivity.
The results presented here clearly demonstrate that several factors are important to achieve sensitive PCR detection of specific target microorganisms in environmental samples. The purity of the DNA from contaminants (data not shown), the amount of template DNA added to the PCR mixture (Fig. 4), the amount of background DNA present in the soil sample (Table 4), and the condition of the extracted DNA (Fig. 5) can significantly affect the achieved detection limit. It is therefore important to determine the range of detection limits one can expect from different environments. The variability in detection sensitivity due to uncontrollable factors, such as background DNA and inhibiting materials that coextract with the DNA, demonstrates the need for internal controls in all samples. Including a spiked organism in a suspect sample will reveal the detection limit that can be achieved with the sample. For investigations involving environmental samples, knowledge of the effects of these factors on detection success or failure is essential to interpreting results.
ACKNOWLEDGMENTS
We thank Dan Martin for providing B. globigii spores and Craig Liddell for providing the N.Mex. soil. We thank Lawrence Ticknor for assistance with the statistical analysis, and we thank John Dunbar, Susan Barns, and Arlene Wise for helpful comments on the manuscript.
This work was supported in part by and was conducted under the auspices of the U.S. Department of Energy.
REFERENCES
- 1.Burlage R S, Hooper S W, Sayler G S. The TOL (pWW0) catabolic plasmid. Appl Environ Microbiol. 1989;55:1323–1328. doi: 10.1128/aem.55.6.1323-1328.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carl M, Hawkins R, Coulson N, Lowe J, Robertson D L, Nelson W M, Titball R W, Woody J N. Detection of spores of Bacillus anthracis using the polymerase chain reaction. J Infect Dis. 1992;165:1145–1148. doi: 10.1093/infdis/165.6.1145. [DOI] [PubMed] [Google Scholar]
- 3.Cobb N S, Mopper S, Gehring C A, Caouette M, Christensen K M, Whitham T G. Increased moth herbivory associated with environmental stress of pinyon pine at local and regional levels. Oecologia. 1997;109:389–397. doi: 10.1007/s004420050098. [DOI] [PubMed] [Google Scholar]
- 4.Erb R W, Wagner-Döbler I. Detection of polychlorinated biphenyl degradation genes in polluted sediments by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1993;59:4065–4073. doi: 10.1128/aem.59.12.4065-4073.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardes M, Bruns T. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 6.Herrick J B, Madsen E L, Batt C A, Ghiorse W C. Polymerase chain reaction amplification of naphthalene-catabolic and 16S rRNA gene sequences from indigenous sediment bacteria. Appl Environ Microbiol. 1993;59:687–694. doi: 10.1128/aem.59.3.687-694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Hopkins T R. Physical and chemical cell disruption for the recovery of intracellular proteins. In: Seetharam R, Sharma S K, editors. Purification and analysis of recombinant proteins. New York, N.Y: Marcel Dekker, Inc.; 1991. [PubMed] [Google Scholar]
- 7.Johns M, Harrington L, Titball R W, Leslie D L. Improved methods for the detection of Bacillus anthracis spores by the polymerase chain reaction. Lett Appl Microbiol. 1994;18:236–238. [Google Scholar]
- 8.Kuske, C. R., K. K. Hill, and P. J. Jackson. Unpublished data.
- 9.Leff L G, Dana J R, McArthur J V, Shimkets L J. Comparison of methods of DNA extraction from stream sediments. Appl Environ Microbiol. 1995;61:1141–1143. doi: 10.1128/aem.61.3.1141-1143.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liddell, C. (New Mexico State University). Personal communication.
- 11.Moré M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Northrup M A, Benett B, Hadley D, Landre P, Lehew S, Richards J, Stratton P. A miniature analytical instrument for nucleic acids based on micromachined silicon reaction chambers. Anal Chem. 1997;70:918–922. doi: 10.1021/ac970486a. [DOI] [PubMed] [Google Scholar]
- 13.Ogram A, Sayler G S, Barkay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 14.Picard C, Ponsonnet C, Paget E, Nesme X, Simonet P. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1992;58:2717–2722. doi: 10.1128/aem.58.9.2717-2722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porteus L R, Armstrong J L. A simple mini-method to extract DNA directly from soil for use with polymerase chain reaction amplification. Curr Microbiol. 1993;27:115–118. doi: 10.1007/BF01570868. [DOI] [PubMed] [Google Scholar]
- 16.Porteus L R, Armstrong J L, Seider R J, Watrud L S. An effective method to extract DNA from environmental samples for polymerase chain reaction amplification and DNA fingerprint analysis. Curr Microbiol. 1994;29:301–307. doi: 10.1007/BF01577445. [DOI] [PubMed] [Google Scholar]
- 17.Reif T C, Johns M, Pillai S D, Carl M. Identification of capsule-forming Bacillus anthracis spores with the PCR and a novel dual-probe hybridization format. Appl Environ Microbiol. 1994;60:1622–1625. doi: 10.1128/aem.60.5.1622-1625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selenska S, Klingmuller W. DNA recovery and direct detection of Tn5 sequences in soil. Lett Appl Microbiol. 1991;13:24–31. doi: 10.1111/j.1472-765x.1991.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 19.Setlow P. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu Rev Microbiol. 1995;49:29–54. doi: 10.1146/annurev.mi.49.100195.000333. [DOI] [PubMed] [Google Scholar]
- 20.Smalla K, Cresswell N, Mendonca-Hagler L C, Wolters A, van Elsas J D. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol. 1993;74:78–85. [Google Scholar]
- 21.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 22.Stark, P. C., C. R. Kuske, and K. Mullen. Unpublished data.
- 23.Steffan R J, Goksoyr J, Bej A K, Atlas R M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988;54:2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai Y-L, Olson B H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volossiouk T, Robb E J, Nazar R N. Direct DNA extraction for PCR-mediated assays of soil organisms. Appl Environ Microbiol. 1995;61:3972–3976. doi: 10.1128/aem.61.11.3972-3976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols, a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 29.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson, K. H. (Duke University). Personal communication.
- 31.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson K H, Blitchington R B, Green R C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wipat A, Wellington E M H, Saunders V A. Streptomyces marker plasmids for monitoring survival and spread of streptomycetes in soil. Appl Environ Microbiol. 1991;57:3322–3330. doi: 10.1128/aem.57.11.3322-3330.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woolley A T, Hadley D, Landre P, deMello A J, Mathies R A, Northrup M A. Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device. Anal Chem. 1996;68:4081–4086. doi: 10.1021/ac960718q. [DOI] [PubMed] [Google Scholar]
- 35.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young C C, Burghoff R L, Keim L G, Minak-Bernero B, Lute J R, Hinton S M. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA from soils. Appl Environ Microbiol. 1993;59:1972–1974. doi: 10.1128/aem.59.6.1972-1974.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]