Abstract
Natural products synthesized by plants have substantial industrial and medicinal values and are therefore attracting increasing interest in various related industries. Among the key enzyme families involved in the biosynthesis of natural products, uridine diphosphate-dependent glycosyltransferases (UGTs) play a crucial role in plants. In recent years, significant efforts have been made to elucidate the catalytic mechanisms and substrate recognition of plant UGTs and to improve them for desired functions. In this review, we presented a comprehensive overview of all currently published structures of plant UGTs, along with in-depth analyses of the corresponding catalytic and substrate recognition mechanisms. In addition, we summarized and evaluated the protein engineering strategies applied to improve the catalytic activities of plant UGTs, with a particular focus on high-throughput screening methods. The primary objective of this review is to provide readers with a comprehensive understanding of plant UGTs and to serve as a valuable reference for the latest techniques used to improve their activities.
Keywords: Glycosyltransferase, UGT, Structure, Mechanism, Protein engineering
Graphical Abstract
1. Introduction
Natural products derived from plants have significant values in both industrial and pharmaceutical applications. These products are synthesized through diverse metabolic pathways that include various modifications such as glycosylation, phosphorylation, acetylation, and methylation. Among these modifications, glycosylation is a ubiquitous and vital processes [1]. Glycosylation reactions are catalyzed by glycosyltransferases, which transfer sugar moieties from active sugar donor molecules to acceptor substrates. Depending on how the sugar binds to the acceptor, glycosylation is typically categorized into four types, i.e., O-glycosylation, C-glycosylation, N-glycosylation and S-glycosylation. These reactions play a crucial role in the biosynthesis of natural products, e.g., in facilitating their storage and accumulation in plant cells, as well as in modulating their chemical properties and biological activities [1], [2], [3]. Glycosyltransferases have been classified into 116 different families based on the sequence similarity (CAZy, http://www.cazy.org/GlycosylTransferase-family), and this number continues to increase as new members are identified.
Uridine diphosphate glycosyltransferases (UDP-glycosyltransferase, UGTs), belonging to the family 1 of the glycosyltransferase superfamily, catalyze sugar transfer from UDP-activated sugar donors to various substrates. Plant genomes contain a large number of UGTs. For example, 122 UGT genes have been identified in Arabidopsis, and a total of 339 putative UGT genes have been identified in the genome of Eupatorium pubescens, which contains the largest known family of UGT genes [4], [5]. Plant UGTs are widely involved in the biosynthesis of natural compounds such as flavonoids, terpenoids, etc. Flavonoids, which affect the growth and development of plant organs [6], [7], have significant pharmaceutical potential for human health. For instance, the catalytic product of Arabidopsis thaliana UGT78D1 (AtUGT78D1), quercetin 3-O-glycoside, showed therapeutic effects against various diseases including e.g. cardiovascular diseases, diabetes and cancer [8], [9], [10], [11]. Silibinin glycosides synthesized by Zea mays UGT706F8 (ZmUGT706F8) showed hepatoprotective and anticancer activities [12], [13]. In addition, terpenoids, the largest group of natural products with more than 40,000 members [14], can be widely used as industrial raw materials such as toners, flavorings and spices [15]. Certain terpenoids, similar to flavonoids, have shown anti-tumor, anti-malarial, sterilizing and insecticidal effects [16].
Plant UGTs are not only involved in the biosynthesis of compounds for industrial and pharmaceutical applications, but also play a crucial role in regulating protective responses to environmental stresses. For instance, Camellia sinensis UGT78A14 (CsUGT78A14) was highly upregulated under cold stress in tea plants. Suppression of the expression of CsUGT78A14 led to decreased levels of flavonols and reactive oxygen species, which subsequently weakened the cold stress tolerance of tea plants [17]. Another representative example is Arabidopsis thaliana UGT89C1 (AtUGT89C1), which catalyzes the biosynthesis of flavonoid substrates that protect cells from UV damage [18]. In addition, plant UGTs are involved in maintaining the homeostasis of auxins, which are essential plant hormones and are tightly regulated for plant growth. For example, Arabidopsis thaliana UGT76F1 (AtUGT76F1) regulates auxin homeostasis through glycosylating the growth hormone indole-3-pyruvic acid in response to environmental factors such as light intensity and temperature [19].
The production of natural products from plants is often costly, primarily due to the low natural yields and the long lifespan of the plants. Moreover, chemical synthesis as an alternative approach faces major challenges because of the typically complex molecular structures and chemical-physical properties of these compounds. Reactions suffer from poor regioselectivity and often require intense effort and aggressive solvents or conditions, etc. As a result, in the last decade, there has been a growing interest in developing biosynthetic processes of natural products using microorganisms.
Advances in biotechnologies such as gene mining, synthetic biology, microbiology and protein engineering have contributed significantly to the field of natural product biosynthesis [20], [21], [22]. For example, the rare ginsenoside Rh2 was successfully synthesized with yeast cells in 2013 by introducing the UGTPg45 gene into a yeast chassis that produced protopanaxadiol [23]. The product yield was later significantly improved after a number of optimizations, including increasing the copy number of UGTPg45, engineering its promoter for high-level expression, substituting UGTPg45 with more efficient homologs from other plants or mutants obtained through directed evolution, and overexpressing the metabolic precursors [24]. In a 10- L fed-batch fermentation, the yeast factory achieved an impressive Rh2 titer of 2.25 g/L. However, the production of natural products with microbes still faces major challenges. One critical factor is the limited catalytic activity or poor regioselectivity of the key enzyme(s), resulting in a low yield of the desired product [20], [25]. The development of efficient enzymes and protein engineering are essential to overcome this bottleneck.
As key enzymes involved in the biosynthesis of many natural products, plant UGTs have been extensively studied and used in the establishment of microbial cell factories [20], [21], [22]. In this review, we provided a comprehensive analysis of the recent structures of plant UGTs and addressed the relationship between their structures and biological activities. Additionally, we systematically discussed the approaches for engineering and optimizing plant UGTs to enhance their performance or achieve new features.
2. Structural insights into plant UGTs
2.1. Structures of plant UGTs
The substrate recognition and catalytic activity of plant UGTs depend on their three-dimensional structures. Currently, structures of 30 plant UGTs have been released in the Protein Data Bank (PDB, https://www.rcsb.org/; accessed in May 2023), all of which were resolved using X-ray crystallography (Table 1). These structures shared a conserved GT-B fold, consisting of two Rossmann-like domains. Each domain comprises a central β-sheet flanked by α-helices, forming a substrate-binding pocket. The C-terminal tail forms an elongated helix that curves and extends back into the N-terminal domain (NTD) to stabilize the bi-lobed structure. Structures that were co-crystallized with ligands have shown that the NTD and C-terminal domain (CTD) are responsible for the recognition and binding of the acceptor and donor, respectively. The NTD and CTD are connected by an extended linker, which typically consists of 14–30 amino acids. In some UGTs such as Sorghum bicolor UGT85B1, this linker contains a small helical structure. However, in most UGTs, this region forms a flexible loop, and a portion of it cannot be modeled.
Table 1.
The Plant UGTs with solved structures. The sources of these structures were retrieved from the Protein Data Bank in May 2023. Abbreviations: UDP, uridine diphosphate; UDP-2FGlc, UDP-2-deoxy-2-fluoro glucose; DCA, 3,4-dichloroaniline; TCP, 2,4,5-trichlorophenol; DTX, digitoxigenin; SA, salicylic acid; 2-BA, 2-bromobenzoic acid (an SA analogue); UDP-Glc, UDP-glucose; UDP-Gal, UDP-galactose; UDP-GlcA, UDP-glucuronic acid; UDP-Xyl, UDP-xylose; UDP-Arap, UDP- arabinopyranose; UDP-Rha, UDP-rhamnose; UDP-GlcNAc, UDP-N-acetylglucosamine; D3G, deoxynivalenol 3-O-glucoside; DCP,3,4-dichlorophenol; DCT,3,4-dichlorothiophenol;Sm, steviolmonoside; Reb E (D), rebaudioside E (D); ST, steviol; STB, a glycoside of steviol.
| UGT (Species) | Subfamily | Sugar donor* | Acceptor# | Apo/complex (PDB ID) | Reference |
|---|---|---|---|---|---|
|
LpCGTb (Landoltia punctata) |
UGT71 | UDP-Arap UDP-Xyl UDP-Gal (20%)* * |
2-hydroxyflavanone | Apo (6LFN) | [26] |
|
SbCGTb (Scutellaria baicalensis) |
UGT71 | UDP-Arap UDP-Xyl UDP-Glc |
2-hydroxyflavanone | UDP-Glc (6LFZ) | [26] |
|
GgCGT (Glycyrrhiza glabra) |
UGT71 | UDP-Glc UDP-Xyl UDP-Gal UDP-Arap |
Phenolic compounds (flavonoids, simple aromatic compound) |
UDP-Glc (6L5P) UDP-Gal (6L5Q) UDP, phloretin (6L5R,6L5S) UDP, nothofagin (6L7H) |
[27] |
|
LpCGTa (Landoltia punctata) |
UGT71 | UDP-Glc UDP-Arap UDP-Gal UDP-Xyl |
2-hydroxyflavanone | UDP (6LG1) | [26] |
| MiCGT## (Tetranychus Urticae) |
UGT71 | UDP-Glc UDP-Xyl |
Flavonoids (quercetin, maclurin) Acetophenones (2,4,6-trihydroxyacetophenone) DCA |
UDP-Glc (7VA8) | [28], [29] |
|
Os79## (Oryza sativa Japonica Group) |
UGT74 | UDP-Glc | Tricyclic sesquiterpenoid epoxides (deoxynivalenol nivalenol, isotrichodermol, HT-2 toxin) |
UDP (5TMB, 5TME) UDP-2FGlc, trichothecene (5TMD) UDP, D3G (6BK3) |
[30], [31] |
|
PaGT2 (Phytolacca americana) |
UGT72 | UDP-Glc | Polyphenols (flavonoids, stilbenoids) |
Apo (6JEL) UDP-2FGlc, resveratrol (6JEM) UDP-2FGlc, pterostilbene (6JEN) |
[32] |
|
PaGT3 (Phytolacca americana) |
UGT73 | UDP-Glc | Polyphenols (flavonoids, anthocyanins, stilbenoids) |
15-crown-5 ether (6LZX) 18-crown-6 ether (6LZY) UDP-2FGlc (7VEL) UDP-2FGlc, capsaicin (7VEK) UDP-2FGlc, kaempferol (7VEJ) |
[33], [34] |
|
PtUGT1 (Persicaria tinctoria) |
UGT72 | UDP-Glc | Indoxyl DCP DCA DCT |
Indoxyl sulfate (5NLM) UDP-Glc (6SU6) DCA (6SU7) |
[35], [36] |
|
SbCGTa (Scutellaria baicalensis) |
UHT71 | UDP-Glc UDP-Arap UDP-Gal UDP-Xyl |
2-hydroxyflavanone | UDP (6LG0) | [26] |
|
TcCGT1 (Trollius chinensis) |
UGT72 | UDP-Glc UDP-Xyl UDP-Gal UDP-Arap |
Aromatic compounds (flavonoids, hydroxynaphthalenes, stilbenes) |
UDP (6JTD) | [37] |
| UGT706F8 (Zea mays) |
UGT71 | UDP-Glc | Flavonoids (silybin) |
UDP (7Q3S) | [13] |
| UGT708C1 (Fagopyrum esculentum) |
UGT71 | UDP-Glc UDP-Gal |
Flavonoids (phloretin) Acetophenones (2′,4′,6′-trihydroxyacetophenone) |
Apo (6LLG) UDP (6LLW) UDP-Glc (6LLZ) BrUTP (7CYW) |
[38] |
| UGT71G1 (Medicago truncatula) |
UGT71 | UDP-Glc | Triterpenoids (medicagenic acid, hederagenin) Flavonoids (quercetin) Isoflavonoids (genistein, biochanin A) |
UDP (2ACV) UDP-Glc (2ACW) |
[39] |
| UGT72B1 (Arabidopsis thaliana) |
UGT72 | UDP-Glc UDP-Xyl UDP-Gal (weak)* * |
DCA TCP |
UDP-2FGlc, TCP (2VCE) UDP (2VCH) UDP-Tris (2VG8) |
[40] |
| UGT74AC1## (Siraitia grosvenorii) |
UGT74 | UDP-Glc | Triterpenoids (mogrol) Flavonoids (quercetin) |
Apo (6L90) UDP-Glc (6L8Z) |
[41] |
| UGT74AC2 (Siraitia grosvenorii) |
UGT74 | UDP-Glc | Flavonoids (silybin) |
UDP (7BV3) | [42] |
| UGT74AN2 (Calotropis gigantea) |
UGT74 | UDP-Glc UDP-GlcNAc (13%)* * UDP-Gal (6%)* * |
Aromatic compound (flavonoids, stilbenes) Steroids Terpenoids |
Apo (7W09) UDP (7W0K) UDP-Glc (7W1H) UDP, sibufogenin (7W0Z) UDP, bufalin (7W10) UDP, DTX (7W1B) UDP, DTXG (7W11) |
[43] |
| UGT74F2## (Arabidopsis thaliana) |
UGT74 | UDP-Glc | Benzoic acid and derivatives (benzoic acid, SA) |
UDP, SA (5U6M) UDP, 2-BA (5U6S) |
[44] |
| UGT78G1 (Medicago truncatula) |
UGT78 | UDP-Glc | Flavonoids (kaempferol, myricetin) Isoflavonoids (formononetin) Anthocyanidins (pelargonidin, cyanidin) |
UDP (3HBJ) UDP, myricetin (3HBF) |
[45] |
| UGT78K6 (Clitoria ternatea) |
UGT78 | UDP-Glc | Anthocyanidins (delphinidin, petunidin) Flavonoids (kaempferol) |
Apo (3WC4) UDP (4WHM) Kaempferol (4REL) Delphinidin (4REM) Petunidin (4REN) |
[46], [47] |
| UGT85B1 (Sorghum bicolor) |
UGT85 | UDP-Glc | Cyanohydrins (P-hydroxymandelonitrile) Terpenoids (geraniol) Aromatic compound (mandelonitrile, benzyl alcohol) Hexanol derivatives (1-hexanol) |
UDP (7ZER) UDP, p-hydroxmandelonitrile (7ZF0) |
[48], [49] |
| UGT85H2 (Medicago truncatula) |
UGT85 | UDP-Glc | Flavonoids (kaempferol) Isoflavonoids (biochanin A) Chalcones (isoliquiritigenin) |
Apo (2PQ6) | [50] |
| UGT91C1 (Oryza sativa Japonica Group) |
UGT91 | UDP-Glc | Diterpenoids (Steviol glycosides) |
Apo (7ERY) UDP, Reb E (7ES0) UDP, ST (7ES1) UDP, STB (7ERX) UDP, Reb D (7ES2) |
[51] |
|
VvGT1 (Vitis vinifera) |
UGT78 | UDP-Glc UDP-GlcNAc (weak) UDP-Gal (weak) * * UDP-Xyl (weak) * * |
Flavonoids (quercetin, kaempferol) Anthocyanidins (cyanidin) |
UDP (2C1X) UDP-2FGlc, kaempferol (2C1Z) UDP, quercetin (2C9Z) |
[52] |
|
ZmCGTa (Zea mays) |
UGT71 | UDP-Glc UDP-Arap UDP-Gal UDP-Xyl |
2-hydroxyflavanone | UDP (6LF6) | [26] |
| UGT73P12 (Glycyrrhiza uralensis) |
UGT73 | UDP-GlcA UDP-Glc (14%)* * UDP-gal (1.3%)* * |
Triterpenoids (glycyrrhetinic acid 3-O-monoglucuronide) |
UDP, glycyrrhetinic acid 3-O- monoglucuronide (7C2X) | - |
| UGT89C1 (Arabidopsis thaliana) |
UGT89 | UDP-Rha | Flavonoids (quercetin, kaempferol) |
Apo (6IJ7) UDP (6IJ9) UDP-Rha (6IJA) Quercetin (6IJD) |
[53] |
| UGT76G1## (Stevia rebaudiana) |
UGT76 | UDP-Xyl UDP-Glc |
Diterpenoids (steviol glycosides) |
UDP (6INF, 6KVI, 6O87) UDP-Xyl (6KVJ) UDP, Sm (6KVK) UDP, Reb B (6KVL,6INI,6O88) |
[54], [55], [56] |
|
ZjOGT3 (Ziziphus jujuba var. spinosa.) |
UGT84 | UDP-Glc UDP-Xyl UDP-GlcNAc (22%)* * UDP-Arap (19%)* * |
Flavonoids Isoflavonoids Chalcones |
UDP (8INH) | [57] |
*The sugar donors are listed in order of preference.
* * Percentage represents the conversion rate, whereas “weak” represents low catalytic activity.
# Specific compounds are listed in parentheses.
## Mutant structures are not listed.
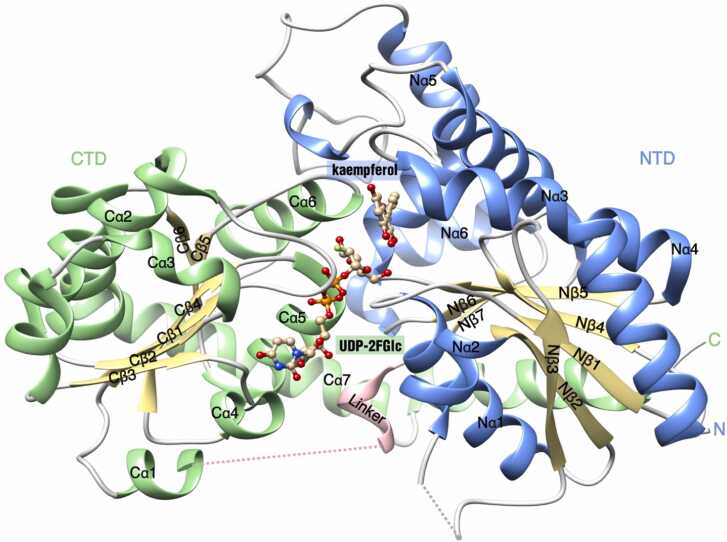
The typical plant UGT structure can be exemplified by the UDP-Glucose:flavonoid 3-O-glycosyltransferase (VvGT1) of Vitis vinifera, which represents a plant UGT structure in its ‘Michaelis’ complex with both a non-hydrolysable UDP-2-deoxy-2-fluoro glucose donor (UDP-2FGlc) and the acceptor kaempferol (Fig. 1) [52].
Fig. 1.
Structure ofVitis viniferaUDP-Glucose:flavonoid 3-O-glycosyltransferase (VvGT1) in Michaelis complex with UDP-2-deoxy-2-fluoro glucose (UDP-2FGlc, non-hydrolysable donor) and kaempferol (acceptor) (PDB ID: 2C1Z). The cartoon shows the overall structure of VvGT1, with the ligands displayed in ‘ball-and-stick' representation. The β-sheets in both N-terminal domain (NTD, residues 1–242) and C-terminal domain (CTD, residues 260–456) are shown in yellow, while the α-helices in the NTD and CTD are depicted in blue and green, respectively. Loops and the linker connecting the NTD and CTD are presented in gray and pink, respectively. The ligand atoms are color-coded using the CPK scheme, with oxygen in red, nitrogen in blue, fluorine in green and phosphorus in orange. All α-helices and β-strands are labeled to facilitate subsequent analysis. This figure was generated using UCSF Chimera.
The similarity of amino acid sequence among plant UGTs is typically less than 30%, even though they have similar overall structures. For instance, when comparing Medicago truncatula UGT85H2 (MtUGT85H2) and VvGT1, the root-mean-square deviation (R.M.S.D.) for 382 Cα atoms was just 1.7 Å, even though only ∼25% of their amino acid sequences were identical [50]. These results strongly suggest that a structure-based mechanism underlies the biological functions of plant UGTs.
2.2. Mechanism of glycosylation by plant UGTs
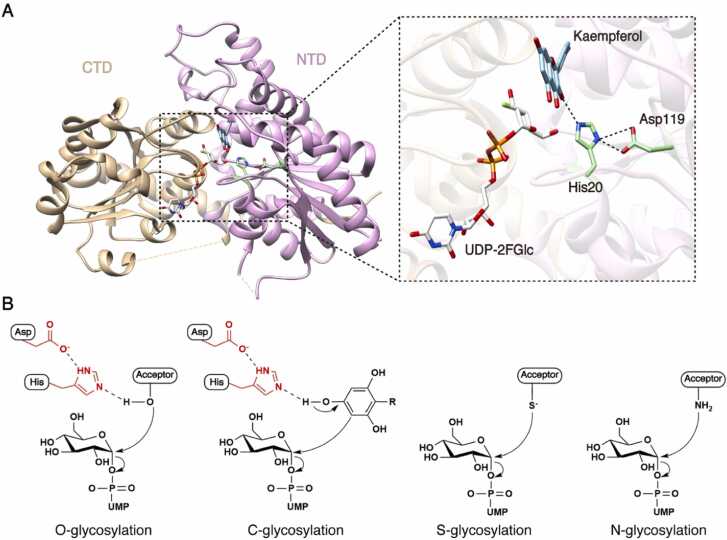
Plant UGTs are glycosyltransferases that use an inverting mechanism. It typically utilizes SN2-like direct displacement to catalyze glycosyl transfer and reverse the configuration at the anomeric carbon of the UDP-sugar. This process led to the formation of glycoside products. Extensive structural, docking and simulation studies have developed a model for the catalytic mechanism of plant UGTs [16], [58], [59], which consists of two steps: i) deprotonation of the catalytic atom of the acceptor molecule results in nucleophilic properties; and subsequently ii) the nucleophilic catalytic atom initiates a direct displacement reaction by attacking the C1 anomeric carbon of the UDP-sugar. While all plant UGTs follow to a common mechanism for the second step, they exhibit variations in the first step, as the deprotonation process is influenced by the different physicochemical properties of acceptor molecules involved in O-, C-, N-, and/or S-glycosylation.
O-Glycosylation is the predominant type of glycosylation catalyzed by plant UGTs, and its catalytic mechanism has been studied extensively. In most cases of O-glycosylation, deprotonation of the catalytic hydroxyl group of the acceptor molecule is initiated by a highly conserved histidine located at the N-terminus (Fig. 2A). A conserved aspartate residue (Fig. 2A) in the immediate vicinity of the histidine residue further stabilizes the deprotonated state of the acceptor by balancing the charge on histidine and forming a catalytic Acceptor-Histidine-Aspartate triad (Fig. 2B). After deprotonation, the acceptor molecule directly performs a nucleophilic attack on the anomeric carbon of the glucose moiety, replacing the UDP moiety and leading to the formation of the final glycoside product [39]. This model mechanism was demonstrated experimentally, as mutations of the histidine or aspartate residue to alanine or asparagine eliminated or significant decreased the catalytic activity of O-glycosylation [28], [43].
Fig. 2.
Proposed catalytic mechanisms of plant UGTs. A. The Histidine-Aspartate dyad of VvGT1 (PDB ID: 2C1Z). The NTD and CTD are represented in pink and tan, respectively. Proposed hydrogen bonds are depicted as dashed lines. B. Schematic diagram illustrating the SN2-like direct displacement catalytic mechanisms of plant UGTs. In O- and C-glycosylation, the Histidine-Aspartate dyad deprotonates the hydroxyl group and the ortho-aromatic carbon of the acceptor molecules, respectively. In S- and N-glycosylation, the deprotonation of acceptor molecules occurs spontaneously. Subsequently, the deprotonated acceptor molecules undergo nucleophilic attack on the anomeric carbon of the sugar donors, ultimately forming the inverted glycoside products.
However, there are exceptions to this model, including the catalytic mechanisms for the Ziziphus jujuba flavonoid 7,4′-di-O-glycosyltransferase (ZjOGT3) [57], Trollius chinensis O-/C-glycosyltransferase (TcCGT1) [37], Phytolacca americana polyphenol glycosyltransferase (PaGT2) [32], Zea mays silibinin glucosyltransferase UGT706F8 [13], and AtUGT89C1 [53]. In the O-glycosylation reactions catalyzed by ZjOGT3 and TcCGT1, spontaneous deprotonation of the catalytic hydroxyl group under physiological conditions was proposed [37], [57]. The conserved histidine residue in these two glycosyltransferases probably facilitated the reaction by forming (and thus stabilizing) hydrogen bonds with the deprotonated substrates or glycoside products. In the case of PaGT2, His81, in addition to the canonical Histidine-Aspartate dyad, was able to independently deprotonate the acceptor molecule [32]. In contrast, both ZmUGT706F8 and AtUGT89C1 lack the conserved aspartate residue, and the histidine residue was able to deprotonate the acceptor molecule to create a nucleophile itself [13], [53].
Unlike that of the O-glycosylation, the catalytic mechanism of C-glycosylation is controversial. Recently, a relatively generally accepted mechanistic model based on the structures of several plant C-glycosyltransferases (CGTs) has been proposed (Fig. 2B) [27], [38], [60]. The typical acceptor substrates for C-glycosylation are flavonoids, benzophenone, trihydroxyacetophenone, and other phenolic compounds. The catalytic aromatic carbon is normally in an ortho-position to a hydroxyl group. According to the proposed model, the highly conserved Histidine-Aspartate dyad facilitates the deprotonation of the phenolic hydroxyl group of the acceptor. Through resonance, the ortho-aromatic carbon is deprotonated, allowing it to carry out a nucleophilic attack on the anomeric carbon of the sugar donor, ultimately leading to the formation of the C-glycoside products. An exception to this model is the multifunctional glycosyltransferase TcCGT1 [37]. Similar to the O-glycosylation reaction catalyzed by TcCGT1, the acceptor substrate flavone performed spontaneous deprotonation to facilitate the C-glycosylation. In this case, deprotonation occurred at the hydroxyl group in the ortho-position, resulting in a negatively-charged catalytic aromatic carbon due to the electron rearrangement in the aryl ring. The negatively-charged catalytic carbon then attacked the anomeric carbon of the sugar donor, completing the C-glycosylation reaction [37]. Since both C- and O-glycosylation activities of TcCGT1 shared a common catalytic mechanism, the binding orientation of the substrate determined whether TcCGT1 acts as a CGT or OGT. In particular, the proximity of the negatively-charged catalytic atoms to the anomeric carbon of the sugar donor was critical for the mode of glycosylation [37].
In contrast to C- and O-glycosylation, which require the Histidine-Aspartate catalytic dyad for the deprotonation of the acceptor substrate, N- and S-glycosylation are normally initiated through spontaneous deprotonation (Fig. 2B) [36], [40]. The catalytic reactions of N- and S-glycosylation depend on the relative positioning from the acceptor substrate to the donor molecule. However, the conserved histidine residue may still play a role in the control and orientation of the nucleophilic attack from the acceptor substrates [36], [40]. For the Arabidopsis thaliana bifunctional O-/N-glycosyltransferase UGT72B1 (AtUGT72B1), mutation of the histidine residue to alanine (His19Asn) only led to a slight decrease in N-glycosyltransfer activity compared to the wildtype enzyme [40]. Furthermore, the multifunctional enzyme PtUGT1 from Polygonum tinctorium showed comparable kinetics of O-, N-, and S-glycosylation. However, acceptor specificity can be significantly altered by a mutation of the histidine residue (His26). Noteworthy, PtUGT1-His26Ala reached exclusively N-glycosylation selectivity, while PtUGT1-Asp122Asn exclusively for S-glycosylation. The PtUGT1-His26Glu performed improved N-glycosylation activity, likely due to the changes in the positioning and orientation of the acceptor molecules in the mutant enzyme [36].
2.3. Sugar donor recognition and specificity
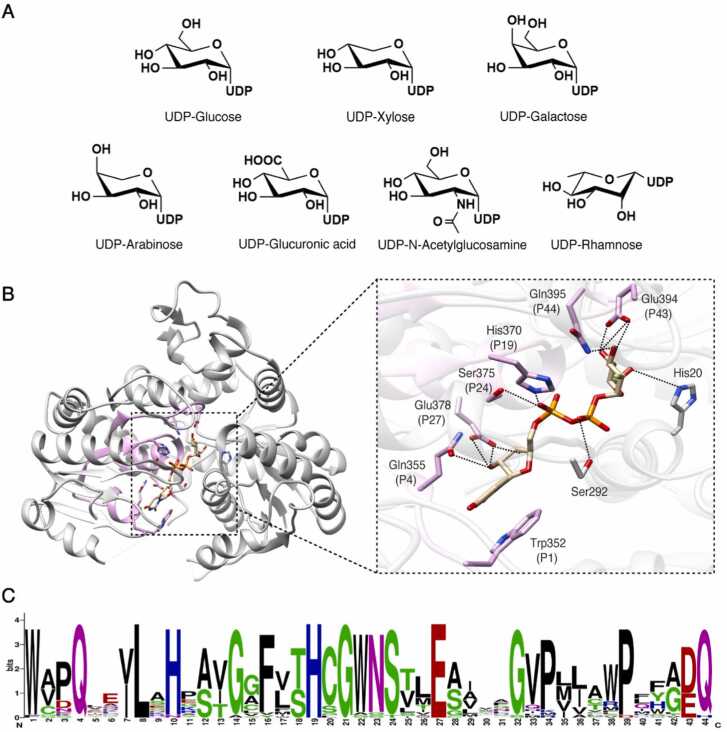
Sugar donors of UGTs mainly include UDP-glucose (UDP-Glc), UDP-glucuronic acid (UDP-GlcA), UDP-galactose (UDP-Gal), UDP-xylose (UDP-Xyl), UDP-arabinose (UDP-Ara), UDP-rhamnose (UDP-Rha) and UDP-N-acetylglucosamine (UDP-GlcNAc) (Fig. 3A). They all share a common UDP base but differ in their sugar ligands, including hexose, pentose and their derivatives. Among them, UDP-Glc is the most commonly used sugar donor for plant UGTs, while UDP-Gal, UDP-Ara and UDP-Xyl are also frequently observed.
Fig. 3.
Sugar donor recognition and specificity of plant UGTs. A. Structures of common sugar donors utilized by plant UGTs. B. The sugar donor-binding site for the structure of Phytolacca americana Glycosyltransferase 3 (PaGT3, a flavonoid/triterpene glycosyltransferase, PDB ID: 7VEJ) in Michaelis complex with UDP-2FGlc. The plant secondary product glycosyltransferase (PSPG) motif is highlighted in pink. Proposed hydrogen bonds are indicated by dashed lines. C. Consensus of the PSPG motif generated by WebLogo (https://weblogo.berkeley.edu/) from the alignment of the 30 plant UGTs (Table 1). The height of symbols within the stack represents the relative frequency of each amino acid at that position.
2.3.1. Sugar donor recognition
The CTD of plant UGTs includes the sugar donor binding pocket and plays a critical role in the recognition and binding of sugar donors. Sequence alignment analysis of CTDs has revealed certain sequence and structural variations, particularly in the loops connecting the α-helices and β-strands (e.g. the Cβ2-Cα2 and Cβ6-Cα7 linkers) (Fig. S1). Nevertheless, the CTDs are generally highly conserved in all UGTs (Fig. S1), likely due to the similarity of chemical structures of the sugar donors they recognize. Within the CTD, the most conserved region is known as the plant secondary product glycosyltransferase (PSPG) motif, which spans 44 amino acids and is located in the center of the CTD [1] (Fig. 3B and Fig. S1). The PSPG motif forms one side of the sugar donor binding pocket and primarily contributes to the interaction with the sugar donor (Fig. 3B).
Multiple amino acids within the PSPG motif play a crucial role in the recognition of sugar donors. The first amino acid in the PSPG motif, tryptophan (Trp352 in Figs. 3B and 3C), forms a parallel π-stacking interaction with the uracil ring of the UDP-sugar molecule via its indole ring. The glutamine at the 4th position (Gln355 in Figs. 3B and 3C) and the glutamate at the 27th position (Glu378 in Figs. 3B and 3C) of the PSPG motif form hydrogen bonds with the ribose-hydroxyl groups. In addition, the histidine at the 19th position (His370 in Figs. 3B and 3C) and the serine at the 24th position (Ser375 in Figs. 3B and 3C) interact with the phosphoric acid of the sugar donor [61]. Experimental evidence has shown that mutations at these specific positions led to the loss of glycosyl transfer activities in plant UGTs [36], [43], [51].
Apart from the highly conserved amino acids, other residues within the PSPG motif, which may have less conservation or even lack conservation, could also be essential for the substrate recognition. This is exemplified by the residue that is located at the 30th position of the PSPG motif in the curcumin glucosyltransferase CaUGT2 from Catharanthus roseus (Fig. 3C) [62]. Replacing the PSPG motif of CaUGT2 with that of NtGT1b (a phenolic glucosyltransferase from tobacco) led to a complete loss of activity, although only 14 out of 44 residues differ between the two motifs [62]. However, when Arg378 (the 30th residue in the PSPG motif of NtGT1b) was replaced with cysteine (the corresponding amino acid in the PSPG motif of CaUGT2), the catalytic activity of the chimeric enzyme was restored. Further site-directed mutagenesis experiments indicated that the size of the amino acid side chain at this particular position was critical for the catalytic activity of CaUGT2 [62].
2.3.2. Sugar donor specificity
Plant UGTs generally have broad specificity. Of the 30 plant UGTs with known structures, half were observed to accept two or more sugar donors (Table 1). Nevertheless, they also show specific preferences for particular sugar donors, as supported by their common preference for UDP-Glc over UDP-Gal.
The last two residues of the PSPG motif are critical in the selectivity of UGTs towards different sugar donors. The side chains of these residues interact directly with the sugar moiety of the donor molecules [39]. For instance, the 43rd residue of the PSPG motif is typically aspartate or glutamate (Fig. 3C), which can form hydrogen bonds with the hydroxyl group of the sugar moiety of the sugar donors. These hydrogen bonds contribute to the preference of UDP-Glc over UDP-Gal: the aspartate or glutamate can form hydrogen bonds with both 3-OH and 4-OH of the glucose moiety of UDP-Glc [27], [30], [39], [40], [52]; however, the spatial orientation of the galactose moiety in UDP-Gal prevents the access to its 4-OH group. Mutations of this residue to other amino acids could lead to the loss of catalytic activity [27], [38]. However, replacing it with the other conserved residue can have varied effects on different UGTs. For instance, the mutation of Asp382 (the 43rd residue in the PSPG motif) to glutamate decreased the catalytic efficiency of Fagopyrum esculentum UGT708C1 (FeUGT708C1) towards UDP-Glc [38]. However, the same mutation in Glycyrrhiza glabra di-C-glycosyltransferase (GgCGT) did not cause any detectable loss of enzyme activity [27].
The 44th amino acid of the PSPG motif in almost all plant UGTs with known structures is glutamine, except for AtUGT89C1 that has histidine at this site (Fig. S1). The histidine residue is crucial for AtUGT89C1 to specifically recognize the sugar donor UDP-Rha. The mutation to glutamine (His357Gln) allowed AtUGT89C1 to accept both UDP-Rha and UDP-Glc as sugar donors [53]. However, the substitution of histidine for glutamine allowed FeUGT708C1 to recognize UDP-Glc and UDP-Gal as the donors but not to accept UDP-Rha as a sugar donor [38]. Moreover, Landoltia punctata C-glycosyltransferase (lpCGTb) does not recognize UDP-Glc as a sugar donor, although it does contain the conserved glutamine at this site [26]. Interestingly, the apiosyltransferase UGT94 AX1 of Apium graveolens (AgUGT94AX1), which had strict substrate specificity for the sugar donor UDP-apiose, had a leucine at the 44th position (Leu356). Mutations in this residue led to a significant reduction or even complete loss of the catalytic activity of AgUGT94AX1 for UDP-apiose, demonstrating the critical role of Leu356 in the recognition of UDP-apiose [63].
Furthermore, additional residues beyond the PSPG motif also contribute to the specificity of sugar donor. For instance, the Calotropis gigantea steroid glycosyltransferase UGT74AN2 (CgUGT74AN2) showed the ability to convert up to 98% of UDP-Glc, while only 13% for UDP-GlcNAc and 6% for UDP-Gal [43]. Structural analysis revealed that in CgUGT74AN2, Asp392/Gln393 (the last two residues of the PSPG motif), Trp371 (the 22nd residue of the PSPG motif) and Thr141/Gln142 (outside the PSPG motif) form hydrogen bonds with the 3-OH, 4-OH and 6-OH of the glucose moiety of UDP-Glc, respectively. However, the 4-OH of galactose points away from Trp371, inhibiting the formation of hydrogen bonds, and the N-acetyl group of UDP-GlcNAc causes significant steric obstruction with CgUGT74AN2 [43]. Another example was the specific recognition of UDP-GlcA by Glycyrrhiza uralensis UGT73P12 (GuUGT73P12) [64]. GuUGT73P12 showed a high affinity for UDP-GlcA, the only sugar donor with a negatively charged carboxyl group in its sugar moiety. The Arg32 residue of GuUGT73P12 can form an electrostatic interaction with the carboxyl group of UDP-GlcA, thereby stabilizing its binding. The mutation of the arginine residue to serine increased the conversion efficiencies of GuUGT73P12 to UDP-Glc and UDP-Gal by 7-fold and 73-fold, respectively, while the utilization of UDP-GlcA was reduced by 11-fold [64].
Homology modeling in combination with biochemical analysis have highlighted the central role of an isoleucine residue around position 140 in pentosyltransferases in the recognition of UDP-pentose (including UDP-xylose, UDP-arabinofranose, UDP-apiose) [63], [65], [66]. This can be attributed to a steric hindrance resulting from the side chain of isoleucine, which prevents the entry of larger UDP-hexose into the donor recognition pocket. Remarkably, the mutation of Ile141 to Ser in Camellia sinensis UGT94P1 (CsUGT94P1) transformed it from a xylosyltransferase into a highly specific glucosyltransferase [65].
2.4. Acceptor recognition and specificity
The plant UGTs possess the ability to transfer sugar moieties to a diverse range of acceptor molecules, including phenols, flavonoids, terpenoids, saponins, sterols and alkaloids (Fig. 4A). Certain UGTs can specifically glycosylate glycosides, which are called glycoside-specific glycosyltransferases, as represented by Oryza sativa UGT91C1 (OsUGT91C1) and Stevia rebaudiana UGT76G1 (SrUGT76G1). Unlike sugar donors that share a common UDP base, the acceptors exhibit significant heterogeneity in terms of size, shape, hydrophobicity and other properties. Some plant UGTs show a pronounced substrate specificity that directs glycosylation reactions to specific acceptor molecules and their derivatives, thereby yielding distinct glycosylated products. Conversely, some UGTs exhibit specificity for specific acceptor molecules while facilitating the generation of different products through glycosylation at multiple sites of the same acceptor molecule. Moreover, a significant cohort of UGTs exhibits broad substrate promiscuity, allowing them to recognize and modify different acceptor molecules from different chemical groups.
Fig. 4.
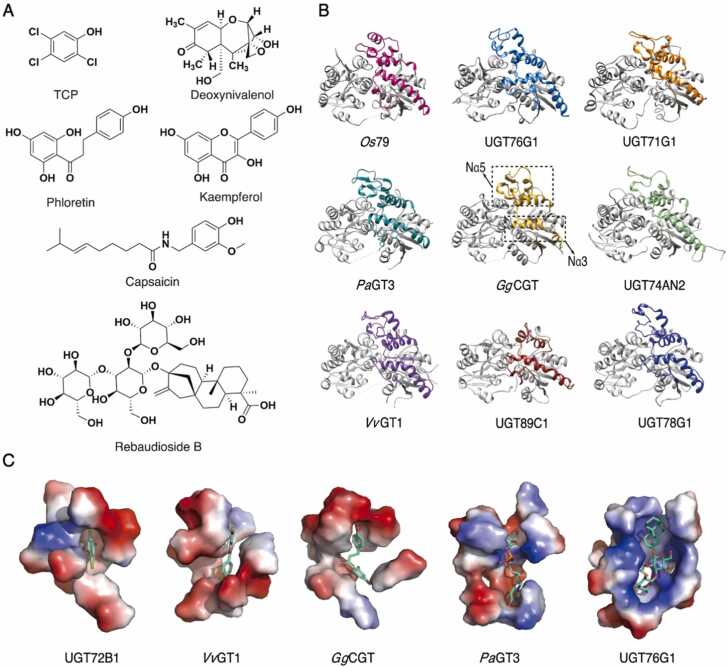
Acceptor recognition and specificity of plant UGTs. A. Structures of representative acceptor molecules catalyzed by plant UGTs. TCP: 2,4,5-chlorophenol. B. Variations in the N3α and N5α among different UGTs. The structures of nine UGTs are aligned and displayed in the same orientation. The Nα3 and Nα5 of different UGTs are highlighted in distinct color. UGTs in the first row are able to catalyze terpenoids, those in the second row exhibit high sugar acceptor promiscuity, and those in the third row can catalyze flavonoids. C. The representative sugar acceptor binding pockets of plant UGTs. The binding substrates of AtUGT72B1, VvGT1, GgCGT (i.e. Glycyrrhiza glabra CGT), PaGT3 and SrUGT76G1 are TCP, kaempferol, phloretin, capsaicin and Rebaudioside B, respectively. All substrates are shown in stick representation.
2.4.1. Acceptor binding pocket
The acceptor binding pocket is primarily located within the NTD of plant UGTs. Unlike the highly conserved CTD, NTD has a significant variety of amino acid sequences (Fig. S2). This heterogeneity reflects the NTD’s ability to recognize and bind acceptors with different structures and physiochemical properties. Despite the sequence diversity, however, the NTDs do share a common Rossmann fold in their three-dimensional structures (Fig. 1). Through sequence alignment of the 30 plant UGTs (Table 1), it can be observed that the β-strands in the center have a higher conservation compared to the surrounding helices (Fig. S2). The best-conserved region of the NTD is the N-terminus of Nα1, which hosts the histidine residue in the catalytic center.
The acceptor binding pocket consists of mainly the N-terminus of Nα1 and Nα3, the small loop that connects Nβ4 and Nα4, and the N- and C-termini of Nα5 (Fig. S2). The N-terminus of Cα6, which includes the last two residues of the PSPG motif, also contributes to the formation of the acceptor binding pocket. Among these structural elements, Nα3 and Nα5 show greater diversity and comprise the main part (i.e. the top) of the acceptor binding pocket (Fig. 4B). In most plant UGT structures, these two regions are highly flexible, as supported by their high B-factors. This indicates their potential mobility during the catalytic process. This mobility implies a putative role as a gate that controls the entry of the acceptor and the exit of the catalytic products. This hypothesis was supported by the structural variation observed in two copies of TcCGT1 within an asymmetric unit of its crystal [37]. Although the two structures were highly similar overall, they showed significant differences in the Nα3 region, indicating conformational changes of Nα3 during glycosylation that may be involved in substrate entry and product exit.
2.4.2. Acceptor recognition and specificity
The size of the acceptor binding pocket plays a crucial role in the recognition and specificity of the substrate. SrUGT76G1, GgCGT and PaGT3, which can catalyze large substrates, have significantly larger acceptor binding pockets compared to VvGT1 and AtUGT72B1, which prefer smaller molecules (Fig. 4C). Certain UGTs, such as AtUGT89C1 and Arabidopsis thaliana UGT74F2 (At UGT74F2), have a shorter Nα5 region and can only glycosylate specific small substrates (Fig. S2). This suggests a correlation between the length of Nα5 and the size of the acceptor binding pocket, thereby influencing substrate specificity. A shorter Nα5 region can result in a smaller binding pocket that is only suitable for specific small substrates. In addition, the composition of Nα5, especially the residues at its N-terminus, also influences the size of the acceptor binding pocket and thus the acceptor selectivity. Sequence analysis indicates that plant UGTs involved in the glycosylation of small substrates often have large hydrophobic residues such as phenylalanine and tyrosine at the 8th position of Nα5 (marked by a red triangle in Fig. S2). This position is located at the back of the acceptor binding pocket. In contrast, UGTs that catalyze complex molecules tend to contain smaller residues (e.g. methionine, valine, etc) at this position. Apart from Nα5, Nα3 forms the other side of the acceptor binding pocket and shows a higher frequency of phenylalanine (marked by dot in Fig. S2) in flavonoid-favored UGTs compared to UGTs that catalyze larger compounds like triterpenes (Fig. S2).
In addition to the size, the shape of the acceptor binding pocket also influences substrate specificity. For example, the acceptor binding pocket of FeUGT708C1 has a curved "L" shape that allows the open-chain form of phloretin to enter while excluding the rigid kaempferol molecule [38] (Fig. 4A).
Furthermore, hydrogen bonds and hydrophobic interactions are also important for acceptor recognition and stabilization. In PaGT2, Glu82 forms a hydrogen bond with piceatannol, and Tyr102 of FeUGT708C1 forms a hydrogen bond with phloretin. Disruption of these hydrogen bonds decreased or even eliminated the enzymatic activities of PaGT2 and FeUGT708C1 to their respective substrates [32], [38]. In addition, the activity of PaGT3 toward capsaicin or kaempferol was higher than that toward smaller substrates like salicylic acid due to the formation of more hydrogen bonds [67]. In the case of CgUGT74AN2, its interaction with digitoxigenin was primarily mediated by hydrophobic interactions [43].
The size and shape of the acceptor binding pocket also play a crucial role in determining the regioselectivity of UGTs. The influence of size on the regioselectivity of the substrate is well-represented by Medicago truncatula UGT71G1 (MtUGT71G1) and VvGT1 [39], [52]. They both glycosylate quercetin. MtUGT71G1 with a larger acceptor binding pocket can catalyze the glycosylation of all five hydroxyl groups of quercetin, resulting in the formation of five isomeric quercetin glucosides. In contrast, VvGT1 is limited to producing only the 3-O-glucoside of quercetin. The impact of shape on regioselectivity can be illustrated by the example of CgUGT74AN2. CgUGT74AN2 has a considerable hydrophobic pocket and exhibits significant substrate promiscuity. However, the V-shaped structure of its acceptor binding pocket restricts the orientation of the acceptor digitoxigenin and aligns its C3-OH group with the catalytic histidine residue. Consequently, CgUGT74AN2 selectively produces produced cardiotonic steroid 3-O-glycoside [43].
3. Engineering of plant UGTs
The efficiency of native enzymes is often insufficient to meet the industrial requirements. Protein engineering has demonstrated its unique ability to enhance catalytic efficiency and create novel enzymes for specific purposes [68]. This approach has proven to be very successful in various industrial cases, including the production of Atorvastatin (Pfizer, Lipitor™) and Sitagliptin (Merck, Januvia™) [69], [70]. Over the past decades, protein engineering efforts targeting plant UGTs have experienced exponential growth. Based on the rationale of the methodology, these approaches can generally be categorized into three groups: rational design, directed evolution, and semi-rational design.
3.1. Rational design
Rational design is an effective strategy when comprehensive knowledge of the structure and functional mechanism of a protein is available. Recent advances in structural biology, exemplified by breakthroughs such as AlphaFold 2, a deep learning-based system known for its highly reliable predictions of protein structures, have significantly advanced this field. Rational design involves minimal screening, focusing on generating a small number of variants. Common techniques for rational design include structure-guided site-directed mutagenesis, activity-based sequence conservative analysis (ASCA) and domain swapping.
Structure-guided site-directed mutagenesis is a commonly used rational design method in protein engineering. Structural analysis of plant UGT-substrate complexes can identify essential residues responsible for substrate binding and catalytic activity. Based on these findings, targeted mutations can be developed to refine the properties of enzymes for specific purposes. Numerous studies have focused on modulating the substrate specificity and regioselectivity of plant UGTs using this approach. For example, the terpenoid-favored SrUGT76G1 was successfully engineered to accept flavonoid as substrates by introducing three mutations (Gly87, lle199 and Leu204 to phenylalanine) [56]. Similarly, a triple mutation (H122A/L123A/Q202L) in the Oryza sativa deoxynivalenol:UDP-glucosyltransferase Os79 was developed to increase the volume of the acceptor binding pocket, leading to increased catalytic activity towards C4-acetylated compounds [31]. In addition, the L369/H373Q mutation led to a transformation of Glycyrrhiza uralensis UGT79B74, making it from an exclusive catalytic activity of UDP-apiose to a near-100% activity towards UDP-Xyl [[71]. The improvement of regioselectivity by structure-guided site-directed mutagenesis is exemplified by Siraitia grosvenorii UGT74AC2 (SgUGT74AC2). By introducing a single mutation (L200W), the size of the acceptor binding pocket was reduced, preventing the approach of the C3-OH group of silybin A to the catalytic center. This change significantly improved the regioselectivity of SgUGT74AC2 and increased its preference for the C7-OH from 39% to 92% [42]. Although structure-guided site-directed mutagenesis is an effective method in protein engineering, it often leads to only moderate improvements in catalytic efficiency. This limitation is likely due to the influence of other residues in the binding pocket, which remains to be further investigated. In addition, this approach heavily relies on the availability of a resolved structure of UGT-substrate complexes, which currently limits its wider application.
ASCA is the method that utilizes consensus residues to replace non-consensus residues at each position after multiple sequence alignment has been performed [41]. The rationale of this method is that amino acids which promote the protein functions are conserved during evolution, as suggested by the Darwinian theory of evolution. This was further supported by the fact that conserved amino acids surrounding the catalytic center and the substrate binding pocket have been observed in enzymes with similar substrate spectra [72]. Therefore, performing multiple sequence alignment on these enzymes can provide valuable insights into the crucial residue positions and guide mutation design. The ASCA approach has been shown to be successful in enhancing protein thermostability [73], and shows promising potential in modifying the activity of plant UGTs. By implementing the ASCA method, 19 and 24 mutations were identified for the NTD and CTD of Siraitia grosvenorii UGTMS1 (SgUGTMS1), respectively [72]. Among the 19 mutations in the NTD, three have been shown to increase enzyme activity. Remarkably, the combination of these three mutations (SgUGTMS1-M3) resulted in a 34-fold increase in catalytic efficiency. Sequential incorporation of the effective CTD mutations into SgUGTMS1-M3 led to the development of an optimal mutant, exhibiting a remarkable 351-fold improvement in catalytic efficiency compared to the wild-type enzyme. Another example is the application of ASCA to engineer the activity of Siraitia grosvenorii UGT74AC1 (SgUGT74AC1) [41]. Using the ASCA approach, eight mutations were designed, two of which resulted in a two-fold and three-fold increase in enzyme activity, respectively. Combining these two effective mutations further improved the enzymatic activity by over seven times.
Domain swapping is a widely utilized protein engineering strategy when two enzymes exhibit high sequence similarity but different functions [40], [74]. In this technique, domains or structural motifs/variations are exchanged between the enzymes, resulting in chimeric or mutated enzymes with altered functionalities. For instance, pteridine glycosyltransferases (PGTs) from Arthrospira platensis CY-007 and Arthrospira maxima CY-049 catalyzed glucose transfer and xylose transfer, respectively. By swapping the CTDs of the two enzymes, the sugar donor specificity of the resulting chimeric enzymes was also altered [75]. Similar phenomenon was also reported for the case of Arabidopsis thaliana UGT78D2 (AtUGT78D2) (which exclusively accepts UDP-glucose) and Arabidopsis thaliana UGT78D3 (AtUGT78D3) (that specifically recognizes UDP-arabinose). Various chimeric proteins were constructed by swapping different domains between the two enzymes. Five of these chimeras were capable of utilizing both UDP-glucose and UDP-arabinose as sugar donors [76]. Another successful example of domain swapping is Scutellaria baicalensis C-glycosyltransferases SbCGTa and SbCGTb enzymes involved in the di-C-glycosylation for the biosynthesis of (iso)schaftosides. SbCGTa catalyzes glucose transfer to 2-hydroxynaringenin, while SbCGTb transfers arabinose to the mono-C-glucoside of 2-hydroxynaringenin. By swapping 11 amino acids around the substrate pocket, the resulting SbCGTb-11aa mutant showed functions similar to SbCGTa while losing its native ability to catalyze mono-C-glycosides [26].
Overall, rational design approaches, including structure-guided site-directed mutagenesis, ASCA, and domain swapping, provide effective tools for the development of plant UGTs to achieve desired modifications in enzyme properties, if the mechanistic knowledges of the protein functions were (even partially) available.
3.2. Directed evolution
Directed evolution is a powerful tool for enzyme engineering that requires minimal knowledge beyond the enzyme's function. It involves iterative cycles of engineering and screening until a desired protein variant is obtained. However, compared to rational design, directed evolution often proves to be labor-intensive, as mutant libraries must be generated and subsequently screened for the biological functions of the target protein. The mutation of the protein in the desired direction and the efficient screening of the large number of candidate mutants are two crucial aspects of directed evolution. Various random mutagenesis methods, such as error-prone PCR and DNA shuffling, have been developed and tailored to generate mutant libraries of plant UGTs. However, the establishment of high-throughput screening methods remains a challenging step to facilitate the targeted evolution of plant UGTs. Recent advances in screening methods of plant UGTs are summarized in Table 2. Typically, substrates (i.e. sugar donors and acceptors) as well as glycosylation products and by-products (i.e. UDP and proton) have been used as markers to evaluate the performance of plant UGTs. Among these, UDP is the most widely used and serves as a universal reference compound for screening, although other markers might be better solutions in certain scenarios.
Table 2.
Summary of high-throughput screening methods for plant UGTs.
| Screening target | Method | Application scenarios | Screening sample | Reference |
|---|---|---|---|---|
| Products | Transfer a radiolabeled sugar to the acceptor, and then remove the unreacted sugar donors and detect the labeled products | All UGTs | Purified protein, cell lysate | [77] |
| Products | Utilize cell-permeable fluorescence-labeled acceptors to generate cell-impermeable fluorescence-labeled product, and then screen by FACS. | Specific UGTs | Whole cell | [78], [79] |
| UDP | Synthesize ATP with UDP, then transfer ATP into bioluminescence signal with UDP-Glo™ | All UGTs | Purified protein | [80] |
| UDP | Synthesize Glucose-6-phospate with UDP step by step, then measure Glucose-6-phospate with NADP and resazurin. | All UGTs | Purified protein | [81], [82] |
| UDP | Utilize OleD to catalyze production of colorimetric 2-chloro-4-nitrophenolate with UDP | All UGTs | Purified protein, cell lysate | [41], [83], [84], [85] |
| UDP | Use rYND1 to hydrolyze UDP into UMP and Pi, then use a phosphorus molybdenum blue chromogenic reaction to detect Pi. | All UGTs | Purified protein | [86] |
| UDP | Release Pi from UDP with ectonucleoside triphosphate diphosphohydrolase, and then detect Pi with colorimetric malachite-based reagents | All UGTs | Purified protein | [87] |
| UDP | Utilize a xanthene-based Zn(II) complex fluorescent chemosensor to detect UDP | All UGTs | Purified protein, cell lysate |
[88], [89], [90] |
| UDP | Utilize a binuclear zinc complex fluorescent chemosensor to detect UDP | All UGTs | Purified protein, cell lysate |
[91] |
| UDP | Utilize an anthracene-based Zn(II) complex fluorescent chemosensor to detect UDP | All UGTs | Purified protein, cell lysate |
[92] |
| UDP | Conduct reactions on an immobilized acceptor array, and detect production of UDP with mass spectrometry | All UGTs | Purified protein | [93] |
| UDP | Establish a UDP-antibody based FRET system, and detect UDP via FRET signal decrease caused by UDP displacement of the UDP-tracer | All UGTs | Purified protein | [94] |
| Proton | Utilize pH to detect proton release during glycosylation reaction | All UGTs | Purified protein, cell lysate, whole cell | [95], [96], [97], [98] |
| Acceptor | Detect the fluorescence of the acceptor 4-methylumbelliferone, which is non-fluorescent after glycosylation | Specific UGTs | Purified protein, cell lysate | [99] |
Rapid measurement of UDP is usually achieved by developing a UDP-dependent response that can be tracked using spectrophotometry approaches. These analytical methods were normally based on fluorescent, luminescent, or colorimetric signals (Table 2). Some of the reported methods, such as the UDP-Glo™ assay [80] and the NADP-resazurin assay [81], require purified UGT protein as they are sensitive to background signals from living cells or cell lysates. While these methods are suitable for screening chemical inhibitors targeting specific UGT proteins, they do not meet the high-throughput requirements for the selection of novel UGTs from mutation libraries.
Two UDP-targeting assays designed for high-throughput screening of UGT mutation libraries are the Oled-coupled colorimetric assay and the UDP chemosensor-based fluorescent assay [83]. The Oled-coupled colorimetric assay involves the coupling of UDP with the production of 2-chloro-4-nitrophenolate, a compound that can be quantitatively measured at 410 nm. This method was successfully used to screen an SgUGT74AC1 mutation library containing ∼5000 mutants generated by error-prone PCR [41]. Several derivatives with single-point mutations showed 2- to 8-fold increases in catalytic activity compared to the wild type. Further optimization by combining these mutations resulted in a mutant with a catalytic efficiency of 200-times higher than that of the wild type. Another highly specific and sensitive method for direct detection of UDP is the use of tailor-made chemosensors. These chemosensors, which are based on binuclear zinc complexes, show fluorescence after binding to the diphosphate moiety of UDP [92]. The cost of chemosensors is currently the limiting factor for the widespread use of this method and needs to be drastically decreased. Both approaches can be performed in microplates to enable high-throughput testing. Some of the most time-consuming steps include picking up colonies, amplifying and harvesting biomass, and preparing cell lysates for testing. Automated pipelines are available on the market (e.g. automated colony pickers from Kbiosystems) to replace human labor for these tedious tasks and enable the screening of large libraries (>104 mutants). However, the costs associated with these automated systems are still unaffordable for most laboratories. Therefore, these methods are often used for screening small to medium-sized libraries (<104 mutants).
In addition to UDP, protons can also be used as targets for screening UGT mutant libraries. The production of protons can be easily detected by measuring the pH drop using optodes or color-change reagents. The pH change was used as a selection marker for screening a domain-swapping library containing 106-107 colonies [98]. To screen such a large library, colonies were replicated on LB agar plates and transferred to filter papers by stamping. The filters were then immersed in a glycosylation reaction solution containing the pH-indicator cresol red and incubated for 5 h at 37 °C. Colonies containing functional glycosyltransferase displayed a yellow color, while negative controls turned purple. By this method, a chimeric glycosyltransferase was obtained with approximately 4-fold increased catalytic activity toward 2-deoxystreptamine compared to the chimera generated via rational design. Furthermore, the new mutant was able to act on seven other NDP-sugars in addition to dTDP-glucose [98]. However, the use of proton as an indicator for screening tests has the disadvantage of untargeted pH changes in the glycosylation reaction systems, resulting in high background noise when using whole cells or cell lysates. Furthermore, due to the labor-intensive process of handling colonies, this method is mostly suitable for screening small to medium-sized libraries.
Fluorescently labeled sugar donors and acceptors were used for the screening of specific UGTs. An example of this method was the fluorescence-activated cell sorter (FACS)-based screening system for the directed evolution of sialyltransferases [78]. In this system, the acceptor molecule was labeled with a fluorophore, while its permeability through the cell membrane was not affected. However, the fluorescent product could not be secreted out of the cells after glycosylation. As a result, colonies containing active enzymes were labeled with fluorescence and could be sorted using FACS. In a follow-up study from the same group, a two-color screening protocol was established to minimize false-positive signals. The optimized method enabled the rapid identification of a CgtB variant from a library of over 107 mutants. This variant had a broader substrate tolerance and 300-fold higher activity than the wild-type enzyme [79]. Alternatively, fluorescence quenching has also been employed in screening tests. For instance, after glycosylation, 4-methylumbelliferone loses its fluorescence signal, which was used to screen and obtain a target Oled enzyme from a library of ∼1000 colonies generated by error-prone PCR [99]. These fluorescence-based screening methods are efficient and sensitive, but they are typically applicable to only specific UGTs. The selection and design of fluorescent molecules is required on a case-by-case basis. Nevertheless, they still provide a good alternative strategy for the development of new high-throughput screening protocols for plant UGTs engineering.
3.3. Semi-rational design
Semi-rational design, also known as focused directed evolution, is an approach that combines rational design and directed evolution to increase efficiency in UGT engineering. Instead of performing mutagenesis for the entire sequence, this method selectively targets potential key residues to generate a mutation library. As a result, the screening efforts for the identification of the target variants of interest could be significantly reduced. In this context, even low-throughput analytic methods such as HPLC and mass spectrometry might be sufficient for the screening tasks [85], [100], [101].
The first step in semi-rational design is the identification of putative functional residues. This can be achieved by sequence or structure analysis followed by alanine screening. Typically, solved or predicted structures of desired UGTs are used as templates. Substrates (sugar donors or acceptors) are docked into the structure using software such as Autodock Vina or Glide in Schrödinger. Residues that interact with the docked molecules or are located at a certain distance from them are selected as candidates for further mutation. Another in silico approach for selecting key residues is to use software like HotSpot and SEARCHGTr [85], [96], [102]. These tools predict substrate binding residues based on comprehensive sequence/structure alignment with reported crystal structures of glycosyltransferase-acceptor complexes. In addition, important residues reported and identified in previous studies can be used to concentrate the research focus on specific residues [41], [97]. After the initial selection of potential key residues, each residue is individually mutated to alanine, and the resulting variants are examined for changes in catalytic activity compared to the wild type. Residues that have at least 60% activity of the wild-type enzyme after alanine mutation are then selected as targets for generation of mutagenesis library and further screening [103].
Once the initial selection is complete, the mutagenesis library can be constructed using iterative saturation mutation (ISM). In the saturation mutation, each residue is mutated into the other nineteen residues using degenerative NNK genetic codons. However, the exponential increase in library size and screening efforts can be overwhelming. To address this challenge, researchers have developed ISM methods that can be performed sequentially or in clusters. In sequential ISM, saturation mutation is performed one position at a time in a specific order, while the mutation with best performance at one position serves as the template for the next mutation [85], [104]. Clustered ISM is a faster approach where candidate residues are divided into several groups based on neighborhood or location on the same secondary structure element, with 2–4 residues in each group. Sequential ISM is then performed in parallel for each group, and the mutations with the most significant effects from each group are combined for further testing [96].
Limiting the size of the mutation library helps to increase the screening efficiency, making the semi-rational design an attractive and effective approach in plant UGT engineering. It has been successfully applied to improve catalytic efficiency, substrate recognition, and regioselectivity of numerous plant UGTs. For example, ISM was performed on nine important residues of ScUGT51, resulting in a library of ∼2800 colonies [85]. Screening of the library led to the identification of a variant that outperformed the wild-type enzyme by about 1800-fold in the conversion of protopanaxadiol to ginsenoside Rh2. Similar approaches have been applied to enhance the regioselectivity of UGTBL1 [100] and MiCGT [28].
However, the semi-rational design also faces some challenges due to the effort required for screening. To address this problem and further reduce the number of mutations, a remarkable protocol called focused rational iterative site-specific mutagenesis (FRISM) has recently been developed [101]. Instead of performing saturated mutagenesis, FRISM uses multiple approaches to minimize the number of rational mutations for each functional residue. These approaches include phylogenetic analysis, structural information, Rosetta design, and other techniques commonly used in rational design. This approach has been successfully demonstrated in several studies. For instance, SgUGT74AC2 variants with high regioselectivity for different hydroxyl groups of silybin A were obtained [35]. By combining ASCA, acceptor binding pocket analysis, Rosseta design, and iterative mutagenesis, a small library consisting of ∼115 mutations was built and analyzed. Ultimately, three variants were identified that had a regioselectivity of 94%, > 99%, and > 99% on the C3-OH, C7-OH and C3,7-O-diglycoside of silybin A, respectively, while the original SgUGT74AC2 yielded a mixture of 22%:39%:39% of the three products.
4. Conclusions
Plant UGTs are essential for the biosynthesis of natural products. The reconstitution of biosynthesis pathways of natural products from plants into microorganism offers a more sustainable production alternative to these high-value compounds. This includes the expression of key enzymes including UGTs. Recent advances in molecular biology, biochemistry, plant biology, and structural biology have led to the discovery and characterization of numerous plant UGTs in terms of structures and biological functions. These findings pave the way for protein engineering of UGTs for better and/or desired performance, e.g. catalytic efficiency, substrate promiscuity and regioselectivity.
The efficient manipulation of plant UGTs requires a deep understanding of their functional mechanisms. So far, the crystal structures of 30 plant UGTs have been elucidated in their apo- or substrate-bound forms. These structures, integrated into structure analysis and mutagenesis studies, have provided significant insights into the catalytic mechanism, the recognition of sugar donor and acceptor of plant UGTs. This knowledge enabled the development of various UGTs through rational design and semi-rational design approaches. However, the knowledge obtained so far is still too limited to fully uncover the substrate recognition mechanisms of many key enzymes involved in the biosynthesis of important natural products, considering the high substrate diversity among different plant UGTs. Since some of the solved structures are in the apo form and most acceptors in complex structures are small molecules, it is crucial to resolve more structures of plant UGTs that bind complex substrates like ginsenosides, anthocyanins and mogrosides, which are natural products of therapeutic or industrial interests.
Various strategies have been developed for the development of plant UGTs. Semi-rational design, which combines the benefits of rational design and directed evolution, has emerged as the most widely utilized approach in plant UGTs engineering. It has proven itself with great potential in various applications. To facilitate the engineering efficiency, smaller mutation libraries are usually preferred because of the less screening effort. But this requires good knowledge of substrate binding obtained from structural studies. In addition, certain residues located far from the substrate binding pocket can affect catalytic efficiency and substrate recognition. In this case, directed evolution becomes crucial to screen these sites. However, efficient screening methods for large mutation libraries of plant UGTs remains a critical challenge, and the development of (high-throughput) screening methods is essential in the near future.
CRediT authorship contribution statement
K.M. and B.L. conceived the idea and initiated the writing. M.W, Q.J., Y.L. and K.M. collected and analyzed the data and drafted the manuscript. K.M., B.L. and M.W critically reviewed and edited the manuscript. All authors read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare no conflict of interests to the contents of this paper.
Acknowledgements
K.M. thanks the financial support from the National Science Foundation of China (91954112 and 31900501) and the Young Elite Scientists Sponsorship Program of Tianjin (TJSQNTJ‐2020–19). B.L. is financially supported by the Federal Ministry of Education and Research, Germany (Grant Nr. 031B1273).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.10.046.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Gachon C.M.M., Langlois-Meurinne M., Saindrenan P. Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 2005;10:542–549. doi: 10.1016/j.tplants.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Jones P., Vogt T. Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta. 2001;213:164–174. doi: 10.1007/s004250000492. [DOI] [PubMed] [Google Scholar]
- 3.Bowles D., Isayenkova J., Lim E.K., Poppenberger B. Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol. 2005;8:254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Rehman H.M., Nawaz M.A., Shah Z.H., Ludwig-Müller J., Chung G., Ahmad M.Q., et al. Comparative genomic and transcriptomic analyses of family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci Rep. 2018;8 doi: 10.1038/s41598-018-19535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao Y., Gu J., Luo Y., Wang Y., Pang Y., Shen G., et al. Genome-wide analysis of UGT gene family identified key gene for the biosynthesis of bioactive flavonol glycosides in Epimedium pubescens Maxim. Synth Syst Biotechnol. 2022;7:1095–1107. doi: 10.1016/j.synbio.2022.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn B.M., Errafi S., Bucher R., Dobrev P., Geisler M., Bigler L., et al. 7-rhamnosylated flavonols modulate homeostasis of the plant hormone auxin and affect plant development. J Biol Chem. 2016;291:5385–5395. doi: 10.1074/jbc.M115.701565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baksi R., Singh D.P., Borse S.P., Rana R., Sharma V., Nivsarkar M. In vitro and in vivo anticancer efficacy potential of quercetin loaded polymeric nanoparticles. Biomed Pharmacother. 2018;106:1513–1526. doi: 10.1016/j.biopha.2018.07.106. [DOI] [PubMed] [Google Scholar]
- 8.Ren G., Hou J., Fang Q., Sun H., Liu X., Zhang L., et al. Synthesis of flavonol 3-O-glycoside by UGT78D1. Glycoconj J. 2012;29:425–432. doi: 10.1007/s10719-012-9410-5. [DOI] [PubMed] [Google Scholar]
- 9.Lee J., Lee J., Kim S.J., Kim J.H. Quercetin-3-O-glucoside suppresses pancreatic cancer cell migration induced by tumor-deteriorated growth factors in vitro. Oncol Rep. 2016;35:2473–2479. doi: 10.3892/or.2016.4598. [DOI] [PubMed] [Google Scholar]
- 10.Lee S., Lee J., Lee H., Sung J. Relative protective activities of quercetin, quercetin-3-glucoside, and rutin in alcohol-induced liver injury. J Food Biochem. 2019;43 doi: 10.1111/jfbc.13002. [DOI] [PubMed] [Google Scholar]
- 11.Patel R.V., Mistry B.M., Shinde S.K., Syed R., Singh V., Shin H.S. Therapeutic potential of quercetin as a cardiovascular agent. Eur J Med Chem. 2018;155:889–904. doi: 10.1016/j.ejmech.2018.06.053. [DOI] [PubMed] [Google Scholar]
- 12.Biedermann D., Vavříková E., Cvak L., Křen V. Chemistry of silybin. Nat Prod Rep. 2014;31:1138–1157. doi: 10.1039/c3np70122k. [DOI] [PubMed] [Google Scholar]
- 13.Bidart G.N., Putkaradze N., Fredslund F., Kjeldsen C., Ruiz A.G., Duus J., et al. Family 1 glycosyltransferase UGT706F8 from Zea mays selectively catalyzes the synthesis of silibinin 7-O-β-d-glucoside. ACS Sustain. Chem. Eng. 2022;10:5078–5083. doi: 10.1021/acssuschemeng.1c07593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts S.C. Production and engineering of terpenoids in plant cell culture. Nat Chem Biol. 2007;3:387–395. doi: 10.1038/nchembio.2007.8. [DOI] [PubMed] [Google Scholar]
- 15.Zheng R., Zhu Z., Wang Y., Hu S., Xi W., Xiao W., et al. UGT85A84 catalyzes the glycosylation of aromatic monoterpenes in Osmanthus fragrans Lour. flowers. Front Plant Sci. 2019;10 doi: 10.3389/fpls.2019.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurze E., Wüst M., Liao J., McGraphery K., Hoffmann T., Song C., et al. Structure-function relationship of terpenoid glycosyltransferases from plants. Nat Prod Rep. 2022;39:389–409. doi: 10.1039/D1NP00038A. [DOI] [PubMed] [Google Scholar]
- 17.Zhao M., Jin J., Gao T., Zhang N., Jing T., Wang J., et al. Glucosyltransferase CsUGT78A14 regulates flavonols accumulation and reactive oxygen species scavenging in response to cold stress in Camellia sinensis. Front Plant Sci. 2019;10 doi: 10.3389/fpls.2019.01675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hectors K., van Oevelen S., Geuns J., Guisez Y., Jansen M.A.K., Prinsen E. Dynamic changes in plant secondary metabolites during UV acclimation in Arabidopsis thaliana. Physiol Plant. 2014;152:219–230. doi: 10.1111/ppl.12168. [DOI] [PubMed] [Google Scholar]
- 19.Chen L., Huang X.X., Zhao S.M., Xiao D.W., Xiao L.T., Tong J.H., et al. IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA. 2020;117:6910–6917. doi: 10.1073/pnas.2000172117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J., Sun W., Zhang G., Lv B., Li C. High efficient production of plant flavonoids by microbial cell factories: challenges and opportunities. Metab Eng. 2022;70:143–154. doi: 10.1016/j.ymben.2022.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Li Y., Wang J., Li L., Song W., Li M., Hua X., et al. Natural products of pentacyclic triterpenoids: from discovery to heterologous biosynthesis. Nat Prod Rep. 2022 doi: 10.1039/d2np00063f. [DOI] [PubMed] [Google Scholar]
- 22.Hou M., Wang R., Zhao S., Wang Z. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm Sin B. 2021;11:1813–1834. doi: 10.1016/j.apsb.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P., Wei Y., Fan Y., Liu Q., Wei W., Yang C., et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Wang P., Wei W., Ye W., Li X., Zhao W., Yang C., et al. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 2019;5 doi: 10.1038/s41421-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Y., Jiang Y., Chen L., Li C., Wang Y. Applications of protein engineering in the microbial synthesis of plant triterpenoids. Synth Syst Biotechnol. 2023;8:20–32. doi: 10.1016/j.synbio.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z.L., Gao H.M., Wang S., Zhang M., Chen K., Zhang Y.Q., et al. Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc Natl Acad Sci USA. 2020;117:30816–30823. doi: 10.1073/pnas.2012745117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M., Li F.D., Li K., Wang Z.L., Wang Y.X., He J.B., et al. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J Am Chem Soc. 2020;142:3506–3512. doi: 10.1021/jacs.9b12211. [DOI] [PubMed] [Google Scholar]
- 28.Wen Z., Zhang Z.M., Zhong L., Fan J., Li M., Ma Y., et al. Directed evolution of a plant glycosyltransferase for chemo- and regioselective glycosylation of pharmaceutically significant flavonoids. ACS Catal. 2021;11:14781–14790. doi: 10.1021/acscatal.1c04191. [DOI] [Google Scholar]
- 29.Chen D., Chen R., Wang R., Li J., Xie K., Bian C., et al. Probing the catalytic promiscuity of a regio- and stereospecific C-glycosyltransferase from Mangifera indica. Angew Chem Int Ed Engl. 2015;127:12869–12873. doi: 10.1002/ange.201506505. [DOI] [PubMed] [Google Scholar]
- 30.Wetterhorn K.M., Newmister S.A., Caniza R.K., Busman M., McCormick S.P., Berthiller F., et al. Crystal structure of Os79 (Os04g0206600) from Oryza sativa: a UDP-glucosyltransferase involved in the detoxification of deoxynivalenol. Biochemistry. 2016;55:6175–6186. doi: 10.1021/acs.biochem.6b00709. [DOI] [PubMed] [Google Scholar]
- 31.Wetterhorn K.M., Gabardi K., Michlmayr H., Malachova A., Busman M., McCormick S.P., et al. Determinants and expansion of specificity in a trichothecene UDP-Glucosyltransferase from Oryza sativa. Biochemistry. 2017;56:6585–6596. doi: 10.1021/acs.biochem.7b01007. [DOI] [PubMed] [Google Scholar]
- 32.Maharjan R., Fukuda Y., Shimomura N., Nakayama T., Okimoto Y., Kawakami K., et al. An ambidextrous polyphenol glycosyltransferase PaGT2 from Phytolacca americana. Biochemistry. 2020;59:2551–2561. doi: 10.1021/acs.biochem.0c00224. [DOI] [PubMed] [Google Scholar]
- 33.Maharjan R., Fukuda Y., Nakayama T., Nakayama T., Hamada H., Ozaki S.I., et al. Crown-ether-mediated crystal structures of the glycosyltransferase PaGT3 from Phytolacca americana. Acta Crystallogr D Struct Biol. 2020;76:521–530. doi: 10.1107/S2059798320005306. [DOI] [PubMed] [Google Scholar]
- 34.Maharjan R., Fukuda Y., Nakayama T., Nakayama T., Hamada H., Ozaki S., et al. Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3. Acta Crystallogr D Struct Biol. 2022;78:379–389. doi: 10.1107/S2059798322000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu T.M., Welner D.H., Russ Z.N., Cervantes B., Prathuri R.L., Adams P.D., et al. Employing a biochemical protecting group for a sustainable indigo dyeing strategy. Nat Chem Biol. 2018;14:256–261. doi: 10.1038/nchembio.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teze D., Coines J., Fredslund F., Dubey K.D., Bidart G.N., Adams P.D., et al. O-/ N-/ S-specificity in glycosyltransferase catalysis: from mechanistic understanding to engineering. ACS Catal. 2021;11:1810–1815. doi: 10.1021/acscatal.0c04171. [DOI] [Google Scholar]
- 37.He J., Zhao P., Hu Z., Liu S., Kuang Y., Zhang M., et al. Molecular and structural characterization of a promiscuous C‐glycosyltransferase from Trollius chinensis. Angew Chem Int Ed Engl. 2019;58:11513–11520. doi: 10.1002/anie.201905505. [DOI] [PubMed] [Google Scholar]
- 38.Liu M., Wang D., Li Y., Li X., Zong G., Fei S., et al. Crystal structures of the C-glycosyltransferase UGT708C1 from Buckwheat provide insights into the mechanism of C-glycosylation. Plant Cell. 2020;32:2917–2931. doi: 10.1105/tpc.20.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao H., He X., Achnine L., Blount J.W., Dixon R.A., Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17:3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brazier-Hicks M., Offen W.A., Gershater M.C., Revett T.J., Lim E.K., Bowles D.J., et al. Characterization and engineering of the bifunctional N-and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci USA. 2007;104:20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Yang J., Mu S., Shang N., Liu C., Zhu Y., et al. Efficient O-glycosylation of triterpenes enabled by protein engineering of plant glycosyltransferase UGT74AC1. ACS Catal. 2020;10:3629–3639. doi: 10.1021/acscatal.9b05232. [DOI] [Google Scholar]
- 42.Li J., Qu G., Shang N., Chen P., Men Y., Liu W., et al. Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases. Green Synth Catal. 2021;2:45–53. doi: 10.1016/j.gresc.2021.01.005. [DOI] [Google Scholar]
- 43.Huang W., He Y., Jiang R., Deng Z., Long F. Functional and structural dissection of a plant steroid 3-O-glycosyltransferase facilitated the engineering enhancement of sugar donor promiscuity. ACS Catal. 2022;12:2927–2937. doi: 10.1021/acscatal.1c05729. [DOI] [Google Scholar]
- 44.George Thompson A.M., Iancu C.V., Neet K.E., Dean J.V., Choe J. Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci Rep. 2017;7 doi: 10.1038/srep46629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modolo L.V., Li L., Pan H., Blount J.W., Dixon R.A., Wang X. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids. J Mol Biol. 2009;392:1292–1302. doi: 10.1016/j.jmb.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Hiromoto T., Honjo E., Tamada T., Noda N., Kazuma K., Suzuki M., et al. Crystal structure of UDP-glucose:anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. J Synchrotron Radiat. 2013;20:894–898. doi: 10.1107/S0909049513020712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hiromoto T., Honjo E., Noda N., Tamada T., Kazuma K., Suzuki M., et al. Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. Protein Sci. 2015;24:395–407. doi: 10.1002/pro.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Del Giudice R., Putkaradze N., dos Santos B.M., Hansen C.C., Crocoll C., Motawia M.S., et al. Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis. Plant J. 2022;111:1539–1549. doi: 10.1111/tpj.15904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen K.S., Kristensen C., Tattersall D.B., Jones P.R., Olsen C.E., Bak S., et al. The in vitro substrate regiospecificity of recombinant UGT85B1, the cyanohydrin glucosyltransferase from Sorghum bicolor. Phytochemistry. 2003;64:143–151. doi: 10.1016/S0031-9422(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 50.Li L., Modolo L.V., Escamilla-Trevino L.L., Achnine L., Dixon R.A., Wang X. Crystal structure of Medicago truncatula UGT85H2-insights into the structural basis of a multifunctional (iso)flavonoid glycosyltransferase. J Mol Biol. 2007;370:951–963. doi: 10.1016/j.jmb.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J., Tang M., Chen Y., Ke D., Zhou J., Xu X., et al. Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides. Nat Commun. 2021;12 doi: 10.1038/s41467-021-27144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B.G., Tarling C.A., et al. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25:1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zong G., Fei S., Liu X., Li J., Gao Y., Yang X., et al. Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-β-l-rhamnose and rhamnosylation mechanism. Plant J. 2019;99:257–269. doi: 10.1111/tpj.14321. [DOI] [PubMed] [Google Scholar]
- 54.Yang T., Zhang J., Ke D., Yang W., Tang M., Jiang J., et al. Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations. Nat Commun. 2019;10 doi: 10.1038/s41467-019-11154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee S.G., Salomon E., Yu O., Jez J.M. Molecular basis for branched steviol glucoside biosynthesis. Proc Natl Acad Sci USA. 2019;116:13131–13136. doi: 10.1073/pnas.1902104116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Z., Li J., Sun Y., Zhang P., Wang Y. Structural insights into the catalytic mechanism of a plant diterpene glycosyltransferase SrUGT76G1. Plant Commun. 2020;1 doi: 10.1016/j.xplc.2019.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z.L., Wei W., Wang H.D., Zhou J.J., Wang H.T., Chen K., et al. Functional characterization, structural basis, and regio-selectivity control of a promiscuous flavonoid 7,4′-di-O-glycosyltransferase from Ziziphus jujuba var. spinosa. Chem Sci. 2023 doi: 10.1039/d2sc06504e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X. Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 2009;583:3303–3309. doi: 10.1016/j.febslet.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 59.Yao J., Xing X., Yu L., Wang Y., Zhang X., Zhang L. Structure-function relationships in plant UDP-glycosyltransferases. Ind Crops Prod. 2022;189 doi: 10.1016/j.indcrop.2022.115784. [DOI] [Google Scholar]
- 60.Wang Z.L., Gao H.M., Wang S., Zhang M., Chen K., Zhang Y.Q., et al. Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc Natl Acad Sci USA. 2020;117:30816–30823. doi: 10.1073/pnas.2012745117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Louveau T., Osbourn A. The sweet side of plant-specialized metabolism. Cold Spring Harb Perspect Biol. 2019;11 doi: 10.1101/cshperspect.a034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masada S., Terasaka K., Mizukami H. A single amino acid in the PSPG-box plays an important role in the catalytic function of CaUGT2 (Curcumin glucosyltransferase), a group d family 1 glucosyltransferase from Catharanthus roseus. FEBS Lett. 2007;581:2605–2610. doi: 10.1016/j.febslet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Yamashita M., Fujimori T., An S., Iguchi S., Takenaka Y., Kajiura H., et al. The apiosyltransferase celery UGT94AX1 catalyzes the biosynthesis of the flavone glycoside apiin. Plant Physiol. 2023 doi: 10.1093/plphys/kiad402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nomura Y., Seki H., Suzuki T., Ohyama K., Mizutani M., Kaku T., et al. Functional specialization of UDP-glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 2019;99:1127–1143. doi: 10.1111/tpj.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohgami S., Ono E., Horikawa M., Murata J., Totsuka K., Toyonaga H., et al. Volatile glycosylation in tea plants: Sequential glycosylations for the biosynthesis of aroma β-primeverosides are catalyzed by two Camellia sinensis glycosyltransferases. Plant Physiol. 2015;168:464–477. doi: 10.1104/pp.15.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugimoto K., Ono E., Inaba T., Tsukahara T., Matsui K., Horikawa M., et al. Identification of a tomato UDP-arabinosyltransferase for airborne volatile reception. Nat Commun. 2023;14 doi: 10.1038/s41467-023-36381-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noguchi A., Kunikane S., Homma H., Liu W., Sekiya T., Hosoya M., et al. Identification of an inducible glucosyltransferase from Phytolacca americana L. cells that are capable of glucosylating capsaicin. Plant Biotechnol J. 2009;26:285–292. doi: 10.5511/plantbiotechnology.26.285. [DOI] [Google Scholar]
- 68.Chen K.Q., Arnold F.H. Enzyme engineering for nonaqueous solvents: random mutagenesis to enhance activity of subtilisin E in polar organic media. Biotechnology. 1991;(11):1073–1077. doi: 10.1038/nbt1191-1073. [DOI] [PubMed] [Google Scholar]
- 69.Savile C.K., Janey J.M., Mundorff E.C., Moore J.C., Tam S., Jarvis W.R., et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329:305–309. doi: 10.1126/science.1189106. [DOI] [PubMed] [Google Scholar]
- 70.Patel J.M. Biocatalytic synthesis of atorvastatin intermediates. J Mol Catal B Enzym. 2009;61:123–128. doi: 10.1016/j.molcatb.2009.07.004. [DOI] [Google Scholar]
- 71.Wang H.T., Wang Z.L., Chen K., Yao M.J., Zhang M., Wang R.S., et al. Discovery, mechanisms, and engineering of the missing apiosylation step in the biosynthesis of apiosides in Leguminosae. Plants (Prepr) 2023 doi: 10.21203/rs.3.rs-2748229/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J., Mu S., Yang J., Liu C., Zhang Y., Chen P., et al. Glycosyltransferase engineering and multi-glycosylation routes development facilitating synthesis of high-intensity sweetener mogrosides. IScience. 2022;25 doi: 10.1016/j.isci.2022.105222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Modarres H.P., Mofrad M.R., Sanati-Nezhad A. Protein thermostability engineering. RSC Adv. 2016;6:115252–115270. doi: 10.1039/c6ra16992a. [DOI] [Google Scholar]
- 74.Cartwright A.M., Lim E.K., Kleanthous C., Bowles D.J. A kinetic analysis of regiospecific glucosylation by two glycosyltransferases of Arabidopsis thaliana. J Biol Chem. 2008;283:15724–15731. doi: 10.1074/jbc.M801983200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim H.L., Kim A.H., Park M.B., Lee S.W., Park Y.S. Altered sugar donor specificity and catalytic activity of pteridine glycosyltransferases by domain swapping or site-directed mutagenesis. BMB Rep. 2013;46:37–40. doi: 10.5483/BMBRep.2013.46.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim H.S., Kim B.G., Sung S., Kim M., Mok H., Chong Y., et al. Engineering flavonoid glycosyltransferases for enhanced catalytic efficiency and extended sugar-donor selectivity. Planta. 2013;238:683–693. doi: 10.1007/s00425-013-1922-0. [DOI] [PubMed] [Google Scholar]
- 77.Wagner G.K., Pesnot T. Glycosyltransferases and their assays. ChemBioChem. 2010;11:1939–1949. doi: 10.1002/cbic.201000201. [DOI] [PubMed] [Google Scholar]
- 78.Aharoni A., Thieme K., Chiu C.P.C., Buchini S., Lairson L.L., Chen H., et al. High-throughput screening methodology for the directed evolution of glycosyltransferases. Nat Methods. 2006;3:609–614. doi: 10.1038/nmeth899. [DOI] [PubMed] [Google Scholar]
- 79.Yang G., Rich J.R., Gilbert M., Wakarchuk W.W., Feng Y., Withers S.G. Fluorescence activated cell sorting as a general ultra-high-throughput screening method for directed evolution of glycosyltransferases. J Am Chem Soc. 2010;132:10570–10577. doi: 10.1021/ja104167y. [DOI] [PubMed] [Google Scholar]
- 80.Engel L., Alves J., Hennek J., Goueli S.A., Zegzouti H. Utility of bioluminescent homogeneous nucleotide detection assays in measuring activities of nucleotide-sugar dependent glycosyltransferases and studying their inhibitors. Molecules. 2021;26:6230. doi: 10.3390/molecules26206230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumagai K., Kojima H., Okabe T., Nagano T. Development of a highly sensitive, high-throughput assay for glycosyltransferases using enzyme-coupled fluorescence detection. Anal Biochem. 2014;447:146–155. doi: 10.1016/j.ab.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 82.Fu H., Pan W., Vincent S.P. Pyruvate-kinase-coupled glycosyltransferase assays: limitations, struggles and problem resolution. ChemBioChem. 2017;18:2129–2136. doi: 10.1002/cbic.201700326. [DOI] [PubMed] [Google Scholar]
- 83.Gantt R.W., Peltier-Pain P., Cournoyer W.J., Thorson J.S. Using simple donors to drive the equilibria of glycosyltransferase- catalyzed reactions. Nat Chem Biol. 2011;7:685–691. doi: 10.1038/nchembio.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gantt R.W., Peltier-Pain P., Singh S., Zhou M., Thorson J.S. Broadening the scope of glycosyltransferase-catalyzed sugar nucleotide synthesis. Proc Natl Acad Sci USA. 2013;110:7648–7653. doi: 10.1073/pnas.1220220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhuang Y., Yang G.Y., Chen X., Liu Q., Zhang X., Deng Z., et al. Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme. Metab Eng. 2017;42:25–32. doi: 10.1016/j.ymben.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 86.Li Y., Hou J., Wang F., Sheng J. High-throughput assays of leloir-glycosyltransferase reactions: The applications of rYND1 in glycotechnology. J Biotechnol. 2016;227:10–18. doi: 10.1016/j.jbiotec.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 87.Wu Z.L., Ethen C.M., Prather B., MacHacek M., Jiang W. Universal phosphatase-coupled glycosyltransferase assay. Glycobiology. 2011;21:727–733. doi: 10.1093/glycob/cwq187. [DOI] [PubMed] [Google Scholar]
- 88.Chung S.Y., Nam S.W., Lim J., Park S., Yoon J. An “off-on” type UTP/UDP selective fluorescent probe and its application to monitor glycosylation process. Org Lett. 2009;11:2181–2184. doi: 10.1021/ol9004849. [DOI] [PubMed] [Google Scholar]
- 89.Lee H.S., Thorson J.S. Development of a universal glycosyltransferase assay amenable to high-throughput formats. Anal Biochem. 2011;418:85–88. doi: 10.1016/j.ab.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ojida A., Takashima I., Kohira T., Nonaka H., Hamachi I. Turn-on fluorescence sensing of nucleoside polyphosphates using a xanthene-based Zn(II) complex chemosensor. J Am Chem Soc. 2008;130:12095–12101. doi: 10.1021/ja803262w. [DOI] [PubMed] [Google Scholar]
- 91.Wongkongkatep J., Miyahara Y., Ojida A., Hamachi I. Label-free, real-time glycosyltransferase assay based on a fluorescent artificial chemosensor. Angew Chem Int Ed Engl. 2006;45:665–668. doi: 10.1002/anie.200503107. [DOI] [PubMed] [Google Scholar]
- 92.Ryu J., Eom M.S., Ko W., Han M.S., Lee H.S. A fluorescence-based glycosyltransferase assay for high-throughput screening. Bioorg Med Chem. 2014;22:2571–2575. doi: 10.1016/j.bmc.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 93.Ban L., Pettit N., Li L., Stuparu A.D., Cai L., Chen W., et al. Discovery of glycosyltransferases using carbohydrate arrays and mass spectrometry. Nat Chem Biol. 2012;8:769–773. doi: 10.1038/nchembio.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zielinski T., Reichman M., Donover P.S., Lowery R.G. Development and validation of a universal high-throughput UDP-glycosyltransferase assay with a time-resolved FRET signal. Assay Drug Dev Technol. 2016;14:240–251. doi: 10.1089/adt.2016.711. [DOI] [PubMed] [Google Scholar]
- 95.Deng C., Chen R.R. A pH-sensitive assay for galactosyltransferase. Anal Biochem. 2004;330:219–226. doi: 10.1016/j.ab.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 96.Choi Y.H., Kim J.H., Park B.S., Kim B.G. Solubilization and iterative saturation mutagenesis of a1,3-fucosyltransferase from helicobacter pylori to enhance its catalytic efficiency. Biotechnol Bioeng. 2016;113:1666–1675. doi: 10.1002/bit.25944/abstract. [DOI] [PubMed] [Google Scholar]
- 97.Persson M., Palcic M.M. A high-throughput pH indicator assay for screening glycosyltransferase saturation mutagenesis libraries. Anal Biochem. 2008;378:1–7. doi: 10.1016/j.ab.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 98.Park S.H., Park H.Y., Sohng J.K., Lee H.C., Liou K., Yoon Y.J., et al. Expanding substrate specificity of GT-B fold glycosyltransferase via domain swapping and high-throughput screening. Biotechnol Bioeng. 2009;102:988–994. doi: 10.1002/bit.22150. [DOI] [PubMed] [Google Scholar]
- 99.Williams G.J., Zhang C., Thorson J.S. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat Chem Biol. 2007;3:657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- 100.Fan B., Dong W., Chen T., Chu J., He B. Switching glycosyltransferase UGTBL1 regioselectivity toward polydatin synthesis using a semi-rational design. Org Biomol Chem. 2018;16:2464–2469. doi: 10.1039/c8ob00376a. [DOI] [PubMed] [Google Scholar]
- 101.Qu G., Li A., Acevedo-Rocha C.G., Sun Z., Reetz M.T. The crucial role of methodology development in directed evolution of selective enzymes. Angew Chem Int Ed Engl. 2020;59:13204–13231. doi: 10.1002/anie.201901491. [DOI] [PubMed] [Google Scholar]
- 102.Kamra P., Gokhale R.S., Mohanty D. SEARCHGTr: a program for analysis of glycosyltransferases involved in glycosylation of secondary metabolites. Nucleic Acids Res. 2005;33:W220–W225. doi: 10.1093/nar/gki449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Choi Y.H., Kim J.H., Park J.H., Lee N., Kim D.H., Jang K.S., et al. Protein engineering of α2,3/2,6-sialyltransferase to improve the yield and productivity of in vitro sialyllactose synthesis. Glycobiology. 2014;24:159–169. doi: 10.1093/glycob/cwt092. [DOI] [PubMed] [Google Scholar]
- 104.Reetz M.T., Carballeira J.D. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat Protoc. 2007;2:891–903. doi: 10.1038/nprot.2007.72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material