Abstract
Bacteria that metabolize p-nitrophenol (PNP) oxidize the substrate to 3-ketoadipic acid via either hydroquinone or 1,2,4-trihydroxybenzene (THB); however, initial steps in the pathway for PNP biodegradation via THB are unclear. The product of initial hydroxylation of PNP could be either 4-nitrocatechol or 4-nitroresorcinol. Here we describe the complete pathway for aerobic PNP degradation by Bacillus sphaericus JS905 that was isolated by selective enrichment from an agricultural soil in India. Washed cells of PNP-grown JS905 released nitrite in stoichiometric amounts from PNP and 4-nitrocatechol. Experiments with extracts obtained from PNP-grown cells revealed that the initial reaction is a hydroxylation of PNP to yield 4-nitrocatechol. 4-Nitrocatechol is subsequently oxidized to THB with the concomitant removal of the nitro group as nitrite. The enzyme that catalyzed the two sequential monooxygenations of PNP was partially purified and separated into two components by anion-exchange chromatography and size exclusion chromatography. Both components were required for NADH-dependent oxidative release of nitrite from PNP or 4-nitrocatechol. One of the components was identified as a reductase based on its ability to catalyze the NAD(P)H-dependent reduction of 2,6-dichlorophenolindophenol and nitroblue tetrazolium. Nitrite release from either PNP or 4-nitrocatechol was inhibited by the flavoprotein inhibitor methimazole. Our results indicate that the two monooxygenations of PNP to THB are catalyzed by a single two-component enzyme system comprising a flavoprotein reductase and an oxygenase.
p-Nitrophenol (PNP) is a priority environmental pollutant (10), occurring in industrial effluents (20) and in the soil as a hydrolytic product of parathion (14) or methyl parathion (17, 21). Several aerobic pure cultures of bacteria belonging to species of Flavobacterium, Pseudomonas, Moraxella, Nocardia, and Arthrobacter metabolize PNP with removal of the nitro group as nitrite (7, 8, 19, 22, 26). Two alternative pathways that convert PNP to maleylacetate have been elucidated for aerobic PNP degradation (24). The first pathway is more common in gram-negative isolates and results in the formation of hydroquinone from PNP, probably via 1,4-benzoquinone, with concomitant nitrite release. Hydroquinone is oxidized by a ring-cleaving dioxygenase to yield γ-hydroxymuconic semialdehyde, which is subsequently transformed to maleylacetate (26). In the second catabolic pathway, an Arthrobacter sp. hydroxylates PNP to produce either 4-nitrocatechol or 4-nitroresorcinol. Subsequent oxidative removal of the nitro group yields 1,2,4-trihydroxybenzene (THB) with concomitant release of nitrite. The THB is oxidized by a ring cleavage dioxygenase to yield maleylacetate, which is converted enzymatically to 3-ketoadipate (8). While a complete pathway for PNP degradation via hydroquinone has been described in detail, the initial steps in the pathway involving conversion of PNP to THB are not fully understood.
Oxidative removal of the nitro groups from nitroaromatic compounds has been described for several degradative pathways (24). A preliminary characterization of p-nitrophenol-2-hydroxylase, which catalyzes the conversion of PNP to 4-nitrocatechol, was reported in cell extracts from a Nocardia sp. (13). A particulate monooxygenase from a Moraxella sp. that releases nitrite from PNP has been partially purified (26). Zeyer and Kocher (32) purified a soluble nitrophenol oxygenase from Pseudomonas putida B2 that converts ortho-nitrophenol to catechol and nitrite. Ecker et al. (4) suggested the involvement of a dioxygenase attack in the displacement of a nitro group from 2,6-dinitrophenol. Recently, 4-methyl-5-nitrocatechol oxygenase has been purified from Burkholderia sp. strain DNT (6). 4-Methyl-5-nitrocatechol oxygenase oxidizes 4-methyl-5-nitrocatechol to a quinone with concomitant release of nitrite.
We report here a preliminary characterization of a novel monooxygenase from Bacillus sphaericus JS905 that catalyzes the first two steps in the degradation of PNP via 4-nitrocatechol and THB. The enzyme consists of two components, a flavoprotein reductase and an oxygenase, and catalyzes two sequential monooxygenation reactions that convert PNP to THB. The first reaction converts PNP to 4-nitrocatechol, and the second removes the nitro group. The reactions are very specific, and the enzyme does not release nitrite directly from PNP.
MATERIALS AND METHODS
Organism and culture conditions.
A gram-positive, motile rod with round terminal spores, lacking fluorescent pigments, was isolated by selective enrichment with PNP from an agricultural soil in India with a history of methyl parathion application. The strain, identified as B. sphaericus JS905, based on morphological and biochemical characteristics (Institute of Microbial Technology, Chandigarh, India), was maintained on minimal salts medium (MSB) (25) containing 15 mg of PNP, 200 mg of yeast extract, and 18 g of agar per liter. For induction with PNP, cells grown in 0.75% (wt/vol) tryptic soy broth (TSB) were harvested by filtration, washed, and suspended in MSB containing PNP (150 μM) and yeast extract (0.1%). The cultures were incubated at 37°C with shaking (300 rpm), and the disappearance of PNP was monitored by high-performance liquid chromatography (HPLC). For preparation of cell extracts, cells were cultivated in 4 liters of TSB overnight, harvested by centrifugation at 7,000 × g, and suspended in MSB containing PNP (150 μM) to an A600 of 1.5. The cell suspension was incubated with shaking at 250 rpm. Additional PNP (150 μM) was added when the yellow color of the PNP disappeared, and the sequence was repeated six to eight times over 3 h. Ice was added to the cell suspensions, and the culture was harvested by centrifugation. The cell pellet was washed with Tris-HCl (50 mM, pH 7.6) and stored at −70°C until further use.
Respirometry.
Cells were grown for induction as described above, harvested by centrifugation, and suspended in MSB. Uninduced, TSB-grown cells served as controls. Oxygen uptake was measured polarographically at 25°C with a Clark-type oxygen electrode.
Preparation of cell extract.
Frozen cells (18 g [wet weight]) were suspended in 2 volumes (wt/vol) of lysis buffer that consisted of Tris-HCl (50 mM, pH 7.6), ethanol (2.5%, vol/vol), glycerol (2.5%, vol/vol), and flavin adenine dinucleotide (FAD; 10 μM). Cells were broken by two passages through a French pressure cell at 20,000 lb/in2. The resulting lysate was centrifuged at 100,000 × g for 1 h at 4°C, and the supernatant was used immediately.
Partial purification of PNP monooxygenase.
All procedures were carried out at 4°C unless otherwise specified. The clarified cell extract was loaded onto a DEAE-Sepharose fast-flow column (2.5 by 14 cm; Pharmacia Biotech, Piscataway, N.J.) that had been equilibrated with TEF buffer (50 mM Tris-HCl [pH 7.6], 0.25% [vol/vol] ethanol, 2 μM FAD). The column was washed with 150 mM NaCl in TEF buffer at a flow rate of 1.5 ml/min. Bound proteins were eluted with a 270-ml linear NaCl gradient (150 to 500 mM). Fractions (3 ml each) exhibiting maximal nitrite release from PNP or 4-nitrocatechol were pooled and concentrated over a Centriplus 100 (Amicon, Danvers, Mass.) concentrator to a final volume of 3 ml. The protein solution was diluted 1:3 in TEF buffer and applied to a Q-Sepharose fast-flow column (1.0 by 10 cm; Pharmacia). The column was washed with 100 ml of 200 mM NaCl in TEF buffer, and the adsorbed proteins were eluted with a linear NaCl gradient (80 ml, 200 to 400 mM) at a flow rate of 1.0 ml/min. The fractions containing the enzyme activity were pooled and concentrated to 1.5 ml over a Centriplus 100 filter. Glycerol (10%, vol/vol) was added, and the sample was applied to a Sephacryl S-300 column (1.5 by 107 cm; Pharmacia) preequilibrated with 100 mM NaCl in TEF buffer. The proteins were resolved by ascending chromatography at a flow rate of 1.0 ml/min with the same buffer.
Enzyme assays.
PNP monooxygenase activity was determined by measuring the nitrite released from the substrate (PNP or 4-nitrocatechol) at 30°C. The standard reaction mixture contained in 1 ml of TE buffer (50 mM Tris-HCl, 0.25% ethanol, pH 8), 0.2 mM NADH, 0.02 mM FAD, 1 mM MgSO4, and various amounts of protein. The reaction was initiated by addition of either PNP or 4-nitrocatechol (0.08 mM). The substrates were omitted from the control reaction mixtures. After 30 min, nitrite was determined by the method adapted by Daniels et al. (3).
1,2,4-Trihydroxybenzene 1,2-dioxygenase activity was measured either spectrophotometrically or polarographically at 25°C in reaction mixtures containing phosphate buffer (20 mM, pH 6.8) and protein. The reaction was started by addition of 100 μM THB.
NADH–2,6-dichlorophenolindophenol reduction, cytochrome c reduction, and nitroblue tetrazolium reduction were measured spectrophotometrically (5). The 1.0-ml reaction mixture contained 50 mM TE buffer (pH 8.0), substrate (0.1 mM 2,6-dichlorophenolindophenol, 0.1 mM nitroblue tetrazolium, or 0.05 mM cytochrome c) and 2 to 100 μg of protein. Concentrations of reduced 2,6-dichlorophenolindophenol were calculated based on the A600 using a molar extinction coefficient of 17,000 M−1 cm−1.
Molecular weight determination.
The relative molecular masses of the native proteins were determined by gel filtration on a Sephacryl S-300 column (1.5 by 107 cm; Pharmacia) at a flow rate of 1.0 ml/min with 100 mM NaCl in TEF buffer. The calibration standards were ferritin, catalase, aldolase, and ovalbumin (Pharmacia) (12). Partially purified proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (11), and the gels were stained with Coomassie brilliant blue to visualize proteins.
Inhibition studies.
PNP-induced cells were suspended to an A600 of 1.5 in 50 ml of MSB containing 0.1% yeast extract, 0.2 mM PNP or 4-nitrocatechol, and 1 mM either 2,2′-dipyridyl or o-phenanthroline for inhibition of THB oxidation. Nitrite release was determined, and the THB formed was detected by HPLC. In a separate experiment, cell extracts prepared in phosphate buffer (20 mM, pH 7.2) containing no added FAD were incubated with an inhibitor at 30°C for 15 min prior to the addition of NADH, FAD, MgSO4, and a substrate (PNP or 4-nitrocatechol) to the reaction mixture.
Analytical techniques.
HPLC was performed on a Spherisorb C8 column (5 μm; 250 by 4.6 mm; Altech, Deerfield, Ill.) with an HP 1040A diode array detector (Hewlett-Packard Corp., Palo Alto, Calif.) for detection of PNP or its metabolites. Acetonitrile-water containing 13.5 mM trifluoroacetic acid (40:60) was the mobile phase at a flow rate of 1.0 ml/min. Compounds were identified by comparison of HPLC retention times and UV-visible spectra to those of standards. Protein concentrations were determined by the bicinchoninic acid protein assay (23).
Isolation and characterization of 4-nitrocatechol from PNP hydroxylation.
4-Nitrocatechol was extracted from a reaction mixture with ethyl acetate and characterized by HPLC-mass spectrometry (MS) analysis (HP 1050 LC with an HP 5987 mass selective detector) using a particle beam interface (HP 59980A). All spectra were generated by electron impact. HPLC conditions were as described above.
Chemicals.
Nitrophenols were purchased from Aldrich (Milwaukee, Wis.). Methimazole, miconazole, metyrapone, and α-naphthoflavone were from Sigma. All other chemicals were of the highest purity commercially available.
RESULTS AND DISCUSSION
Biodegradation of PNP.
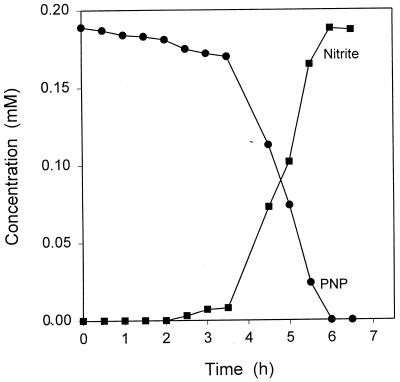
A lag period of 3.5 h preceded rapid degradation of PNP when TSB-grown cells of B. sphaericus JS905 were transferred to media containing PNP (Fig. 1). Nitrite was released in stoichiometric amounts, and no other degradation products were detected by HPLC. PNP-grown cells released stoichiometric amounts of nitrite and produced a purple compound when incubated in media containing PNP or 4-nitrocatechol and iron chelators such as 2,2′-dipyridyl or o-phenanthroline. The color was identical to that produced when 2-hydroxy-1,4-benzoquinone was added to reaction mixtures (8), and HPLC analysis confirmed the presence of the quinone in the culture fluid. 2,2′-Dipyridyl is known to inhibit certain aromatic ring cleavage enzymes that require ferrous ions for their activities (1). It was also shown that PNP-grown Moraxella sp. converted PNP stoichiometrically to hydroquinone in the presence of 2,2′-dipyridyl (26), presumably due to the inhibition of a dioxygenase that catalyzes ring fission of hydroquinone. The reaction mixture was decolorized upon addition of sodium dithionite, indicating the reduction of 2-hydroxy-1,4-benzoquinone to THB (2).
FIG. 1.
Disappearance of PNP and release of nitrite during induction. Cells were grown overnight in 0.75% TSB, washed, and suspended in MSB supplemented with PNP (0.18 mM) to an A600 of 0.87. The suspension was incubated at 37°C with shaking, and culture fluids were analyzed at intervals by HPLC for the disappearance of PNP and by a colorimetric assay for nitrite.
PNP, 4-nitrocatechol, and THB stimulated oxygen consumption by PNP- and 4-nitrocatechol-grown cells (Table 1). 4-Nitroresorcinol stimulated oxygen uptake to a lesser extent. Hydroquinone, 4-aminophenol, catechol, and resorcinol did not stimulate oxygen uptake. Oxygen uptake was not detected with TSB-grown cells. The results indicate that enzymes for PNP degradation are inducible and strongly suggest that 4-nitrocatechol and THB are the intermediates in the catabolic pathway for PNP in B. sphaericus JS905.
TABLE 1.
Substrates oxidized by washed B. sphaericus JS905 cells
| Assay substratea | O2 uptake (nmol/min/mg of protein)b after growth with:
|
||
|---|---|---|---|
| PNP | 4-Nitrocatechol | TSB | |
| PNP | 30 | 25 | <1 |
| 4-Nitrocatechol | 32 | 24 | <1 |
| 4-Nitroresorcinol | 6 | 15 | <1 |
| THB | 50 | 29 | <1 |
| Hydroquinone | <1 | <1 | <1 |
| 4-Aminophenol | <1 | <1 | <1 |
| Catechol | <1 | <1 | <1 |
| Resorcinol | <1 | <1 | <1 |
The concentration of all substrates was 0.05 mM.
Oxygen consumption was measured polarographically. The values are averages of duplicates.
Enzyme activities in cell extracts.
Under aerobic conditions, extracts of PNP-grown cells catalyzed the release of nitrite in stoichiometric amounts from PNP or 4-nitrocatechol. The activity in cell extracts was present in the soluble fraction and represented 75% of the activity observed in intact cells. The rate of nitrite release by cell extracts was linear for at least 30 min and was not directly related to protein concentration (data not shown), which suggested that the oxygenase might be a multicomponent enzyme. The enzymatic activity was dependent on the presence of NAD(P)H and FAD and was not stimulated by the addition of flavin mononucleotide. NADH was the preferred cofactor when residual cofactor was removed from cell extracts by passage over a desalting PD-10 column (Pharmacia) or fractionation with (NH4)2SO4. Magnesium ions enhanced the enzyme activity. Extracts prepared without added FAD lost 52% of their activity towards PNP and 4-nitrocatechol during storage on ice for 48 h. The addition of FAD (2 μM) to chromatography buffers was necessary to maintain full activity. The removal of the nitro group from both substrates was maximum at pH 8 in 50 mM Tris-HCl buffer. Addition of 0.25% ethanol to the buffer stabilized the activity during desalting or dialysis. The presence of β-mercaptoethanol was inhibitory to the enzyme activity. The rate and extent of nitrite release were the same in cell extracts whether PNP or 4-nitrocatechol was used as the substrate.
THB stimulated high rates of oxygen consumption by cell extracts, and the stoichiometry was 0.93 ± 0.02 mol of O2 per mol of substrate. The enzyme catalyzing the oxidation of THB was stable during storage at 4°C. Spectrophotometric assays revealed the disappearance of THB and the appearance of a new compound with an absorbance maximum at 243 nm identical to that of maleylacetate (2, 26). When NADH was included in the reaction mixture, the absorbance peak at 243 nm disappeared, presumably due to the activity of maleylacetate reductase. The results indicate that THB served as a substrate for a ring cleavage dioxygenase, as evidenced by the characteristic spectral changes and the rapid consumption of oxygen with THB. The observed spectral changes suggest an ortho ring fission of THB yielding 3-ketoadipate with maleylacetate as an intermediate (2, 8).
Identification of the products of PNP oxidation.
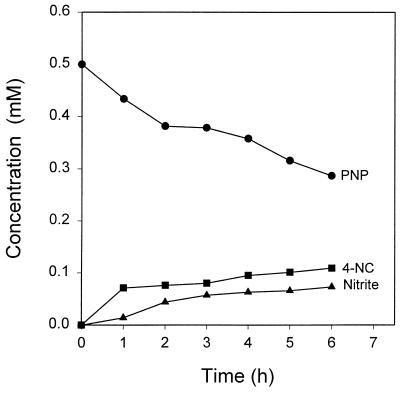
Cell extracts incubated in potassium phosphate buffer with NADPH converted PNP to a yellow metabolite. Addition of 2.5 N NaOH to the reaction mixture after incubation at 30°C gave a deep red solution, which is a characteristic of 4-nitrocatechol (13). A large-scale reaction was performed to confirm the identity of the product. The HPLC retention time, UV spectrum, and MS fragmentation pattern of the isolated compound were identical to those of authentic 4-nitrocatechol. Both a Flavobacterium sp. (19) and a Nocardia sp. (13) have been shown to convert PNP to 4-nitrocatechol, but the details of the oxidative release of nitrite were not presented. A recent study of the conversion of PNP to THB by Arthrobacter sp. strain JS443 (8) did not clarify whether 4-nitrocatechol or 4-nitroresorcinol was the product of the initial reaction of PNP catabolism. The result presented here clearly indicates that the initial reaction in the bacterial degradation of PNP by B. sphaericus JS905 is hydroxylation of the ring at the 2 position to yield 4-nitrocatechol. PNP degradation in the reaction mixtures containing cell extracts in 20 mM potassium phosphate buffer and NADPH resulted in near-stoichiometric accumulation of both 4-nitrocatechol and nitrite (Fig. 2). No other aromatic products accumulated, which indicates that some of the 4-nitrocatechol was further converted with release of nitrite. In contrast, reactions carried out in Tris-HCl yielded negligible amounts of 4-nitrocatechol in the presence of NADPH. When FAD was included in the reaction mixtures, nitrite, but not 4-nitrocatechol, accumulated, regardless of which buffer was used. The results suggest that in assays with phosphate buffer, the rate of oxidation of 4-nitrocatechol from PNP was so slow that 4-nitrocatechol accumulated in the reaction mixture. There was no accumulation of 4-nitrocatechol or release of nitrite from PNP when the reaction mixture was incubated under anaerobic conditions, which confirmed that the hydroxylation of PNP is an oxidation reaction requiring molecular oxygen.
FIG. 2.
Accumulation of 4-nitrocatechol (4-NC) and nitrite in cell extracts.
Cell extracts incubated for 2 h with PNP or 4-nitrocatechol in the presence of 2,2′-dipyridyl or o-phenanthroline accumulated THB or 2-hydroxy-1,4-benzoquinone. This supports earlier observations that intact cells contained oxygenases that converted PNP and/or 4-nitrocatechol to THB. However, it was not clear whether 2-hydroxy-1,4-benzoquinone or THB was the actual product of nitrite release from 4-nitrocatechol. A quinone reductase may have catalyzed the conversion of 2-hydroxy-1,4-benzoquinone to THB, as has been suggested for PNP (27) or o-nitrophenol (32) biodegradation.
Separation of enzyme components.
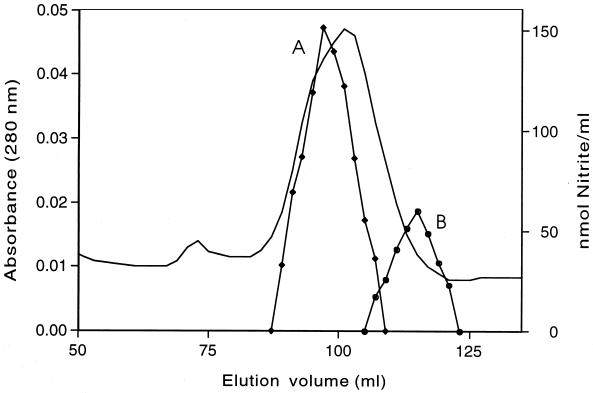
The activities for oxidative removal of the nitro group from PNP or 4-nitrocatechol coeluted during ion-exchange chromatography of cell extract on a DEAE-Sepharose fast-flow column, which suggested that the same enzyme catalyzes both reactions. Further purification by Q-Sepharose chromatography enriched for the enzyme (Table 2); however, size exclusion chromatography on Sephacryl S-300 of Q-Sepharose fractions led to substantial loss of enzymatic activity. The activity was regained when two components that eluted separately (designated A and B in the order of their elution) were combined (Fig. 3). This indicated that the enzyme was a two-component system.
TABLE 2.
Activity of the enzyme system during partial purification
| Purification step | Sp acta (fold purification)
|
|
|---|---|---|
| PNP | 4-Nitrocatechol | |
| Cell extraction | 1.3 | 1.3 |
| DEAE-Sepharose chromatography | 7.5 (6) | 7.1 (5) |
| Q-Sepharose chromatography | 19.4 (15) | 13.5 (10) |
| Sephacryl S-300 chromatographyb | 40.0 (32) | 38.0 (28) |
Nanomoles of nitrite per milligram of protein per minute.
Combination of 104- and 114-ml elution volumes.
FIG. 3.
Resolution of components A and B of PNP monooxygenase by size exclusion chromatography on a Sephacryl S-300 column. Proteins were eluted in 2-ml fractions with 100 mM NaCl in 50 mM TEF buffer. Component A (reductase) was located by measuring the oxidative release of nitrite from PNP in reaction mixtures containing component B (4 μg of protein) (fraction 57). Component B (oxygenase) was located by measuring nitrite release in assay mixtures containing component A (23 μg of protein) (fraction 52). ——, A280.
Fractions containing component A catalyzed NAD(P)H-dependent reduction of 2,6-dichlorophenolindophenol or nitroblue tetrazolium but not cytochrome c. NADPH was also an electron donor for the reductase activity. A flavoprotein and a second protein of the ferredoxin type are required for cytochrome c reductase activity (9). The fact that the reductase component of the monooxygenase reduces 2,6-dichlorophenolindophenol and nitroblue tetrazolium but not cytochrome c and the lack of spectral characteristics of an iron-sulfur center (30) suggest the absence of a second prosthetic group (9). The NADH–2,6-dichlorophenolindophenol reduction activity coeluted with component A from the S-300 column. The fraction with an elution volume of 97 ml which showed maximal nitrite release activity (when combined with component B) also had maximum reductase activity (2.9 μmol/min/mg of protein). No absorption spectrum in the visible region was observed for the dialyzed component A, suggesting that the flavin cofactor was readily removed during dialysis. The addition of FAD, but not flavin mononucleotide, was essential for the NADH-dependent 2,6-dichlorophenolindophenol reduction by the dialyzed component A. The results strongly suggest that component A is a flavoprotein. Component B appears to be the hydroxylase and showed no reductase activity with the electron acceptors tested.
Cytochrome P-450 inhibitors (α-naphthoflavone, miconazole, and metyrapone) at 0.5 mM had very little effect (<5% inhibition) on nitrite release from PNP or 4-nitrocatechol by the reconstituted mixture containing components A and B. Methimazole, a competitive inhibitor of flavin monooxygenases (28), at the same concentration, greatly inhibited (58% inhibition) the enzymatic release of nitrite from the substrates. The above observations argue strongly that the enzyme system contains a flavoprotein reductase.
Based on the elution volumes of the active fractions containing components A and B on the S-300 column, the molecular masses of the native proteins were estimated to be 323 and 146 kDa, respectively. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the fractions containing the active components showed the presence of several bands. Efforts to purify the components to homogeneity failed.
Both components A and B are essential for NADH-dependent oxidative removal of the nitro group from PNP or 4-nitrocatechol (Table 3). THB was not detected in reaction mixtures containing both protein components and PNP or 4-nitrocatechol, even though the nitro group was removed from the substrates. The failure of THB to accumulate can be explained by the presence of THB-1,2-dioxygenase, which was detected in preparations of component A (data not shown). In contrast to the results obtained with cell extracts, NADPH did not support PNP or 4-nitrocatechol oxidation. Since the enzyme system consisted of two components that catalyzed two consecutive monooxygenation reactions, we designated it PNP monooxygenase. The reductase component of PNP monooxygenase has a loosely bound FAD that is readily lost during the purification process. Like most of the hydroxylases (13, 15, 18, 29, 31), PNP monooxygenase uses FAD as the redox chromophore and is NADH dependent. In contrast, nitrophenol monooxygenases (26, 32) and 4-methyl-5-nitrocatechol oxygenase (6) use NADPH as the preferred electron donor.
TABLE 3.
Requirements for PNP monooxygenase activitya
| Constituent(s) of assay mixture | Sp actb
|
|
|---|---|---|
| PNP | 4-Nitrocatechol | |
| Component A | 0 | 0 |
| Component A + NADH | 0 | 0 |
| Component B | 0 | 0 |
| Component B + NADH | 0 | 0 |
| Components A + B | 0 | 0 |
| Components A + B + NADH | 36.5 | 35.6 |
| Components A + B + NADPH | 0 | 0 |
Reaction mixtures (1.0 ml) contained PNP or 4-nitrocatechol (80 μM), FAD (20 μM), component A (10 μg of protein), component B (3.6 μg of protein), and, when indicated, NADH or NADPH (200 μM) in TE buffer (50 mM, pH 8.0).
Nanomoles of nitrite per milligram of protein per minute.
Experiments with the partially purified PNP monooxygenase revealed that the enzyme has a narrow substrate range. 4-Nitroresorcinol also served as a substrate with a rate of nitrite release similar to that observed with the two physiological substrates, PNP and 4-nitrocatechol. o-Nitrophenol, m-nitrophenol, 2-nitroresorcinol, and 2,4-dinitrophenol were not transformed.
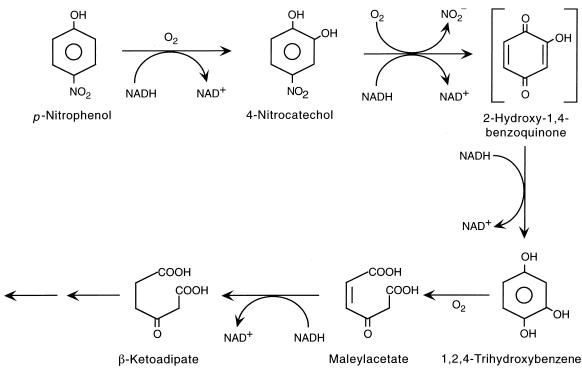
Several lines of evidence indicate that a single monooxygenase system catalyzes both the initial hydroxylation of PNP and subsequent oxidative release of the nitro group from 4-nitrocatechol. (i) The relative activities of the enzyme toward PNP and 4-nitrocatechol remained constant throughout several purification steps and during a variety of manipulations, including inhibition. (ii) Both PNP and 4-nitrocatechol are converted to THB or 2-hydroxy-1,4-benzoquinone. (iii) Enzymes in cell extracts catalyze the conversion of PNP to 4-nitrocatechol in phosphate buffer. The fact that 4-nitrocatechol did not accumulate in reaction mixtures containing partially purified enzyme and Tris buffer suggests that the 4-nitrocatechol is bound to the enzyme until nitrite is released. Based on the accumulation of 2-hydroxy-1,4-benzoquinone during transformation of PNP or 4-nitrocatechol by cells and cell extracts incubated with iron chelators and on analogy with other systems (26, 32), we suggest that 2-hydroxy-1,4-benzoquinone is an intermediate in the pathway (Fig. 4).
FIG. 4.
Proposed pathway for PNP biodegradation by B. sphaericus JS905.
Several bacterial monooxygenase enzymes that attack aromatic compounds are multicomponent systems. 4-Hydroxyphenylacetate 3-hydroxylase, a two-component, NADH-dependent flavomonooxygenase, has been isolated from Escherichia coli W (18). Chlorophenol 4-monooxygenase from B. cepacia AC1100 is another two-component enzyme system (31). A three-component enzyme, toluene 2-monooxygenase, has been purified from B. cepacia G4 (15). Toluene in P. mendocina KR1 is initially hydroxylated by toluene-4-monooxygenase, a three-component enzyme system, to form p-cresol (29). The conversion of phenol to catechol by Pseudomonas strain CF600 is catalyzed by a multicomponent phenol hydroxylase (16).
Sequential hydroxylation by a single enzyme system has been reported in at least two instances. Toluene 2-monooxygenase oxidizes toluene to o-cresol and then to 3-methylcatechol (15). Chlorophenol 4-monooxygenase catalyzes the sequential hydroxylation of 2,4,5-trichlorophenol to 2,5-dichloro-p-hydroquinone and then to 5-chlorohydroxyquinol (31). To our knowledge, PNP monooxygenase from B. sphaericus JS905 is the only known monooxygenase to sequentially hydroxylate a nitroaromatic compound. In contrast to the chlorophenol-4-monooxygenase, the enzyme hydroxylates the ring adjacent to the hydroxyl group first and then displaces the nitro group.
The evolution of the pathway for PNP biodegradation by B. sphaericus JS905 would have been markedly simplified because the first two reactions are catalyzed by a single enzyme system. Such a strategy would also minimize the accumulation of potentially toxic 4-nitrocatechol. The key to the reaction might be the ability of the molecule to form the 1,4-quinone structure upon elimination of the negatively charged leaving group.
ACKNOWLEDGMENTS
We thank Mike Henley for help with HPLC-MS analysis and Billy E. Haigler, Charles C. Somerville, and Urs Lendenmann for useful discussions. Venkateswarlu Kadiyala thanks the Sri Krishnadevaraya University, Anantapur, India, for granting study leave.
This research was supported by the Air Force Office of Scientific Research and the Strategic Environmental Research and Development Program. Venkateswarlu Kadiyala is grateful to the National Research Council for the award of a Senior Research Associateship.
REFERENCES
- 1.Chapman P J, Hooper D J. The bacterial metabolism of 2,4-xylenol. Biochem J. 1968;110:491–498. doi: 10.1042/bj1100491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman P J, Ribbons D W. Metabolism of resorcinylic compounds by bacteria: alternate pathways for resorcinol catabolism in Pseudomonas putida. J Bacteriol. 1976;125:985–998. doi: 10.1128/jb.125.3.985-998.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniels L, Hanson R S, Phillips J A. Chemical analysis. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 512–554. [Google Scholar]
- 4.Ecker S, Knackmuss H-J, Bruhn C. Abstracts of the 89th Annual Meeting of the American Society for Microbiology 1989. Washington, D.C: American Society for Microbiology; 1989. Catabolism of 2,6-dinitrophenol by Alcaligenes eutrophus JMP222, abstr. Q-198; p. 168. [Google Scholar]
- 5.Haigler B E, Gibson D T. Purification and properties of ferredoxinnap, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1990;172:465–468. doi: 10.1128/jb.172.1.465-468.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haigler B E, Suen W-C, Spain J C. Purification and sequence analysis of 4-methyl-5-nitrocatechol oxygenase from Burkholderia sp. strain DNT. J Bacteriol. 1996;178:6019–6024. doi: 10.1128/jb.178.20.6019-6024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanne L F, Kirk L L, Appel S M, Narayan A D, Bains K K. Degradation and induction specificity in actinomycetes that degrade p-nitrophenol. Appl Environ Microbiol. 1993;59:3505–3508. doi: 10.1128/aem.59.10.3505-3508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain R K, Dreisbach J H, Spain J C. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl Environ Microbiol. 1994;60:3030–3032. doi: 10.1128/aem.60.8.3030-3032.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamin H, Lambeth J D, Siegel L M. The role of flavins in electron transfer between two-electron donors and one-electron acceptors. In: Yagi K, Yamano T, editors. Flavins and flavoproteins. Baltimore, Md: University Park Press; 1980. pp. 341–348. [Google Scholar]
- 10.Keith L H, Telliard W A. Priority pollutants. I. A perspective view. Environ Sci Technol. 1979;13:416–423. [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Laurent T C, Killander J. A theory of gel filtration and its experimental verification. J Chromatogr. 1964;14:317–330. [Google Scholar]
- 13.Mitra D, Vaidyanathan C S. A new 4-nitrophenol-2-hydroxylase from a Nocardia sp.: isolation and characterization. Biochem Int. 1984;8:605–615. [PubMed] [Google Scholar]
- 14.Munnecke D M. Enzymatic hydrolysis of organophosphate insecticides, a possible pesticide disposal method. Appl Environ Microbiol. 1976;32:7–13. doi: 10.1128/aem.32.1.7-13.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman L M, Wackett L P. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry. 1995;34:14066–14076. doi: 10.1021/bi00043a012. [DOI] [PubMed] [Google Scholar]
- 16.Nordlund I, Powlowski J, Shingler V. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990;172:6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou L T. Methyl parathion degradation and metabolism in soil: influence of high soil water contents. Soil Biol Biochem. 1985;17:241–243. [Google Scholar]
- 18.Prieto M A, Garcia J L. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. J Biol Chem. 1994;269:22823–22829. [PubMed] [Google Scholar]
- 19.Raymond D G M, Alexander M. Microbial metabolism and cometabolism of nitrophenols. Pestic Biochem Physiol. 1971;1:123–130. [Google Scholar]
- 20.Shackelford W M, Keith L H. Frequency of organic compounds identified in water. EPA 600/4-76-062. U.S. Athens, Ga: Environmental Protection Agency; 1976. [Google Scholar]
- 21.Sharmila M, Ramanand K, Sethunathan N. Effect of yeast extract on the degradation of organophosphorus insecticides by soil enrichment and bacterial cultures. Can J Microbiol. 1989;35:1105–1110. [Google Scholar]
- 22.Simpson J R, Evans W C. The metabolism of nitrophenols by certain bacteria. Biochem J. 1953;55:XXIV. [PubMed] [Google Scholar]
- 23.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 24.Spain J C. Bacterial degradation of nitroaromatic compounds under aerobic conditions. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. Vol. 49. New York, N.Y: Plenum Press; 1995. pp. 19–35. [Google Scholar]
- 25.Spain J C, Nishino S F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987;53:1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spain J C, Gibson D T. Pathway for biodegradation of p-nitrophenol in Moraxella sp. Appl Environ Microbiol. 1991;57:812–819. doi: 10.1128/aem.57.3.812-819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spain J C, Wyss O, Gibson D T. Enzymatic oxidation of p-nitrophenol. Biochem Biophys Res Commun. 1979;88:634–641. doi: 10.1016/0006-291x(79)92095-3. [DOI] [PubMed] [Google Scholar]
- 28.Tomasi I, Artaud I, Bertheau Y, Mansuy D. Metabolism of polychlorinated phenols by Pseudomonas cepacia AC1100: determination of the first two steps and specific inhibitory effect of methimazole. J Bacteriol. 1995;177:307–311. doi: 10.1128/jb.177.2.307-311.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whited G M, Gibson D T. Toluene-4-monooxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J Bacteriol. 1991;173:3010–3016. doi: 10.1128/jb.173.9.3010-3016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xun L. Purification and characterization of chlorophenol 4-monooxygenase from Burkholderia cepacia AC1100. J Bacteriol. 1996;178:2645–2649. doi: 10.1128/jb.178.9.2645-2649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi M, Fujisawa H. Reconstitution of iron-sulfur cluster of NADH-cytochrome c reductase, a component of benzoate 1,2-dioxygenase system from Pseudomonas arvilla C-1. J Biol Chem. 1981;256:6783–6787. [PubMed] [Google Scholar]
- 32.Zeyer J, Kocher H P. Purification and characterization of a bacterial nitrophenol oxygenase which converts ortho-nitrophenol to catechol and nitrite. J Bacteriol. 1988;170:1789–1794. doi: 10.1128/jb.170.4.1789-1794.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]