Abstract
The hydrozoan family Eirenidae is known scientifically for its morphological plasticity and challenges in species identification. We used an integrative taxonomic approach based on morphological, molecular and life history evidence to systematically assess field-collected medusae of Eirene menoni Kramp 1953 and captive raised polyps of both E. menoni and E. lacteoides Kubota and Horita 1992. Following morphological review, we updated the genus description to include the presence of rudimentary bulbs (warts) on the ring canal in at least eight of the 24 valid Eirene species. We propose the potential for the mature E. menoni hydrotheca to develop into a gonotheca. However, this proposal will require additional study for verification. We provide validated distribution records from the Indo-Pacific Ocean for E. menoni,and updated collection records for E. lacteoides from the Yellow and East China Seas, and public aquaria-cultured specimens from Japan and Hawaii, using cytochrome c oxidase I (COI) sequences that we generated and compared with those from GenBank. The COI gene reliably separated four species, each forming a monophyletic clade with strong bootstrap support and low mean intraspecific molecular divergences (≤ 1%) within clades. However, some of the deeper nodes of the tree remained poorly resolved, and our analysis failed to demonstrate monophyly among eirenid genera Eirene and Tima. Our integrative taxonomic approach is essential in confirming species identity within the family Eirenidae and genus Eirene,and we have also identified a likely range expansion of E. lacteoides to Hawaii.
Keywords: Hydrozoa; Leptomedusae; Plasticity; Campanulinida hydroid, COI mtDNA
BACKGROUND
Hydrozoans are well known for their phenotypic plasticity and challenging taxonomic identification (Bouillon et al. 2006; Moura et al. 2008; He et al. 2015; Maronna et al. 2016). Genetic barcoding has provided a useful tool for gelatinous zooplankton identification, enhancing the ability to rapidly assess regional biodiversity, reveal the presence of invasive taxa, and delineate species distributions (Holland et al. 2004; Zhou et al. 2013; Scorrano et al. 2017). However, misidentifications in the GenBank database (www.ncbi. nlm.nih.gov/genbank/) have led to sequences that do not accurately represent the associated binomials (Moura et al. 2008; Lindsay et al. 2017). An additional challenge lies in the fact that at present, DNA sequences are rarely published from species type localities, and we argue that this should be standard taxonomic practice.
DNA barcoding typically relies on 16S rRNA or COI mtDNA sequences for species identification, although both markers have shown polyphyletic and paraphyletic patterns in the hydrozoan genera Eirene and Clytia (Zheng et al. 2014; He et al. 2015; Zhang et al. 2015). Due to the challenges posed by jellyfish species taxonomy and frequent-misidentifications, accurate species recognition should include morphological descriptions of polyps, juvenile and adult medusae, documentation of complete life history, and molecular systematics (Straehler-Pohl et al. 2011; Lindsay et al. 2017; Toshino et al. 2019; Lawley et al. 2021). The 16S gene has been recommended for basal clade and species identification in this group (Zheng et al. 2014; Maronna et al. 2016). However, the COI gene also provides a reliable species-level identification (Laakmann and Holst 2014; He et al. 2015; Zhang et al. 2015; Bucklin et al. 2021; Calder et al. 2021). Folmer et al. (1994) and Bucklin et al. (2021) reported that COI: 1) is broadly applicable due to availability of universal primers, effective in amplifying this fragment from a wide range of phyla; 2) tends to show informative phylogenetic signals over a range of taxonomic levels; 3) evolves rapidly enough to allow for the discrimination of very closely related species; and 4) tends to reveal phylogeographically significant intraspecific variation associated with fine scale distribution patterns.
The hydrozoan family Eirenidae has proven to be a taxonomically difficult group to classify, due to multiple factors, including a lack of comprehensive species descriptions, a scarcity of detailed knowledge of life cycle information for most species, and unusually high evolutionary distances between congeneric species (e.g., Calder et al. 2021). This has resulted in inadequate family level assessment (Maronna et al. 2016), problematic generic relationships among Eirene, Clytia (Schuchert 2017), and Tima (Calder et al. 2021), and even misidentifications in species groupings (He et al. 2015; Maronna et al. 2016).
The Eirene genus comprises three documented species from Japan: E. hexanemalis (Goette, 1886), E. lacteoides (with the type locality being Toba Aquarium), and E. menoni. Additionally, there is an as-yetundescribed species (Kubota and Gravili 2007) within the genus. But since misidentifications continue to plague this group, our objective here is to use historical records to revise morphological descriptions and use molecular tools to improve species identifications. We also recommend that other researchers focusing on hydromedusan biology follow the same approach. We worked with preserved specimens obtained from a regional museum collection, captive bred aquarium samples, and field collected samples. We examined published biogeographic and taxonomic descriptions, medusa and polyp morphology, and phylogenetic analyses of E. menoni and E. lacteoides to verify taxonomic status and distribution. Eirene menoni was originally described from a single hydromedusa collected off of the Great Barrier Reef and was subsequently documented throughout the Indo-Pacific (Kramp 1953; Kramp 1968; Gershwin et al. 2010; Zheng et al. 2014). In Japanese waters, E. menoni has been documented from Kumamoto Prefecture, Kyushu (Sugiura 1979), Enoshima, Kanagawa Prefecture, Honshu (Sakiyama and Adachi 2001), Hamanako (Lake Hamana), Shizuoka Prefecture, Honshu (Okamoto et al. 2016), and northern Oita Prefecture, Kyushu (Iwai 2021). Eirene lacteoides was described from medusae cultured in the Toba Aquarium, Mie Prefecture, Honshu, Japan (Kubota and Horita 1992) and cultured polyps at the Qingdao Marine Science Museum, Qingdao, China (Huang et al. 2009). We reconfirmed the identities of COI sequences of E. lacteoides from GenBank which had been previously misidentified as Tima formosa Agassiz, L. 1862. These sequences had been collected in the field from Chanjiang River Estuary, China [JQ71666-67] and Jiaozhou Bay, China [JQ71616870], and from cultured samples at the Waikiki Aquarium, Oahu, Hawaii. Schuchert (2017) reported 16S [FJ418650] and 18S [FJ418671] sequences from hydromedusan specimens collected in the “South China Sea” (submitted to GenBank by Zheng, exact geographic source unknown) as Eirene lacteoides. We again utilized an integrative taxonomic approach (Dayrat 2005) that included descriptions of the entire life cycle, documentation of geographic sources, phylogenetic reconstruction, and morphological descriptions. Finally, in this study, we verify the phylogeographic distributions of these two species in the Pacific Ocean.
MATERIALS AND METHODS
Collecting and culturing
Wild specimens of Eirene menoni were collected (12 Sep 2021) using a rectangular dipnet (32 cm × 19 cm with 0.5 mm mesh) or an open-top plastic 1.5 L container on a pole from shallow waters (0–1 m) at Shonan Fishing Port, Sagami Bay, Kanagawa Prefecture, Japan. Cultured specimens of E. lacteoides that originated at the Toba Aquarium and produced multiple generations at both the Enoshima Aquarium and Tsuruoka City Kamo Aquarium (Kamo Aquarium) were examined. This species also was discovered in sea water tanks at the Waikiki Aquarium in July 2019. Cultured medusae and polyp samples were placed in small plastic containers (4 L) supplied with aeration to ensure growth and were transferred to (3 L) kreisel aquaria (Hamner 1990; Raskoff et al. 2003). Hydroids and hydromedusae were maintained on a diet of either Vietnamese (Japan) or Great Salt Lake (Hawaii) strains of Artemia nauplii, fed once per day. Mean water temperatures were maintained at 20°C and salinity range was kept between 30–35 psu. Field salinity was measured with a ATAGO® (MASTER-S/milla) refractometer. Water quality was maintained via regular water changes. Medusae and hydroids were preserved in 3% formalin seawater solution for anatomical evaluation and 90% ethanol for DNA extraction and sequencing.
Morphology and systematics
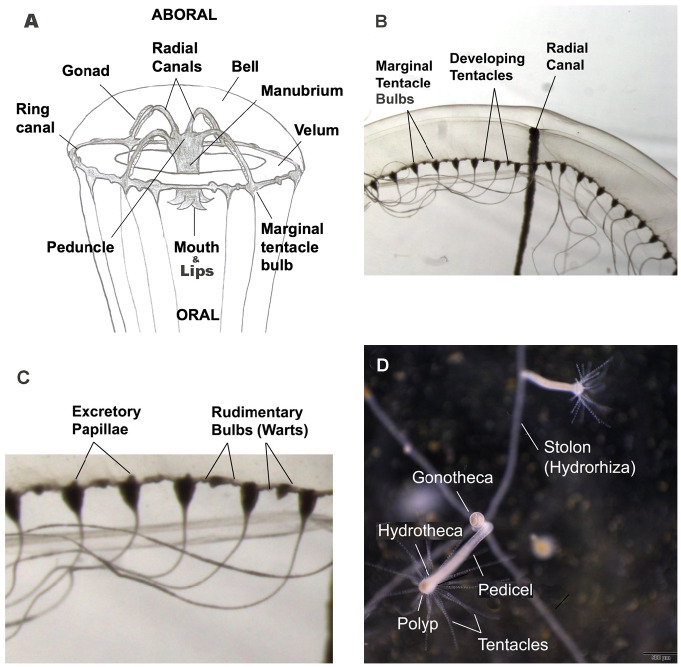
Basic terminology and morphology of medusa and polyps are provided in (Fig. 1). The formalin specimens were morphologically examined to verify species identity as Eirene menoni and Eirene lacteoides and measured with a Scienceware® Bel-Art Products dial caliper to the nearest 0.1 mm.An Olympus SZX16 Stereomicroscope with a Teledyne Lumenera INFINITY3-1®, 1.4 megapixel CCD microscopy camera, was used for morphological descriptions and images. We utilized a 0.5X camera objective with 0.7X to 13.5X magnification for viewing and imaging. Medusa bell size, morphology, and marginal tentacle numbers were compared to the original descriptions for each species. We also examined and imaged hydroids from both the Enoshima and Kamo Aquaria, and used Bouillon et al. (2006) for anatomical terminology to describe the medusae and hydroids for both species.
Museum specimens of E. menoni at the South Australian Museum, Adelaide (SAMA), South Australia were inspected for geographic location, taxonomic verification, and images were recorded. Voucher specimens from Japan for both species were deposited into the National Museum of Nature and Science Tokyo (NSMT), Tsukuba, Japan andsamples of E. lacteoides from Hawaii were deposited into the Bernice P. Bishop Museum (BPBM) Honolulu, Hawaii.
Fig. 1.
Anatomy diagram of a typical Eirenidae medusa and polyp with labels. Some sections cut away. Medusa not to scale. Drawing of medusa (1A) by Brenden Holland, image of ring canal with marginal tentacles and rudimentary bulbs (1B, C) by Gerald Crow, and image of polyps (1D) by Shuhei Ikeda, Tsuruoka City Kamo Aquarium.
DNA extraction, PCR amplification, and DNA sequencing
The hydromedusae specimens from Japan were supplied in an ethanol storage buffer. These specimens were carefully extracted and rinsed with deionized autoclaved water. Genomic DNAs were extracted from each whole organism using a Macherey-Nagel NucleoSpin® Mini kit for DNA from cells and tissue according to the manufacturer’s protocol. Genomic DNA was eluted in 200 µL of deionized autoclaved water and stored at -20°C.
We used existing and additional GenBank COI gene fragments to examine the species level and genus level phylogenetic position of Eirenidae species (Table 1). Fragments of 613 basepairs (bp) of the mitochondrial DNA (mtDNA) cytochrome c oxidase I (COI) gene were amplified by polymerase chain reaction (PCR) using the primers LCO1490/HCO2198 (Folmer et al. 1994). Target fragments were amplified using a MyCycler® Thermal Cycler (Bio-Rad, Hercules, CA, USA) with Conquest PCR Master Mix Optimizing Pack reagents and buffers from Lamba Biotech (catalog #D911-Mix1234). Twenty µL PCR reactions were run at Hawaii Pacific University’s Oceanic Institute, Oahu, Hawaii, under the following conditions: 4 minutes at 94°C, followed by 33 cycles of 94°C for 40 seconds, 52°C for 30 seconds and 72°C for 90 seconds, with a final 72°C extension for 5 minutes.
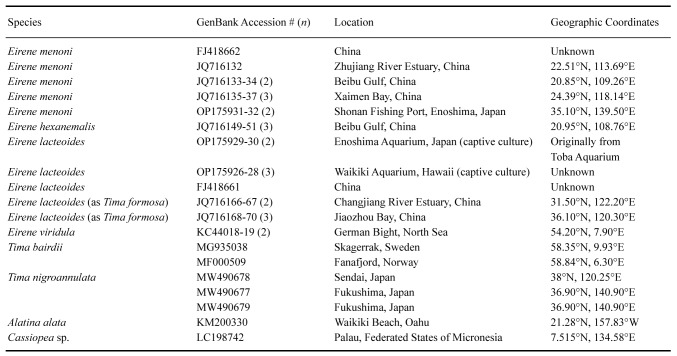
Table 1.
Taxa included in COI phylogenetic analysis, with GenBank accession numbers, sampling locations, and GPS coordinates. Note that geographic source of collection is not always known with confidence for specimens obtained from public aquaria, and for two gene sequences from China. The GenBank entries that lacked geographic source information are marked as unknown. In cases where more than one COI sequence was included from a location, the number of samples is indicated (n)
PCR fragments were purified with MachereyNagel NucleoSpin Gel and PCR clean-up columns, per the manufacturer’s protocol, and visualized via agarose minigel electrophoresis. Amplified mtDNA fragments were sequenced using the forward primer. DNA sequencing was performed at the Advanced Studies in Genomics, Proteomics and Bioinformatics (ASGPB) lab at the University of Hawaii at Manoa, Oahu, Hawaii.
Phylogenetic analysis
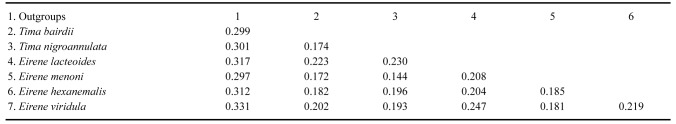
Randomized Axelerated Maximum Likelihood [RAxML] (Stamatakis 2014) and Molecular Evolutionary Analysis [MEGA X 11.0.13] (Tamura et al. 2021) were used for phylogenetic reconstruction (Table 1). The tree was generated with 1000 bootstrap replicates using the maximum-likelihood optimality criterion with the empirically determined best-fit substitution model. The optimal model selected was Tamura-Nei. Non-uniformity of evolutionary rates among sites was modeled using a discrete gamma distribution (+G) with 5 rate categories under the assumption that a specified fraction of sites were evolutionarily invariable (+I), resulting in the model TN93+G+I. This model had 66 parameters, BIC = 6461.115 (Bayesian Information Criterion), AICc = 6461.12 (Akaike Information Criterion, corrected), maximum likelihood score lnL = -2907.703, gamma correction (G) = 1.40, and the proportion of invariant sites (I) = 0.49. Genetic distances among and within clades were determined using the Kimura 2-parameter substitution model. We selected species for this phylogenetic analysis that were clearly morphologically valid, and therefore reflect correct binomial identifications. Intraspecific (within species) and interspecific (between species) differentiation were compared to aid our understanding of phylogenetic relationships.
RESULTS
Taxonomic Account
Phylum Cnidaria Verrill, 1865
Subphylum Medusozoa Petersen, 1979
Class Hydrozoa Owen, 1843
Subclass Hydroidolina Collins, 2000
Order Leptothecata Cornelius, 1992
Family Eirenidae Haeckel, 1879
Genus Eirene Eschscholtz, 1829
There are 24-valid Eirene species in the World Registry of Marine Species [WoRMS database] (www. marinespecies.org/aphia.php?p=taxdetails&id=117080 accessed 17 June 2022). Amended Genus description based on (Kramp 1961 1968; Bouillon and Boero 2000; Bouillon et al. 2006; Schuchert 2017; this study).
Typically small medusae ≤ 32 mm bell diameter, with a distinct gastric peduncle, no marginal or lateral cirri, with or without excretory pores, numerous statocysts present (may disappear in preserved specimens), with or without rudimentary bulbs (warts), these bulbs are present on the ring canal in at least eight species [E. conica Du, Xu, Huang and Guo, 2010, E.elliceana (Agassiz and Mayer, 1902), E. hexanemalis, E. kambara Agassiz and Mayer, 1899, E. lacteoides, E. macrogonia Huang, Sun and Liu, 2019, E. palkensis Browne, 1905, E. tenuis (Browne, 1905)], with 4–12 simple radial canals, gonads only on subumbrella portions of radial canals that do not extend down the peduncle.
Eirene menoni Kramp, 1953
Menon’s hydromedusa Eirene-Kurage
Type locality. Great Barrier Reef, outside Trinity Harbor, near Cairns, Queensland, Australia (~16°S, 146°E), water temperature 25.7°C, at 32 m depth. One specimen captured 5 Dec 1928. Holotype deposited in Natural History Museum, London, UK [British Museum Natural History BMNH 1954.3.4.559] (Not seen).
Material examined
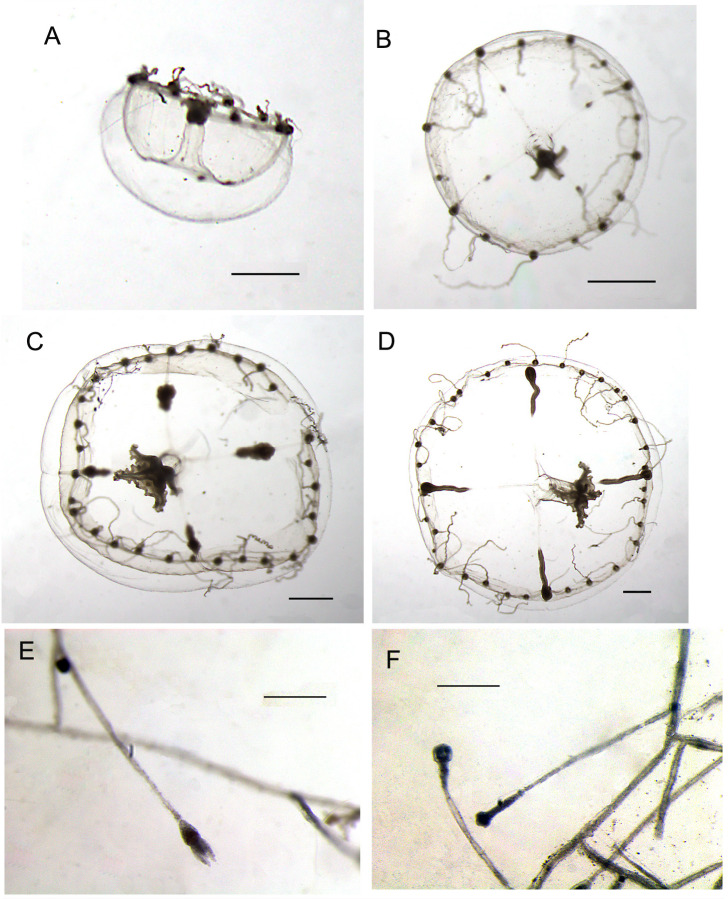
Medusae- SAMA H3614 (4 specimens) wild collected Aldinga Reef, South Australia, 12 Feb 1961 [Fig. 2A](see Kramp 1965). SAMA H3615 (1 specimen, 10.6 mm bell width) wild collected between Hopkins and Thistle Islands, South Australia, 01 Jan 1964 collected by Ronald Vernon Southcott (Fig. 2B). SAMA H1605 (1 specimen, current 13.38 mm bell diameter) wild collected Pumicestone Passage, Moreton Bay, Queensland, Australia (~27.6°S, 153°E), Feb 2000- collected by Puk Petersen. NSMT-Co 1808 (9-specimens) wild collected Shonan Fishing Port, Kanagawa Prefecture, Japan (35.17°N, 139.28°E), collected near surface 12 Sept 2021, Gaku Yamamoto collector. NSMT-Co 1810 (1 specimen) medusa Kamo Aquarium, Shuhei Ikeda collector. NSMT-Co 1811 (1 specimen) medusa Kamo Aquarium, Shuhei Ikeda collector. NSMT-Co 1812 (1 specimen) medusa Kamo Aquarium, S. Ikeda collector.
Polyps- NSMT-Co 1809 cultured colony Kamo Aquarium preserved 1 Oct 2021, transferred from Enoshima Aquarium 31 Jan 2007 [Shuhei Ikeda, pers comm].
GenBank sequences - for E. menoni (see Table 1).
Fig. 2.
Eirene menoni medusa Australia images. (A) SAMA H3614 (current bell width 14.2 mm) collected 12 Feb 1961 at Aldinga Reef, Victoria, South Australia and identified by Paul Kramp. (B) SAMA H3615 (current bell width 10.6 mm) collected 1 Jan 1964 between Hopkins and Thistle Islands, Victoria, South Australia and identified by Ronald V. Southcott. Images courtesy of the South Australian Museum (Shirley Sorokin, Andrea Crowther and Peter Hunt).
Amended species description
After Kramp (1953) original description, modified by Bouillon (1984) description of polyp, Bouillon (1995), and this study. Medusae-Bell more flat than bell shaped; Bell diameter usually 12 mm may reach 20 mm, narrow tubular peduncle extending basically same size down to manubrium (Fig. 3A), manubrium and oral “lips” exceed bell height in living specimens, oral “lips” strap-like in young specimens less than 3.2 mm bell diameter (Fig. 3B), four radial canals with gonads that begin to develop in mid-radial canal and extend to the bell ring canal (Fig. 3B–D), variability of gonad formation was described by Kramp (1953) based on one specimen with gonads extending from the base of the peduncle almost to the ring canal. Kramp (1965) reported that gonadal length was variable and in Victoria, South Australia specimens extends from near the radial ring canal to more than half the distance to the base of the peduncle. Sugiura (1979) illustrated and stated, “gonads begin to be formed on the radial canals neighboring the basal part of the stomachal peduncle and with growth of the medusa they gradually elongate downwards and widen along the radial canal.” Marginal tentacles typically same length, and about 48 in number, may reach 54 with some small developing marginal tentacles. Fully formed tentacle number from Sagami Bay, Japan varies from 16 to 20 in 3.2–5.1 mm bell diameter and 30 to 36 in 5.2–8.0 mm bell diameter specimens. Color of tentacle bulb and between bases of the four oral “lips” varies from green, reddish brown to dark brown. No cirri, excretory papillae or rudimentary bulbs (warts) are present on ring canal, canal contains 1–3 statocysts between successive tentacles (not visible after preservation). Reproduction through a polyp and planula stage. Cnidome composed of elongated fusiform microbasic mastigophores, measuring 10.5 × 3.0 µm to 9.0 × 2.5 µm. Typically found near surface, often near rivers or estuaries, down to a depth of 51 m.
Fig. 3.
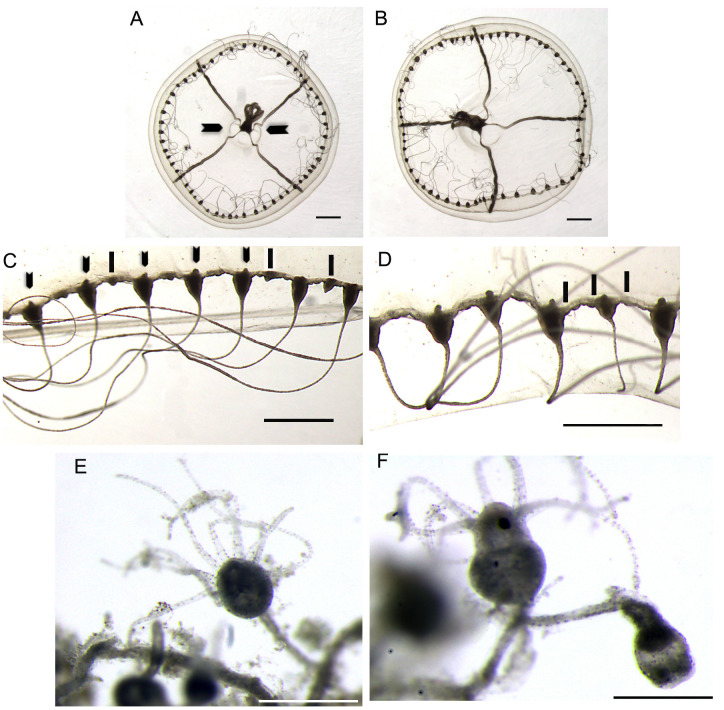
Eirene menoni medusae and polyps Japan images. Medusae (A–D) are images from preserved specimens NMST-Co 1808 originally collected at Shonan Fishing Port, Kanagawa Prefecture, Japan (collected by Gaku Yamamoto). Polyps (E–F) are images of preserved cultured specimens NMST-Co 1809 from the Kamo Aquarium, Japan (collected by Shuhei Ikeda). Image (E) retracted tentacles on the long hydrocaulus and (F) shape of hydrotheca buds. Scale bars: A–D = 1 mm; E–F = 0.125 mm.
Polyps (live and fixed) - Released planula settle at 24 hours and differentiates in polyp stage after three days. Attached planula forms stolonal colonies, arising from creeping hydrorhiza. Hydrorhizal stolons long, slender with distance between polyps (Fig. 3E), hydrothecal pedicels with a thin annulated hydrocaulus that extend in a strong, extensible polyp. The polyp grows erect with a long hydrocaulus, hydranths can be large or reduced, narrowest at base, gastric region typically vase-to slightly club-shaped, constricted short distance below tentacular whorl, constriction usually pronounced but sometimes, indistinct, hydranth expanding again at the distal end, here becoming subspherical to knob-shaped (Fig. 3E, F) supporting whorl of tentacles; tentacles filiform, in one whorl, 10 marginal tentacles; their bases with intertentacular web. Hydroid hydranth tentacles when disturbed or preserved retract almost completely into the hydrotheca (Fig. 3E). Gonotheca unknown. Polyps appear as “campanulinida” type. The polyp cnidome is represented by elliptical atrichs measuring 6.0 × 2.5 µm to 5.0 × 2.0 µm.
Indo-Pacific Ocean biogeographic distribution for 21-records of Eirene menoni [alphabetized by geographic source with map numbers] (Fig. 4).
Fig. 4.
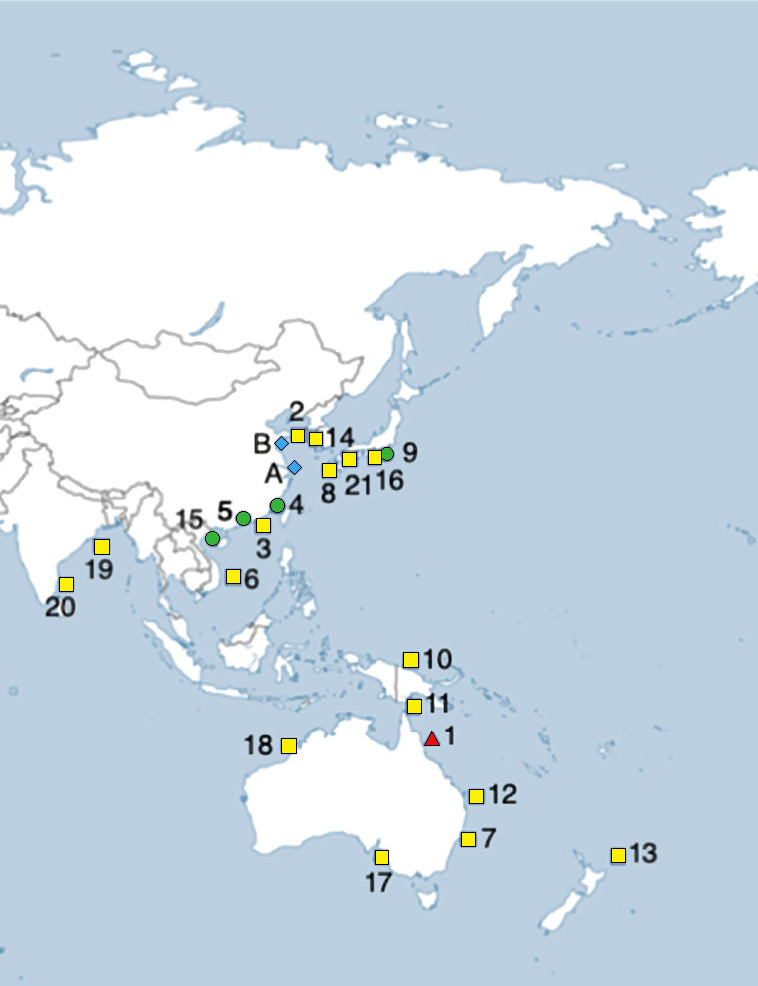
Biogeographical map showing validated wild capture distribution records for Eirenemenoni and Eirene lacteoides. Eirene menoni records that are indicated by red triangle refer to holotype location, green circles refer to localities with COI mtDNA sequences, and yellow squares refer to localities obtained from published studies with sufficient description detail or documentation. Eirene lacteoides records are indicated by a blue diamond. The GPS data for these records is located under the specific species in the text. Specimens with public aquaria captive culture locations are referred to in the manuscript text.
Australia - (1) Trinity Harbor, Cairns [~16°S, 146°E], (7) Sydney, NSW [~33.8°S, 151.2°E], (11) North Cape, Northern Territory [10°S, 143°E], (12) Moreton Bay, Queensland [27.3°S, 153.3°E], (17) Aldinga Reef, Gulf of Saint Vincent [35.16°S, 138.46°E], (18) Broome, North West Territory [17.96°S, 122.24°E], (Kramp 1953; Kramp 1965; Gershwin et al. 2010). China - (2) Chengshan [37.2°N, 122.2°E], (as Phortis lactea) (3) Daya Bay [22.4°N, 114.4°E], (4) Xaimen Bay [24°N, 118°E], (5) Zhujiang River Estuary [36.1°N, 120.3°E], (15) Beibu Gulf [20.85°N, 109.26°E], (Ling 1936–1937; Du et al. 2010; Zheng et al. 2014; Zhang et al. 2015). India - (19, 20) Puri, Odisha [19.8°N, 85.5°E], Chennai [Madras] [13.1°N, 80.2°E] (as Phortis sp.), (Menon 1932; Kramp 1955). Japan - (8) Aitsu Marine Biological Station, Kumamoto Prefecture, Kyushu [32.8°N, 130.7°E]; (9) Shonan Fishing Port, Kanagawa Prefecture, Honshu [35°N, 139.5°E this study], (16) Hamanako, Shizuoka Prefecture, Honshu [34.4°N, 137.3°E], (21) Northern Oita Prefecture, Kyushu [~33.3°N, 131.2°E] (Sugiura 1979; Sakiyama and Adachi 2001; Okamoto et al. 2016; Iwai 2021). PapuaNew Guinea - (10) Laing Island [5.3°S, 147.1°E], (Bouillon 1984). New Zealand - (13) North Island [35.18°S, 174.17°E], (Bouillon 1995). South Korea - (14) Chunjangdae [36.2°N, 126.5°E], Park 1996). Vietnam - (6) Nha Trang [12.3°N, 126.5°E], (Kramp 1962). This species is widespread throughout the Indo-Pacific.
Incertae sedis? Eirene menoni from the Indo-Pacific Basins
?Irene ceylonensis Annandale (1907) Port Channing, Calcutta, India adult medusae 20–25 mm wide, with 100 marginal tentacles.?Phortis lactea Mayer 1910 from Tortugas, Florida, USA. Geographic distance and width of peduncle base. ?Phortis lactea var. chiaochowensis n. var. Kao et al. (1958) two specimens off Tsingtao, China, 15 Jun 1956. Medusae with 60 or more marginal vesicles. Stomach short, situated on conical peduncle, never extending beyond umbrella margin. Illustration with wide peduncle base, illustration may be that of Phortis lactea from Mayer (1910) original drawing. ?Eirene menoni Kramp (1955) from Khal, Dakhnidari Canal, near Calcutta collected 13 May 1926 at 7–8 m depth. Specimens 1–13 mm with 16 to 72 tentacles. ?Eirene menoni Santhakumari and Vannucci (1971) Cochin Harbor, India no description. ?Eirene menoni Thomas and Chhapgar (1977) Maharashtra, India 46 marginal tentacles with two rudimentary bulbs between tentacles. ?Eirene menoni Santhakumari et al. (1997) Bombay Harbor, India no description. ?Eirene menoni SAMA H1605 (non “Eirene menoni” specimen with no peduncle and more than 4 oral “lips”) Moreton Bay, Australia. ?Eirene menoni (sensu Eirene hexanemalis) Buecher et al. (2005) from eastern South Africa (Algoa Bay to Tugela River mouth, Indian Ocean), up to 10 mm bell diameter with up to 30 marginal tentacles, and up to 6 radial canals.
Eirene lacteoides Kubota and Horita, 1992
Kobu-eirene-kurage
Type locality:Captive culture-Toba Aquarium, Toba, Japan. Holotype ZIHU-498 deposited in the Zoological Institute, Faculty of Science, Hokkaido University, Sapporo, Japan (not observed).
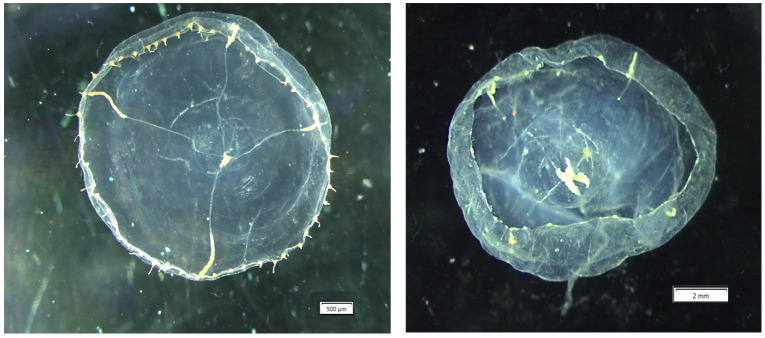
Material examined: Medusae - preserved cultured NSMT-Co 1813 (3 specimens) 17 Feb 2002 bell diameter mm (12.5,14.5, 22.3, Fig. 5A–D) Enoshima Aquarium via Toba Aquarium polyps), Gaku Yamamoto. NSMT-Co 1814 (2 specimens) 9 Feb 2022 Enoshima Aquarium, Gaku Yamamoto. BPBM-D2777 (8 specimens) 6 Aug 2022 Waikiki Aquarium cultured 8- grown out in monoculture tanks, Kelley Niide collector. Polyps - preserved cultured: NSMT-Co 1815 colony 4 Mar 2022 Enoshima Aquarium via Toba Aquarium polyps, G. Yamamoto collector Fig. 5E–F). BPBM-D2777 colony 6 Aug 2022 Waikiki Aquarium, Kelley Niide collector. GenBank sequences - for E. lacteoides (see Table 1).
Morphological description: Amended species description. After Kubota and Horita (1992); Huang et al. (2009) with polyp description; this study.
Medusae - bell wider than high and the umbrella apex is as thick as the length of the peduncle, bell diameter reaches 32 mm with up to 160 marginal tentacle bulbs (fully formed and initial presentation as small tentacle bulbs), typically with four radial canals, rarely five, gonads start to develop near center of radial canals and move distally to the ring canal, but do not reach the ring canal (Fig. 5A, B), never extending to the peduncle, the peduncle is fairly wide at proximal base and is somewhat cone shaped towards the distal end (Fig. 5B), with well-developed gastric peduncle, without marginal and lateral cirri, with adaxial excretory papillae (visible onlarger specimens) (Fig. 5C), rudimentary bulbs (marginal warts) range from 2–7 per quadrant reaching up to 17 in medusae (Fig. 5D) and statocysts (not visible in preserved specimens); four conical projections present in larger specimens (11–31 mm bell diameter) on the interradial distal end of the peduncle (Fig. 5A), four oral “lips” extend beyond the bell and become highly crenulated and folded with maturity, oral “lips” in preserved larger specimens appear “talon-like” (Fig. 5A, B).
Polyps - colonies stolonal, arising from creeping hydrorhiza, hydrorhizal stolons medium length giving rise to hydrothecal pedicels of short lengths, hydrothecal pedicels (Fig. 5E) with hydrocaulus that branch with pedicellate gonotheca forming a single bud (Fig. 5F), hydranth expands at distal end becoming subspherical to knob shaped, with a whorl of tentacles (6–18), tentacles filiform, with intertentacular web. Polyps appear “campanulinida” type.
Current validated wild biogeographic distribution of E. lacteoides - Jiaozhou Bay [Yellow Sea] (36.12°N, 120.25°E), and Changjiang River Estuary [East China Sea] (31.52°N, 122.15°E), China, Pacific Ocean [Zheng et al. 2014] (Fig. 3). Expected presence in the waters of Japan and Hawaii.
Fig. 5.
Eirene lacteoides cultured medusae image of NMST-Co 1813 originally captive culture Enoshima Aquarium specimen obtained by Gaku Yamamoto. Images (A) arrows- indicate conical projections, (C) arrows- adaxial excretory pores, (D) lines- rudimentary bulbs (warts). Images of polyps NMST-Co 1815 originally cultured at Enoshima Aquarium (Gaku Yamamoto). Image (F) shows a hydrotheca and a gonotheca. Scale bars: A– C = 1 mm, check on D–F = 0.25 mm.
Molecular phylogenetic results
The COI gene sequences provided consistent species separation for all Eirene species within the phylogenetic tree, including E. menoni and E.lacteoides (Fig. 6). Eirene menoniCOI sequences from Japanese and Chinese formed a well-supported monophyletic clade for the nine specimens, comprising seven closely related haplotypes, with three individuals sharing a single haplotype, and differing by less than 1% overall. Likewise, the 11 E. lacteoides COI sequences from Japan, China and Hawaii formed a well-supported monophyletic clade with extremely low pairwise divergence, again with a mean of less than 1%, and consisted of four very closely related haplotypes. Interestingly, the GenBank sequences mislabeled as “Tima formosa” from China matched E. lacteoides sequences forming a third strongly supported monophyletic clade. We conducted a pairwise molecular divergence analysis comparing within and between clade divergences. As evidenced by the low bootstrap support in the deeper nodes of the tree occurring between the six clades, there was no clear pattern of similarity within the genera Tima and Eirene. For example, while the pairwise molecular divergence between the two species of Tima was high, 0.174 (17.4%), the mean divergence among the four species of Eirene was even higher, 0.220 (22%) (Table 2). By comparing the divergence of each species of Tima one at a time to each of the four species of Eirene and taking the mean value, we found T. bairdii had a divergence of 0.195 (19.5%) when compared to all Eirene species, and T. nigroannulata had a divergence of 0.191 (19.1%) when compared to each of the four Eirene clades. By calculating the range of divergences from the lowest to the highest values, we found a minimum value of 0.144 (14.4%) which was actually between two species from the two different genera, T. nigroannulata compared to E. menoni. Likewise, the highest pairwise value between any two species was 0.247 (24.7%), and this was found between two species within the same genus, E. viridula and E. lacteoides. The COI data for species of Eirene and Tima therefore suggest that there are substantial evolutionary differences among species within a genus in the family Eirenidae. Based on the rule of thumb for the estimated substitution rate of the COI gene of about 2% per million years, these within genus divergence values and the overall pattern suggest that the time to a most recent common ancestor within each genus is between 8.7 million years for Tima (17.4% divergence), and 11 million years for Eirene (22% divergence). Despite the high divergence values that this study revealed between species within a genus, sequences from a single species from thousands of kilometers away showed little or no divergence, such as a single haplotype shared between specimens of E. lacteoides from China, Hawaii and Japan.
Fig. 6.
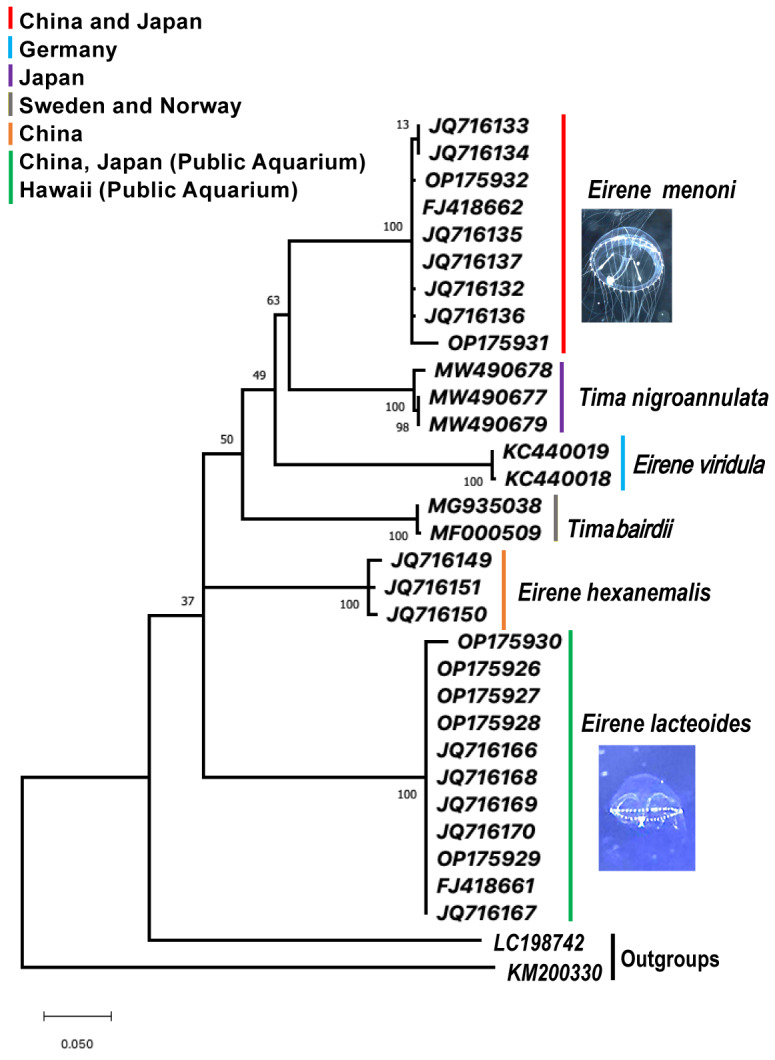
Phylogram based on mitochondrial cytochrome c oxidase I gene fragments from 30 hydromedusan ingroup members of the family Eirenidae, with two outgroup taxa. Specimens were collected from the wild and two public Aquaria. The tree was generated with 1000 bootstrap replicates using the maximum-likelihood optimality criterion with the empirically-determined best-fit substitution model. The optimal model selected was TamuraNei, non-uniformity of evolution rates among sites and this was modeled using a discrete Gamma distribution (+G) with 5 rate categories. Under the assumption that a specified fraction of sites is evolutionary invariable (+l), TN93+G+l. This model had 66 parameters, BIC = 6461.115 (Bayesian Information Criterion), AICc = 6461.12 (Akaike Information Criterion, corrected), maximum likelihood score InL = -2907.703, gamma correction (G) = 1.40, and the proportion of invariant sites (l) = 0.49. Clades are color-coded by species and geographic sampling, including the following general regions by bar color: Red = Eirene menoni, field sampled offshore of China and Japan; Purple = Tima nigroannulata field sample offshore of Japan; Blue = Eirene viridula field sampled offshore of Germany; Gray = Eirene bairdii, field collected offshore Norway and Sweden; Orange = Eirene hexanemalis, field collected offshore China; Green = Eirene lacteoides, field collected offshore of China and captive culture Enoshima Aquarium and Waikiki Aquarium. Outgroup taxa are LC198742 = Cassiopea sp. and KM200330 = Alatinaalata (see Table 1).
Table 2.
Pairwise COI mtDNA sequence divergence matrix, based on the Kimura-2 parameter substitution model and COI gene sequences for family Eirenidae species and two outgroup species. Values in the table are molecular divergence values, each value represents the difference between two clades in the tree shown in figure 4 (to convert molecular divergence values to percentages multiply x 100, i.e., 0.196 = 19.6%)
DISCUSSION
The morphological plasticity present in the Hydrozoa is manifested in intraspecific variation in life cycles, population dynamics, meristic characters, growth, and reproduction (Cunha et al. 2016). Typically, only a single medusa size class is captured during plankton tows, which compounds difficulty in identifying the complete species ontogenetic morphology. As we move forward in classifying the family Eirenidae integrative taxonomy is essential to determine species delineations, boundaries and ranges (Dayrat 2005).
Based on the phylogenetic reconstruction presented in this study, which was generated using aligned mtDNA gene fragments, the genera Eirene and Tima are sister taxa. In spite of this relatively close evolutionary relationship, these two groups exhibit key morphological differences. Divergent morphological traits include size (with adult Tima exceeding 32 mm in bell width), tentacle bases and proximal portion of tentacles that have a longitudinal furrow, and the presence of continuous gonads along the radial canal to stomachal peduncle (Kramp 1936; Bouillon et al. 2006; this study). Upon our examination of the genus Eirene descriptions, we found that rudimentary bulbs (warts) were present in at least eight species. Therefore, we have amended the genus description to reflect this morphological feature. However, more attention needs to be focused on rudimentary bulbs in species that have 60 or more marginal tentacles, as the tentacles tend to exhibit continuous growth. This results in a challenge in differentiating between developing marginal tentacles and permanent rudimentary bulbs. In addition, some COI sequences obtained from GenBank labeled as Clytia sp., matched our EireneCOI data (unpublished data). Thus, genus Eirene warrants a careful systematic re-evaluation.
It should be mentioned that previous phylogenetic studies based on COI sequence data have shown a tendency to lack a sufficient phylogenetic signal to consistently diagnose generic level designations among certain hydromedusan lineages (Zheng et al. 2014). This pattern is evident in the lack of genus-level monophyly for Eirene, Clytia, and Tima in COI phylogenies, and to some degree in 16S-based phylogenies for these taxa (Zheng et al. 2014; Maronna et al. 2016). However, the COI gene fragment is a proven, powerful and accurate tool in species identification and taxonomically matching specimens with GenBank accessions in cases where the GenBank sequences are correctly identified.
The COI gene readily separated E. menoni and E. lacteoides samples and showed well supported monophyletic clades within our phylogenetic tree. However, when we examined family and generic relationships, the COI gene sequences revealed a general lack of ability to resolve basal clade relationships, resulting in generic polytomies and polyphyly with low bootstrap support among the various resolved and supported species clades. Zheng et al. (2014) and Maronna et al. (2016) also reported that COI showed a similar inability to achieve genus level monophyly. Currently, neither 16S nor COI appear to be ideally suited for higher level taxonomic assessment of the genera that comprise Eirenidae.
Based on our analyses, E. menoni is a wideranging species in the Indo-Pacific Ocean and likely has a wider distribution in the Indian Ocean than is currently recognized. The E. menoni hydromedusae from China and Japan formed a well-supported and resolved monophyletic clade (Fig. 6). During this study we encountered multiple misidentifications in the literature, and therefore urge caution when creating species checklists. Eirene menoni is a distinct species that can be identified by COI sequencing and a number of diagnostic morphological features, including a relatively narrow tubular peduncle, marginal tentacle number, and absence of cirri, rudimentary bulbs, and adaxial excretory pores. There is variability in the position of the gonads on the four radial canals from differing geographic locations, and this also warrants further investigation.
This study reflects a clear need for critical analysis of species checklists even when combined with DNA sequence records, which can result in misidentified GenBank entries that can perpetuate hydrozoan taxonomic confusion and errors. For example, we were forced to use incertae sedis for numerous E. menoni published records, a misidentified museum specimen, and unverified checklist records, where the description did not match the original or the amended description of the species in question. Additionally, there was a serious lack of formal descriptions.
The polyp of E. menoni has a long, erect hydrocaulus, with strongly retractable tentacles into the hydranth (Bouillon 1984). An examination of the cultured polyps revealed no direct presence of gonotheca or remnants on the hydrocaulus. The hydrorhiza/stolon also had no presence of gonotheca. Many hydrocaulus pedicels had a clean distal end with no remnants. It is possible that the mature hydranth becomes a gonangia and produces a medusa bud. The absence of obvious gonotheca in E. menoni warrants additional study.
We have confirmed the presence of E. lacteoides in the Yellow and East China Seas. This species likely has a larger range than previously documented, as noted with regards to the cultured samples that appeared in tanks at the Toba Aquarium and the Waikiki Aquarium. Although hydrozoan specimens found in public aquaria may not offer direct information about their geographic distribution, they can serve as indicators of a species’ presence in coastal waters, even if there is no official documentation. This situation raises the possibility of a marine biological introduction. The presence of Tima formosa in the Pacific Ocean cannot be verified with specimens from Japan identified as T. nigroannulata (Calder et al. 2021) or the samples from China recognized as E. lacteoides (this study). The genus Eirene is represented by two readily identified species in the coastal waters of Japan (E.hexanemalis and E. menoni) and likely E. lacteoides. Additional study is needed as 14 Eirene species have been reported from the seas around China (Du et al. 2010; Zheng et al. 2019).
Thorough integrative taxonomic studies focused on the Pacific Eirenidae are warranted. Furthermore, documentation of levels of interspecific morphological variation is essential to the understanding and delineation of biogeographic boundaries and distributional ranges (Cunha et al. 2016). Small planktonic marine species, and particularly those with complex life cycles that include a sessile life stage, such as hydrozoans, tend to be predisposed to anthropogenic introduction via commercial shipping, and longdistance dispersal via rafting on marine debris (Carlton 1987; Choong et al. 2018). The Hawaii specimens shared a single haplotype with China and Japan in the North Pacific Ocean. The presence of E. lacteoides in Hawaiian waters likely represents a previously undetected biological invasion (see Calder 2020).
Effective marine coastal management is enhanced by regular harbor and nearshore surveys, as early detection of invasive marine species, before they can spread and become established is essential to their control. Due to their small size, complex life histories and poorly resolved taxonomic status, marine hydrozoans are not frequently detected or documented as introduced species. The integrative approach used in this study, namely the use of molecular, morphological and life history data together provide an unambiguous basis to identify biological invasions as well as delineate biogeographic patterns, and ultimately to better understand the geographic sources of marine introductions.
CONCLUSIONS
The description of the genus Eirene was modified to include the presence of rudimentary bulbs (warts). Validated distribution records from the Indo-Pacific are provided for E. menoni. The first field collected records are provided for E. lacteoides from the Yellow and East China Seas. The distribution of E. lacteoides may be more extensive than the original Japan record with the Waikiki Aquarium record from Hawaii. Using COI genetic sequencing correctly identifies eirenid species. However, COI sequencing does not resolve family and genus relationships within the Eirenidae. Integrative taxonomy is essential to properly identify Eirene species.
Acknowledgments
This paper is dedicated to Dale R. Calder (1941–2022) based on his multiple decades of particularly comprehensive studies on the Hydrozoa. Dale was much more than just a productive researcher: he was also generous, kind, and a great collaborator. He regularly made time to help other scientists. He mentored and taught numerous young researchers and made everyone he worked with a better scientist. We are grateful to the staff at the Toba Aquarium for sharing polyps from E. lacteoides that enabled the genetic confirmation of taxonomic identity of wild specimens from China. We thank the staff of the Enoshima, Tsuruoka City Kamo and Waikiki Aquarium’s for research support. We appreciate the helpful discussion and comments of Peter Schuchert and are grateful to Andrea Crowther, Shirley Sorokin, and Peter Hunt of the South Australian Museum for providing images. Thanks to Holly Bolick, Bernice P Bishop Museum, Hawaii and Hiroshi Namikawa, National Museum of Nature and Science, Tokyo, Tsubuka, Japan for archiving specimens in their collections. GC and BH acknowledge Ocean Research Explorations for financial support, the WoRMS database for maintaining a comprehensive species list and providing access to obscure literature, Jennifer Crites for medusae and polyp figure creation, and Tina Carvalho, University of Hawaii at Manoa Microscopy Core, supported by NIH NIGMS grants 139753 and P20GM125508 for technical microscope assistance. Support was provided by the Holland Laboratory and the Oceanic Institute at Hawaii Pacific University. This is Ocean Research Explorations publication number 010.
Footnotes
Authors’ contributions: GLC conceived the project and conducted morphological study. GLC and BSH wrote drafts of the manuscript. BSH conducted PCR, gene sequencing and created the phylogenetic tree. GY SI, AA, and KN collected and supplied specimens. All authors reviewed drafts of the manuscript and approved the final manuscript.
Competing interests: The authors declare that they have no conflict of interest.
Availability of data and materials: DNA sequence data generated from this study were deposited into the NCBI GenBank. Specimens from Japan were deposited into the NSMT, Tsukuba and from Hawaii at the BPBM, Honolulu.
Consent for publications: All authors agree to the publication of this work in Zoological Studies.
Ethics approval consent to participate: Not applicable.
References
- Annandale N. 1907. Notes on the freshwater fauna of India, No II, preliminary note on the occurrence of a Madras (Irene ceylonensis Browne) in a brackish pool in the Ganges Delta, and on the hydroid stage of this species. Proc Asiat Soc Bengal N S 3:79.
- Bucklin A, Peljnenburg KTCA, O’Brien TD, Blanco-Bereial L, Cornils A, Falkenhaug T, Hopcroft T, Hosia A, Laakmann S, Li S, Martell L, Questel JM, Wall-Palmer D, Wang M, Wiebe PH. 2021. Toward a global reference database of COI barcodes for marine zooplankton. Mar Biol 168:78. doi:10.1007/s00227-02103887-y.
- Bouillon J. 1984. Hydroméduses de la Mer de Bismark (Papouasie Nouvelle-Guinée. Partie IV: Indo-Malayan Zool 1:25–112.
- Bouillon J. 1995. Hydromedusae of the New Zealand Oceanographic Institute (Hydrozoa, Cnidaria). N Z J Zool 22:223–238. doi:10.1 080/03014223.1995.9518038.
- Bouillon J, Boero F. 2000. Synopsis of the families and genera of the Hydromedusae of the world, with a list of the worldwide species. Thalassia Salentia 24:1–250. doi:10.1285/I15910725V24P47.
- Bouillon J, Gravili C, Pages F, Gili J-M, Boero F. 2006. An introduction to Hydrozoa. Mémoires du Muséum national d’Histoire naturelle Tome 194:1–588.
- Buecher E, Goy J, Gibbons MJ. 2005. Hydromedusae of the Agulhas Current. African Invertebrates 46:27–69.
- Calder DR. 2020. Some leptothecate hydroids (Cnidaria, Hydrozoa) from Hawaii, mostly from inshore and nearshore waters. Zootaxa 4830:201–246. doi:10.11646/zootax.4830.2.1. [DOI] [PubMed]
- Calder DR, Crow GL, Ikeda S, Adachi A, Yamamoto G, Harrington A, Holland BS. 2021. Tima nigroannulata (Cnidaria: Hydrozoa: Eirenidae), a new species of Hydrozoan from Japan. Zool Sci 38:370–382. doi:10.2108/zs210011. [DOI] [PubMed]
- Carlton JT. 1987. Patterns of transoceanic marine biological invasions in the Pacific Ocean. Bull Mar Sci 41:452–465. doi:10.1515/9780824844264-043.
- Choong HHC, Calder DR, Chapman JW, Miller JA, Geller JB, Carlton JT. 2018. Hydroids (Cnidaria: Hydrozoa: Leptothecata and Limnomedusae) on 2011 Japanese marine debris landing in North America and Hawaii, with revisory notes on Hydrodendron Hincks, 1874 and a diagnosis of Plumaleciidae, new family. Aquat Invasions 13:43–70. doi:10.3391/ai.2018.13.1.05.
- Cunha AF, Maronna MM, Marques AC. 2016. Variability on microevoluntionary and macroevolutionary scales: a review on patterns of morphological variation in Cnidaria Medusozoa. Org Divers Evol 16:431–442. doi:10.1007/s13127-016-0276-4.
- Dayrat B. 2005. Towards integrative taxonomy. Biol J Linn Soc Lond 85:407–415. doi:10.1111/j.1095-8312.2005.00503.x.
- Du F, Xu Z, Huang J, Guo D. 2010. New records of medusae (Cnidaria) from Daya Bay, northern South China Sea, with descriptions of four new species. Pro Biol Soc Wash 123:72–86. doi:10.2988/09-18.1.
- Folmer O, Black M, Hoeh, W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cyctochrome c oxidase subunit 1 from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299. . [PubMed]
- Gershwin L, Zeidler W, Davie PJF. 2010. Medusae (Cnidaria) of Moreton Bay, Queensland, Australia. Mem Queensland Mus 54:47–108.
- Hamner WM. 1990. Design developments in the planktonkreisel, a plankton aquarium for ships ships at sea. J Plankton Res 12:397– 402. doi:10.1093/PLANKT/12.2.397.
- He J, Zheng L, Zhang W, Lin Y, Cao W. 2015. Morphology and molecular analyses of a new Clytia species (Cnidaria: Hydrozoa: Campanulariidae) from the East China Sea. J Mar Biol Assoc U K 95:289–300. doi:10.1017/S0025315414000836.
- Holland BS, Dawson MN, Crow GL, Hofmann DK. 2004. Global phylogeography of Cassiopea (Scyphozoa: Rhizostomeae): molecular evidence for cryptic species and multiple invasions of the Hawaiian Islands. Mar Biol 145:1119–1128. doi:10.1007/ s00227-004-1409-4.
- Huang J-Q, Wang W, Guo D-H. 2009. Description and life cycle of a newly recorded Leptomedusa in China, Eirene lacteoides Kubota et Horita, 1992. Acta Zootaxonomica Sinica 34:691–695. (in Chinese with English abstract)
- Iwai M. 2021. Jellyfish found in northern Oita and Beppu Bay. Journal of natural history of Oita Bungoensis 4:92–93. (in Japanese)
- Kao C-S, Li F-L, Chang Ü-M, Li H-L. 1958. On the hydromedusae from Shangtung Coast. J Shangdong Univ (Natural Science) 1:75–118.
- Kramp PL. 1936. On the leptomedusae of the Genera Eirene Eschscholtz and Helgicirrha. Vidensk Medd fra Dansk Foren 99:239–262.
- Kramp PL. 1953. Hydromedusae. Sci Rep Great Barrier Reef Exped 1928–29 6:259–332.
- Kramp PL. 1955. Hydromedusae in the Indian Museum. Rec Indian Mus 53:339–376.
- Kramp PL. 1961. Synopsis of the medusae of the world. J Mar Biol Assoc U K 40:1–409.
- Kramp PL. 1962. Medusae of Vietnam. Vidensk Medd fra Dansk naturh Foren 124:305–366.
- Kramp PL. 1965. Some medusae (mainly Scyphomedusae) from Australian coastal waters. Trans Roy Soc S Aust 89:257–278.
- Kramp PL. 1968. The Hydromedusae of the Pacific and Indian Oceans. Sections II and III. Dana Reports 72:1–200.
- Kubota S, Gravili C. 2007. A list of hydromedusae (excluding Siphonophora, Milleporidae and Actinulidae) in Japan. The Nanki Seibutu, Nanki Biol Soc 49:189–204. (in Japanese)
- Kubota S, Horita T. 1992. A new hydromedusae of the genus Eirene (Leptomedusae: Eirenidae) from Toba, Japan. Zool Sci 9:413– 421.
- Laakmann S, Holst S. 2014. Emphasizing the diversity of North Sea hydromedusae by combined morphological and molecular methods. J Plankton Res 36:64–76. doi:10.1093/plankt/fbt078.
- Lawley JW, Gamero-Mora E, Maronna MM, Chiaverano LM, Stampar SN, Hopcroft RR, Collins AG, Morandini AC. 2021. The importance of molecular characters when morphological variability hinders diagnosability: systematics of the moon jellyfish genus Aurelia (Cnidaria: Scyphozoa). PeerJ 9:e11954. doi:10.7717/peerj.11954. . [DOI] [PMC free article] [PubMed]
- Lindsay DJ, Grossmann MM, Nishikawa J, Bentlage B, Collins AG, Minemizu R, Hopcroft RR, Miyake H, Hidaka-Umetsu M, Nishikawa J. 2017. The perils of online databases: a case study with the ‘monospecific’ genus Aegina (Cnidaria, Hydrozoa, Narcomedusae). Mar Biol Res 13(5):494–512. doi:10.1080/1745 1000.2016.1268261.
- Ling SW. 1936–1937. Studies on Chinese Hydrozoa. I. On some hydromedusae from the Chekiang Coast. Peking Nat Hist Bull 11:357–358.
- Maronna MM, Miranda TP, Pena Cantero AL, Barbeitos MS, Marques AC. 2016. Towards a phylogenetic classification of Leptothecata (Cnidaria, Hydrozoa). Sci Rep 6:18075. doi:10.1038/srep18075. . [DOI] [PMC free article] [PubMed]
- Mayer AG. 1910. Medusae of the World Volume II, The Hydromedusae. Carnegie Institute of Washington, D.C., 498 pp.
- Menon MGK. 1932. The hydromedusae of Madras. Bull Madras Govt Mus, N S nat Hist Sect 3:1–32.
- Moura CJ, Harris DJ, Cunha MR, Rodgers AD. 2008. DNA barcoding reveals cryptic diversity in marine hydroids (Cnidaria, Hydrozoa) from coastal and deep-sea environments. Zool Scripta 37:93–108. doi:10.1111/j.1463-6409.2007.00312.x.
- Okamoto K, Sugimura T, Ohtake J, Toh H, Katoh O. 2016. A checklist of the gelatinous zooplankton collected in Lake Hamana. Bull Shizuoka Pref Res Inst Fish 49:31–33. (in Japanese with English abstract)
- Park JH. 1996. Four hydromedusae (Cnidaria: Hydrozoa) from Korean waters. J Systematic Zool 12:67–77.
- Raskoff KA, Sommer FA, Hamner WM, Cross KM. 2003. Collection and culture techniques for gelatinous plankton. Bio Bull 204:68–80. doi:10.2307/1543497. [DOI] [PubMed]
- Sakiyama T, Adachi A. 2001. Medusae collected in Enoshima-Shonan Port and its adjacent waters-II. Nat Hist Rep Kanagawa 22:69–72. (in Japanese English abstract)
- Santhakumari V, Ramaiahan N, Nair VR. 1997. Ecology of the hydromedusae from Bombay Harbor- Thana and Bassein Creek estuarine complex. Indian J Mar Sci 26:162–168.
- Santhakumari V, Vannucci M. 1971. Monsoonal Fluctuations in the distribution of the hydromedusae in the Cochin backwater, 1968–1969. J Mar Biol Assoc India 13:211–219.
- Schuchert P. 2017. Systematic notes of some leptomedusa species with a description of Neotima galeai n. spec. (Hydrozoa, Cnidaria). Revue suisse de Zoologie 124:351–375. doi:10.5281/ zenodo.893549.
- Scorrano S, Aglieri G, Boero F, Dawson MN, Pirainos S. 2017. Unmasking Aurelia species in the Mediterranean Sea: an integrative morphometric and molecular approach. Zool J Linn Soc Lond 180:243–267. doi:10.1111/ZOJ.12494.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi:10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed]
- Straehler-Pohl I, Widmer C, Morandini AC. 2011. Characterizations of juvenile stages of some semaeostome Scyphozoa (Cnidaria), with recognition of a new family (Phacellophoridae). Zootaxa 274:1–37. doi:10.11646/zootaxa.2741.1.1.
- Sugiura Y. 1979. On a hydromedusa Eirene menoni Kramp from Amakusa, Japan. Proc Jap Soc Syst Zool no 16:5–8.
- Tamura K, Stecher G, Kumar K. 2021. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 38:3022–3027. doi:10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed]
- Thomas J, Chhapgar BF. 1977. Hydrozoa from the coastal waters of Maharashtra. J Bombay Nat Hist Soc 74:581–591.
- Toshino S, Nishikawa J, Srinui K, Taleb S, Miyaki H. 2019. New records of two Cubozoa from Thailand. Plankton Benthos Res 14:143–149. doi:10.3800/PBR.14.143.
- Zhang C, Huang J, Sun X, Zhao Z, Liu Y. 2019. Two new species of Eirene (Leptomedusae, Conica, Eirenidae) from the Zhanjiang Bay of Guangdong, China. Haiyang Xuebao 41:172–176. (in Chinese with English abstract)
- Zhang D, Zheng L, He J, Zhang W, Lin Y, Li Y. 2015. DNA barcoding of hydromedusae in northern Beibu Gulf for species identification. Biodiversity Science 23:50–60. doi:10.17520/ biods.2014089.
- Zheng L, He J, Lin Y, Cao W, Zhang W. 2014. 16S rRNA is a better choice than COI for DNA barcoding hydrozoans in coastal waters of China. Acta Oceanol Sinica 33:55–76. doi:10.1007/s13131- 014-0415-8.
- Zhou K, Zheng L, He J, Lin Y, Cao W, Zhang W. 2013. Detection of a new Clytia species (Cnidaria: Hydrozoa: Campanulariidae) with DNA barcoding and life cycle analyses. J Mar Biol Assoc U K 93:2075–2088. doi:10.1017/S0025315413000969.