Abstract
Over the past decade, alternative RNA splicing has raised a great interest appearing to be of high importance in the generation of expression diversity. This regulatory process plays a critical role in the normal development and its impact on the initiation and development of human disorders as well as on the pharmacological properties of drugs is increasingly being recognized. Only few studies describe specific alternative splicing expression profiling. Microarray strategies have been conceived to address alternative splicing events but with very few experimental data related to their abilities to provide true quantification values. We have developed a specific microarray configuration relying on a few, well optimized probes per splice event. Basically, five probes of 24mer are used to fully characterize a splice event. These probes are of two types, exon probes and junction probes, and are either specific to a splice event or not. The performances of such a ‘splice array’ were validated on synthetic model systems and on complex biological materials. The results indicate that DNA chips based on this design combining exon and junction derived probes enable the detection and, absolute and relative quantification of splice variants. In addition, this strategy is compatible with all the microarrays that use oligonucleotide probes.
INTRODUCTION
The nearly complete coverage of the human genome indicates that it encodes only 20 000–25 000 protein-coding genes (1). This number is significantly below the one estimated from the complexity observed at the messenger RNA (mRNA) level through analyses of expressed sequence tags (ESTs) (between 100 000 and 150 000) (2,3). This ‘gene shortfall’ has raised a great interest in alternative RNA splicing, a cellular mechanism generating different transcripts from a single gene, which could account for this observed discrepancy between the number of genes and mRNAs.
Large-scale bioinformatics analyses of alternative splicing have reported high rates of alternative splicing, with over 60% of all human genes expressing multiple mRNA (2,4) [see also the following databases: EASED (5) (http://eased.bioinf.mdc-berlin.de/statistics.html), ASD (6) (http://www.ebi.ac.uk./asd) and PALS (7), (http://palsdb.ym.edu.tw)]. More recently, large- scale gene expression profiling experiments via microarrays or SAGE surprisingly revealed an unexpected abundance of expressed genomic sequences (8–10). In-depth analysis of the transciptomes generated from human chromosomes 21 and 22 suggested that every human gene is likely to undergo alternative RNA splicing (9). Thus, alternative RNA splicing is a key element in providing functional diversity from the global expression of the human genome.
Alterations of the splicing patterns, by mutation or defects at the level of the spliceosome machinery, can lead to profound cellular deregulations and cause human diseases (11,12). Thus, 10% of point mutations resulting in inherited diseases are disrupting splice site sequences (13) (see also the Human Gene Mutation Database, http://archive.uwcm.ac.uk/uwcm/mg/hgmd0). In addition, mutations in less well-defined exonic and intronic enhancers or suppressors of splicing also affect splicing (14–16). It is now widely demonstrated that RNA splicing can severely impact gene function. Compared with its wild-type counterpart, a splice variant can exhibit different enzymatic or signaling activities, novel subcellular localization and altered protein stability [reviewed in (17)]. Antagonistic effects for isoforms produced by the same pre-mRNA have also largely been described, most notably in the field of apoptosis (18,19).
Cis- or trans-acting mutations at the pre-mRNA level can affect RNA splicing and also alter the ratio between the expression levels of two isoforms. These ratios are thus critical parameters that need to be quantified as some isoforms can exert dominant negative or positive effects. Aggregation of the microtubule-associated tau protein is associated with several neurological disorders, including Alzheimer's disease and frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17). Tau exon 10 encodes one of the four microtubule-binding sites. The correct balance between tau isoforms is critical to neuronal survival. The normal ratio between the isoforms with or without exon 10 is one. Some FTDP-17 associated mutations occur within the exonic and intronic regulatory sequences in and around exon 10 (20,21) and have been shown to alter the ratio of the two tau isoforms in in vitro models. It has also recently been shown that the inclusion of exon 10 is higher in the affected brain regions from Alzheimer's patients (22,23). In addition, correction of alternative splicing of tau exon 10 with antisense oligonucleotides targeting splice junctions has been reported (24). Another striking example of the critical importance of the splice variants ratios lies within the two main determining genes for spinal muscular atrophy (SMA), SMN1 and SMN2. The SMN protein from the SMN1 gene was not expressed in 95% of SMA patients owing to deletions in both alleles. SMN2 is identical to SMN1 except for a single nucleotide difference which attenuates the activity of an exonic enhancer responsible for primarily producing an SMN2 mRNA lacking exon 7. This transcript gets translated into a less or non-functional SMN protein. There is a correlation between the level of full-length SMN protein produced from the SMN2 gene and the severity of SMA as well as duration of the survival (25,26). Redirecting the balance from the exon 7 skipped form towards the full-length has been demonstrated with oligonucleotide based molecules and with small molecular weight compounds (27–31).
Splice variants expression ratios could also provide better diagnostic or prognostic markers than the absolute levels of either variant as it has recently been shown in the association between acetylcholine esterase splice variants and treatment outcome for Alzheimer's disease patients (32). Thus, monitoring the specific level of absolute or relative expression of splice variants could certainly help better define physiological and pathological mechanisms.
The quantification of splice variants requires discriminating approaches that will take into account the presence of specific sequences within the isoforms. The expression value must be specific to the splice variant and associated isoforms should not interfere with this value. Insertional events such as intron retention, novel alternative exons, or certain uses of 5′ or 3′ cryptic sites will generate additional sequence information as compared with a wild-type form. Such sequences could serve as the basis for oligonucleotide design as probes for microarrays or as PCR primers. On the contrary, deletion events caused by exon skipping events or by certain uses of cryptic splice sites do not generate such novel sequences. In this case, one can only rely on novel junction sequences to discriminate from the wild-type counterpart. The design of oligonucleotide primers that span junction sequences for isoform-specific PCR amplification has been described previously (33–36). The reaction conditions of such PCR assays have to be carefully optimized to avoid contaminating signals due to mispriming. Splice junctions specific probes have also been used in the design of an RNA invasive cleavage assay to monitor the expression of FGFR2 splice variants (37). RASL (RNA-mediated annealing, selection and ligation) has also been applied to the study of splice variants (38). In this latter approach, oligonucleotides that are complementary to sequences located on both sides of a splice junction are ligated to be PCR amplified. Fluorescent amplicons can then be applied to microarray for detection. However, the scale-up of these different platforms, i.e. their ability to study several hundreds or thousands of splice events in parallel, is unclear at this time.
DNA microarrays have become a widely used technology for large-scale gene expression studies. Profiling alternative splicing on such platforms has been described in a limited number of studies (4,38–43). These studies describe the feasibility to monitor splicing events on microarray platforms, but quantification methods for the absolute and relative levels of expression of splice variants have not been extensively developed.
The microarray configuration described here provides an efficient splice variant detection platform offering the possibility to measure expression levels using a limited number of probes. A given splice event is represented by a set of five probes specific for exonic sequences and for sequences representing novel junctions being created or deleted by the splicing event. The method is suitable for every type of splicing events: exon skipping, cassette exons, intron retention and alternative 5′ or 3′ cryptic sites. In addition, the balance between different isoforms can be determined using the expression values generated by this specific set of probes. This configuration is compatible with any microarray that uses oligonucleotide probes.
In this study, a splice array was built with a series of probes designed to monitor the expression of 42 isoforms. The ability of this platform to provide absolute and relative quantification of splice variants was evaluated in synthetic model systems and in more complex biological settings.
MATERIALS AND METHODS
Models for experimental validation
The following 18 genes with corresponding isoforms have been selected for this study:
MCL1 (NM_021960), BIN1 (NM_004305), APP (NM_000484), HSPC111 (NM_016391), APEX (NM_001641), MYBL2 (NM_002466), FKBP1A (NM_00801), DUSP3 (NM_004090), PPIB (NM_00942), NUDC (NM_006600), GSTZ1 (NM_001513), ROBO1 (NM_002941), EWSR1 (NM_005243), PRKDC (NM_006904), CDC37 (NM_007065), MGC8721 (NM_016127), hnRNP-A/B (NM_031266) and hnRNPD (NM_031370) (see Supplementary Table S1: listing of the splice events).
For every gene, a pair of primers was designed to PCR amplify a long and a short isoform in the region where the splice event takes place. PCR was done on breast cell line material. All PCR fragments were cloned into pGEMR-T Vector (Promega) or TOPO™ Vector (Invitrogen) and validated by sequencing.
Biological samples
Human breast cell lines MCF7 (ATCC HTB-22), MDA-MB-435 (ATCC HTB-129), T47D (ATCC HTB-133) and BT549 (ATCC HTB-122) obtained from ATCC were maintained following the supplier's recommendations. Total RNA was isolated using Trizol (Invitrogen) according to the manufacturer's suggested protocol.
Human brain total RNA (ref. 7962) was purchased from Ambion. Total RNA from human metastatic melanoma tissue sample was kindly provided by the Dermatology Department, University of Geneva. RNA from patient's cutaneous back metastasis tissue (stage III, nodular malignant melanoma) was extracted using caesium chloride method. All RNA preparations were quality controlled for integrity, using the Agilent Bioanalyser 2100 (Agilent Technologies).
Parameters for probe design
Probes were designed with homogeneous melting temperature and similar lengths to obtain a common thermodynamic profile. For this pilot study, we applied the following selection criteria using Array Designer 2 (Premier Biosoft): (i) %GC: 40–60% for 24 and 30mer, 30–60% for 40mer; (ii) melting temperature: 65 ± 3°C for 24mer, 69 ± 4°C for 30mer and 72 ± 4°C for 40mer; (iii) primer concentration, 50 nM; and (iv) salt concentration, 50 mM. Finally, oligonucleotides with significant hairpin tendencies (ΔG > −7 kcal/mol), self-dimerization tendencies (ΔG > −4 kcal/mol) or presenting more than six identical bases in a row were discarded. Junction probes were preferably centered on the splice site (12–12 bases for a 24mer) but other combinations were also designed for performance study (10–14, 11–13, 13–11 and 14–10 bases). To avoid cross-hybridization, specificity of each probe was evaluated by BLAST analyses against the human EST databases using parameters for short, nearly exact matches (Expected value = 1000, word size = 7 and penalty for mismatch = −1). Finally, a total of 121 oligonucleotide probes were designed to detect and quantify 42 isoforms. In addition, 24mer control probes targeting exons of Arabidopsis thaliana mRNA for photosystem I chlorophyllA/B-binding protein (X56062) and A.thaliana lipid transfer (AF159801) were designed to serve as exogenous controls.
Splice-array preparation
The 24, 30 and 40mer oligonucleotides modified with a 5′-amine-(CH)5-modification for covalent immobilization were printed on three distinct CodeLink™ (Amersham Biosciences) glass slides according to their length, using the Microgrid II Arrayer (Genomic Solution). Desalted probes were brought to a final concentration of 25 nmol/ml in 150 mM sodium phosphate, pH 8.5 before printing on the slides. Each oligonucleotide was spotted eight times per array. Residual reactive groups were blocked with a solution of 0.1 M Tris, 50 mM ethanolamine, pH 9.0 and 0.1% SDS, rinsed with water and washed with 4× SSC/0.1% SDS.
Target preparation and labeling
For synthetic model systems, labeled targets were prepared from pairs of cDNA clones, each representing a wild-type form and a splice variant [also called : long form (LF) + short form (SF)]. About 2 μg of corresponding cRNA was generated using in vitro transcription systems (T7 or SP6 Megascript kit; Ambion).
For breast cell lines, brain and melanoma RNAs, labeling was performed using a standard amplification protocol. Briefly, 5 μg of total RNA were reverse transcribed in 20 μl with 200 U Superscript II (Life Technologies) and 1 μg Oligo(dT)24-T7 [5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′] in 1× first-strand buffer (Life Technologies) at 42°C. Total RNA was spiked with 300 pg of A.thaliana Cab mRNA (Stratagene) for calibration purposes. Second-strand synthesis (SSS) was carried out in 150 μl final volume containing 40 U DNA polymerase I, 2 U of Escherichia coli RNase H, and 10 U of E.coli DNA ligase in 1× second-strand buffer (Life Technologies) by adding 130 μl of ice-cold SSS premix to the 20 μl RT reaction and incubated at 16°C for 2 h. The double-strand DNA synthesis was terminated by adding 10 U of T4 DNA polymerase and incubating for 10 min at 16°C. cDNA was purified with phenol/chloroform/isoamyl-alcohol 25:24:1 (v/v/v) extraction on Phase lock gels (Eppendorf) and precipitated overnight at −20°C with 7.5 M NH4Ac and absolute ethanol.
Transcription of dsDNA was prepared in 20 μl at 37°C for 3 h using MEGAscript T7 Transcription kit (Ambion) (7.5 mM ATP, 7.5 mM CTP, 7.5 mM GTP, 3.75 mM UTP and 3.75 mM aaUTP). Amplified RNA was purified on RNeasy mini columns (Qiagen) according to the manufacturer's protocol. cRNA was quantified by absorbance at 260 nm, and quality controlled with the Agilent RNA Bioanalyser 2100. For biological material, up to 10 μg of amino-allyl-cRNA (aa-cRNA) sample were labeled using Cy5 or Cy3 (Amersham Biosciences) in 50% (v/v) dimethyl sulfoxide and 0.1 M NaHCO3, pH 9.0 for at least 1 h at room temperature. For aa-cRNA derived from cDNA clones, a mixture of each variant was prepared using 1 μg per gene as starting material for the labeling reaction as described above. The reaction was completed by adding 4 μl of hydroxylamine 4 M as a quenching reagent, and targets were purified with two washes on a Microcon-30 (Millipore). The purified labeled aa-cRNAs were fragmented in 1× fragmentation buffer (200 mM Tris-acetate, 100 mM KOAc and 80 mM MgOAc) at 94°C for 35 min and purified on a Microcon-10 (Millipore). For the spiking experiments, 2 μg Drosophila melanogaster cRNA were added to the target as a carrier during fragmentation to create a complex background. Labeled Drosophila cRNA was used in preliminary experiments and no cross-hybridization signal was detected. Therefore, further hybridizations were done with unlabeled Drosophila cRNA.
Hybridization conditions
Either 7–10 μg of fragmented biological material cRNA or 2 μg of Drosophila cRNA spiked with different isoform RNAs at various concentrations were denatured in 15 μl of 0.1 mg/ml salmon sperm DNA, 5× SSC buffer with 0.1% (w/v) SDS for 3 min at 95°C and hybridized on the splice array. Unless mentioned in the text, slides were incubated for 16 h in a 55°C hybridization oven. Washes were performed twice for 5 min in 2× SSC, 0.1% (w/v) SDS at hybridization temperature, then for 1 min in 0.2× SSC and for 1 min in 0.1× SSC at room temperature. Slides were finally spin-dried before reading.
Splice-array analysis
The splice arrays were scanned with a ScanArray 4000 (Packard-Bioscience). For quantification studies using spiked isoforms, the scanner signal was calibrated using the fluorescent intensity values of the common exon probe (Ex1, see Figure 1) after hybridization with a mixture containing 50% LF labeled with Cy5 and 50% SF labeled with Cy3. For biological material analysis, the calibration required oligonucleotides specific to A.thaliana Cab. Scanned microarray images were analyzed using the QuantArray Analysis Software (Packard-Bioscience) and reports were imported into GeneTraffic Duo data management and analysis software (Iobion Informatics) along with TIFF images. Local background intensities, calculated for each feature by QuantArray, were subtracted prior to normalization by GeneTraffic. All probes with raw intensities less than twice the average local background were discarded from the analysis. For spiking experiments, intensities were not further normalized. Hybridizations with biological material were normalized using the global intensity method included in the microarray analysis software. The ratios of normalized fluorescent intensities for each feature were used for subsequent analysis.
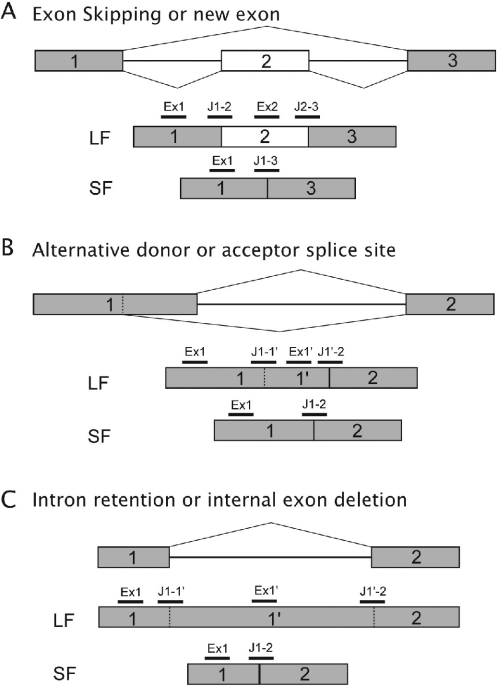
Figure 1.
Alternative splicing events and probe location for splice-array design. One oligonucleotide is specific for the skipped exon or the intron retention (Ex2 or Ex1′) in the spliced isoform and enables the quantification of the long form (LF). One oligonucleotide is specific for the sequence in one of the adjacent exons (Ex1) not involved in the splice and will measure the amount of the LF plus the short isoforms (SF). Different oligonucleotides monitor the different junctions: J1-2, J2-3 and J1-3 (A), J1-1′, J1′-2 (B), J1-1′, J1′-2 and J1-2 (C); two junction probes being LF specific, one junction probe being SF specific.
RESULTS
The splice-array design
The splice array is based on the design of probes located on conserved exons, alternative exons as well as on the constitutive and alternative splice junctions. The different types of splice events (exon skipping, novel exon, internal exon deletion, intron retention or alternative usage of splice donor or acceptor sites) are displayed in Figure 1. Each splice event is associated with two isoforms called LF and SF. The wild-type isoform can either be LF or SF depending upon the nature of the splice event.
For a single splicing event, a set of five oligonucleotides is designed: one probe is specific for one of the conserved exons (not involved in the splice event) to measure the RNA amount of the long and short forms. One probe designed in the additional exonic sequence that differentiates LF and SF and two probes spanning the junctions flanking this additional sequence are used to monitor the LF only. The detection of the short isoform relies on a single junction probe. Note that, theoretically, only one of the three LF-specific probes could be used to detect and quantify the long isoform.
Probe thermodynamics (length, Tm, %GC and secondary structure) were carefully controlled to fall in a range as narrow as possible (see Material and Methods: parameters for probe design). The specificity of the probes was evaluated by BLAST analyses of the oligonucleotide probes against the human EST databases to avoid cross-reactivity issues.
Influence of probe length and validation of probe specificity
Specificity studies were first carried out by evaluating three different probe lengths (24, 30 and 40mer). We restricted our analysis to short oligonucleotides to avoid the potential hybridization of half of the junction probe to a single exon. Independent slides prepared with probes of 24, 30 or 40mer (Figure 2A and B for probe location and sequence) were hybridized at 55, 60 and 65°C, respectively, to increase the stringency and the hybridization specificity. MCG8721 wild-type and isoform targets mixed as an equimolar ratio were hybridized in parallel to the three slides.
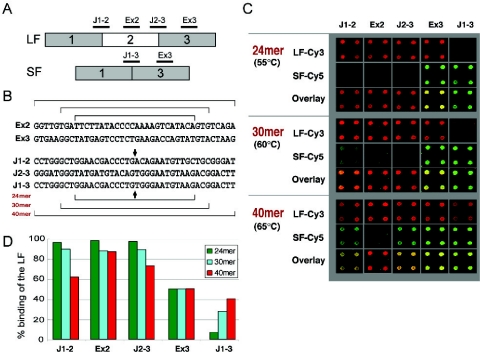
Figure 2.
Probe specificity. (A) Location of the five probes designed to monitor the skipping of exon 2 of the MGC8721 gene (long form, LF; short form SF). (B) Probe sequences [oligonucleotides names are defined in (A)]. Brackets indicate the sequence limits for the 24, 30 and 40mer. The arrows refer to the junction positions. (C) 2.5 ng of the LF transcript was labeled with Cy3, and 2.5 ng of the SF transcript was labeled with Cy5. Hybridization temperatures for the 24, 30 and 40mer were 55, 60 and 65°C, respectively. The overlays of the images are presented for the three probe lengths tested with four replicates for each probe. (D) The graph shows the specificity of the hybridization of the LF on the 24, 30 and 40mer probes after measurements and normalization of the fluorescent hybridization signals derived from (C) in both channels. The percentage of binding of the LF is obtained by measuring the ratio of fluorescent Cy3 signal to the overall signal (Cy3 + Cy5).
The long isoform cRNA was readily detected by the 40mer Jct1-3 oligonucleotide (Figure 2C and D), which should only hybridize to the short isoform junction. Conversely, the short isoform cRNA was detected by the 40mer Jct1-2 and Jct2-3 oligonucleotides, which should only hybridize to the long isoform. The 30mer junction probes also showed a similar behavior, although less pronounced. Only the 24mer provided the specificity required for isoform-specific detection of alternatively spliced events. Similar results have been obtained with two other genes (data not shown).
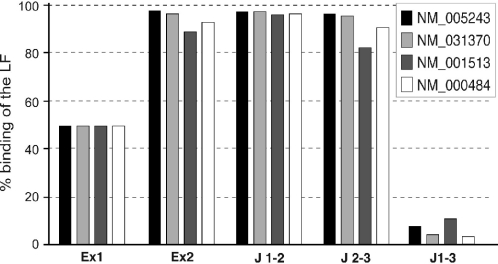
For the subsequent validation steps, we prepared a splice array gathering 121 probes (24mer) designed on 42 isoforms from a set of 18 selected genes. Each probe was spotted in eight replicates. The final density per array was 1176 oligonucleotides including exogenous controls. The validation of the specificity of the 24mer probes was first confirmed with cloned isoforms for four different genes in a complex RNA setting. The long isoforms were labeled with Cy3 and the short isoforms with Cy5. In parallel, D.melanogaster total RNA was amplified and spiked with A.thaliana Cab poly(A). To assess the specificity of the hybridization, equal amounts of long and short isoforms of a given gene were diluted in Drosophila cRNA. As expected, more than 90% of the signal detected for probes Ex2, J1-2 and J2-3 is due to the LF while this isoform is responsible for <10% of the hybridization signal on probe J1-3 (Figure 3).
Figure 3.
Hybridization specificity. The specificity is expressed as a percentage of hybridization of the long isoform with respect to the five oligonucleotides characterizing the splice event. The long form (LF) transcript was labeled with Cy3, the short form (SF) transcript with Cy5. The fluorescent signals were measured in both channels and normalized. The probes are indicated on the x-axis, the y-axis presents the percentage of binding of the LF by determining the ratio of the fluorescent Cy3 signal to the overall signal (Cy3 + Cy5). The results are presented for four different genes.
Quantitative measurements of splice variants
We then performed experiments to simulate a biological system in which a long and a short isoform from the same gene are expressed, the expression of one isoform being altered in one sample compared with a second sample. To this end, we compared two artificial samples made by mixing various molar ratios of cloned isoforms. The goal was to assess whether we could retrieve the expected fold changes for each isoform in the samples. The mixed isoforms were spiked into Drosophila cRNA to generate two samples, A and B. Two sets of experiments have been independently performed and their results are compiled in Table 1. In the first set of experiments (Set 1), sample A contained 80% of long isoforms and 20% of short isoforms (w/w) and both LFs and SFs were labeled with Cy3. Sample B was prepared with 40% long isoforms and 60% short isoforms and both LFs and SFs were labeled with Cy5. In the second set (Set 2), sample A was made with 20% of LF and 80% of SF and sample B with 90% of LF and 10% of SF. Both samples from a given set were co-hybridized on the 24mer splice array, using (i) equal amounts of A and B, (ii) twice the amount of B and (iii) four times the amount of B. Fluorescent hybridization signals were read as described previously and raw data were imported into the GeneTraffic software (Iobion Informatics). We next determined the fold changes (sample B over sample A) for the different spiked genes in each set and for each A:B ratio. RE1, RE2 and RJ were defined as the fold changes calculated for the common exon (with probe Ex1 or Ex3), for the skipped exon (with probe Ex2) and for the short isoform junction (with probe J1-3), respectively. The results are presented in Table 1 (Fold changes section).
Table 1.
Fold changes and relative abundance of long and short isoforms
| Ratio A:B | Isoform | Fold changes (B/A) | Relative abundance | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RE1 | RE2 | RJ | Sample A (Reference) | Sample B (Experimental) | |||||||
| LF + SF | LF | SF | LF (80%) | SF (20%) | SD | LF (40%) | SF (60%) | SD | |||
| Set 1 | |||||||||||
| 1:1 | Theoretical fold changes | 1 | 0.5 | 3 | Isoform quantity (ng) | 4 | 1 | 2 | 3 | ||
| NM_031370 | 0.91 ± 0.11 | 0.49 ± 0.09 | 2.52 ± 0.62 | 77.9 | 22.1 | ±4.6 | 41.4 | 58.6 | ±4.7 | ||
| NM_005243 | 1.26 ± 0.39 | 0.54 ± 0.19 | 3.17 ± 0.87 | 72.7 | 27.3 | ±0.7 | 30.5 | 69.5 | ±1.0 | ||
| NM_001513 | 1.01 ± 0.22 | 0.61 ± 0,15 | 2.26 ± 0.01 | 74.8 | 25.2 | ±7.0 | 44.5 | 55.5 | ±3.1 | ||
| 1:2 | Theoretical fold changes | 2 | 1 | 6 | Isoform quantity (ng) | 4 | 1 | 4 | 6 | ||
| NM_031370 | 1.76 ± 0.17 | 0.94 ± 0.06 | 4.95 ± 0.20 | 79.7 | 20.3 | ±1.9 | 42.9 | 57.1 | ±2.3 | ||
| NM_005243 | 2.33 ± 0.03 | 1.06 ± 0.03 | 5.85 ± 0.08 | 73.5 | 26.5 | ±1.5 | 33.4 | 66.6 | ±2.0 | ||
| NM_001513 | 1.89 ± 0.04 | 1.14 ± 0.02 | 5.25 ± 0.42 | 81.7 | 18.3 | ±1.3 | 49.2 | 50.8 | ±0.7 | ||
| 1:4 | Theoretical fold changes | 4 | 2 | 12 | Isoform quantity (ng) | 4 | 1 | 8 | 12 | ||
| NM_031370 | 3.84 ± 0.28 | 2.10 ± 0.16 | 10.56 ± 0.25 | 79.5 | 20.5 | ±1.2 | 43.5 | 56.5 | ±0.4 | ||
| NM_005243 | 3.52 ± 0.20 | 2.35 ± 0.18 | 9.76 ± 0.98 | 84.0 | 16.0 | ±1.4 | 56.1 | 43.9 | ±2.1 | ||
| NM_001513 | 4.06 ± 0.06 | 2.39 ± 0.03 | 10.80 ± 0.82 | 80.0 | 20.0 | ±1.6 | 47.0 | 53.0 | ±1.0 | ||
| Sample A (Reference) | Sample B (Experimental) | ||||||||||
| LF (20%) | SF (80%) | SD | LF (90%) | SF (10%) | SD | ||||||
| Set 2 | |||||||||||
| 1:1 | Theoretical fold changes | 1 | 4.5 | 0.125 | Isoform quantity (ng) | 2 | 8 | 9 | 1 | ||
| NM_005243 | 1.13 ± 0.24 | 5.97 ± 3.12 | 0.27 ± 0.01 | 18.1 | 81.9 | ±5.7 | 79.5 | 20.5 | ±2.6 | ||
| NM_001513 | 0.86 ± 0.07 | 3.3 ± 0.27 | 0.28 ± 0.15 | 19.3 | 80.7 | ±0.3 | 74.7 | 25.3 | ±11.4 | ||
| NM_000484 | 1.32 ± 0.32 | 3.13 ± 0.57 | 0.23 ± 0.06 | 37.1 | 62.9 | ±2.4 | 89.1 | 10.9 | ±0.2 | ||
| NM_000801 | 1.59 ± 0.58 | 4.89 ± 2.26 | 0.20 ± 0.06 | 31.4 | 68.6 | ±3.6 | 91.2 | 8.8 | ±0.2 | ||
| 1:2 | Theoretical fold changes | 2 | 9 | 0.25 | Isoform quantity (ng) | 2 | 8 | 18 | 2 | ||
| NM_005243 | 1.71 ± 0.68 | 8.88 ± 3.62 | 0.39 ± 0.04 | 15.0 | 85.0 | ±1.1 | 77.8 | 22.2 | ±6.9 | ||
| NM_001513 | 1.44 ± 0.05 | 6.90 ± 0.24 | 0.32 ± 0.07 | 17.1 | 82.9 | ±0.7 | 81.9 | 18.1 | ±3.4 | ||
| NM_000484 | 1.45 ± 0.27 | 5.76 ± 0.49 | 0.24 ± 0.02 | 21.7 | 78.3 | ±2.8 | 86.8 | 13.2 | ±1.9 | ||
| NM_000801 | 2.12 ± 1.01 | 6.25 ± 2.08 | 0.28 ± 0.03 | 28.6 | 71.4 | ±6.5 | 88.1 | 11.9 | ±5.4 | ||
| 1:4 | Theoretical fold changes | 4 | 18 | 0.5 | Isoform quantity (ng) | 1 | 4 | 18 | 2 | ||
| NM_005243 | 2.71 ± 0.86 | 13.3 ± 3.33 | 0.57 ± 0.11 | 16.4 | 83.6 | ±1.7 | 81.7 | 18.3 | ±2.8 | ||
| NM_001513 | 3.13 ± 0.10 | 14.6 ± 2.08 | 0.68 ± 0.14 | 18.0 | 82.0 | ±2.8 | 82.3 | 17.7 | ±3.6 | ||
| NM_000484 | 2.79 ± 0.66 | 11.2 ± 1.25 | 0.51 ± 0.07 | 20.8 | 79.2 | ±3.2 | 85.0 | 15.0 | ±2.1 | ||
| NM_000801 | 3.15 ± 1.40 | 10.1 ± 1.01 | 0.56 ± 0.07 | 26.1 | 73.9 | ±11.3 | 83.9 | 16.1 | ±7.4 | ||
Long and short isoforms for different genes were mixed together in various quantities to generate artificial samples A and B. The expected fold changes and relative abundance (%) are indicated in bold. Set 1 and Set 2 are independent assays with different LF and SF ratios for samples A and B. In each set, three hybridizations were performed with different A:B ratios: (i) equal amount of A and B (1:1), (ii) twice the amount of B (1:2) and (iii) four times the amount of B (1:4). Fold changes and relative abundance values are the mean of two independent experiments performed by two users with the corresponding standard deviation (SD) calculations reported next to values. See Supplementary Table S1 for the LF and SF definition for each transcript.
Theoretical fold changes were anticipated from the initial amounts of LF and SF mixed together in samples A and B (values reported in bold in Table 1). Experimental values were calculated for LF and SF, LF solely and SF solely, using probes Ex1 or Ex3 (specific for the common exon), Ex2 (LF specific) and J1-3 (SF specific), respectively. The experimental fold changes determined for the different genes tested were found to be close to the expected values. Expected fold changes from both sets of Table 1 when plotted against observed values generated an R2 determinant coefficient of 0.945. In set 2, we generated mixtures with larger differences in the amounts of LF and SF to evaluate the limits of the method. Even with ratios of 12 and 0.25, the quantification still provided acceptable results (Table 1). A more significant deviation from the expected values was observed only for the extreme deregulations (0.125 and 18).
The relative abundance between the long and the short isoforms of a given gene in both samples (A and B) can be derived from the following calculation. The composition of samples A and B can be translated into two equations, x being the percentage of long isoform and y the percentage of short isoform. Then:
where
The results obtained for the different spiked isoforms are presented in Table 1 (Relative abundance section) with the expected percentages of LF and SF indicated in bold. For all the genes being evaluated, the experimental values are close to the theoretical values across the hybridizations. Expected relative abundance values from both sets of Table 1 when plotted against observed values generated an R2 determinant coefficient of 0.953. This experiment has been repeated for both sets on different batches of splice arrays by two users with similar results.
Potentially, three probes could serve for the characterization of the longer isoform: Ex2, Jct1-2 and Jct2-3. Calculation performed with the two junction probes, Jct1-2 and Jct2-3 led to similar results (data not shown). Therefore, for further analysis on biological material, a mean value of the ratio corresponding to the three probes (Ex2, Jct1-2 and Jct 2-3) was used for relative abundance calculation.
Sensitivity studies
The accuracy and the sensitivity of the splice variant quantification method were then assessed with spiking experiments using a mixture of six isoforms for the detection of three splice events. Two dilutions of sample A (40% LF, 60% SF, Cy5 labeling) and sample B (80% LF, 20% SF, Cy3 labeling) have been co-hybridized with always twice the amount of spiked cRNA isoforms in sample B than in sample A. The fold changes were determined with GeneTraffic for the sets of hybridizations. The calculation method described above was next applied and the results obtained for different amounts (1.25 and 0.08 ng per isoform in A) of material tested are presented in Table 2.
Table 2.
Quantification of splice events between two situations for sensitivity determination
| Amount of isoforms in samples A:B | Isoform | Fold changes (B/A) | Relative abundance (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RE1 | RE2 | RJ | Sample A (Reference) | Sample B (Experimental) | |||||
| LF + SF | LF | SF | LF (40%) | SF (60%) | LF (80%) | SF (20%) | |||
| 1.25 : 2.5 | Theoretical fold changes | 2 | 4 | 0.66 | Isoform quantity (ng) | 0.5 | 0.75 | 2 | 0.5 |
| NM_006904 | 1.75 | 4.06 | 0.74 | 30.4 | 69.6 | 70.5 | 29.5 | ||
| NM_005243 | 1.89 | 5.43 | 0.45 | 28.9 | 71.1 | 83.1 | 16.9 | ||
| NM_001513 | 1.77 | 3.03 | 0.56 | 48.9 | 51.1 | 83.9 | 16.1 | ||
| 0.08 : 0.16 | Theoretical fold changes | 2 | 4 | 0.66 | Isoform quantity (ng) | 0.03 | 0.05 | 0.12 | 0.03 |
| NM_006904 | 1.61 | 2.57 | 0.71 | 48.3 | 51.7 | 77.2 | 22.8 | ||
| NM_005243 | 1.34 | 2.64 | 0.59 | 36.5 | 63.5 | 72.1 | 27.9 | ||
| NM_001513 | 1.65 | 2.16 | 1.2 | 46.8 | 53.2 | 61.4 | 38.6 | ||
Long forms (LFs) and short forms (SFs) of three different genes have been mixed together at various proportions (40% LF, 60% SF for A; 80% LF, 20% SF for B). In each case, the amount of sample B was twice the amount of sample A. The expected fold changes and relative abundance (%) are reported in bold. Fold changes have been measured and the resulting values were used in our calculation method to determine the relative abundance of the long and short isoforms for each gene.
The experimental values measured for the mixture containing 1.25 ng of sample A are close to the expected values. Diluting the isoforms down to a concentration as low as 0.03 ng (3 pM or 1/10 000 relative abundance) per isoform for the sample A still provide acceptable results taking into account potential errors in consecutive sample dilutions. Although not tested on the different oligoarray platforms, the sensitivity limit of splice variant quantification is likely to correspond to the sensitivity limit of a given platform.
Platform validation with biological material
Additional assays were performed to assess whether we could detect and quantify known isoforms using biological material. Labeled cRNA from MCF7 breast cell line was used as reference for co-hybridizations with cRNA from three other breast cell lines: T47D, MDA435 and BT549. All the hybridizations were repeated with dye swap before performing differential expression and isoform relative abundance calculations.
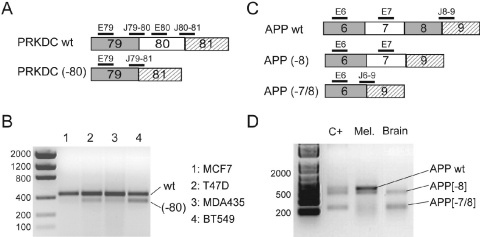
Four isoforms from four genes (out of the 18) were found significantly deregulated at least once in the three hybridizations (fold changes >1.80 or <0.55) (Table 3). Among these genes, we further studied the skipping of exon 80 of PRKDC corresponding to a splice event described in the literature (44) (Figure 4A). The LF of PRKDC is overexpressed in MDA435 and T47D by 2-fold relative to MCF7. The relative abundance calculations indicated that the exon skipped isoform was the minor form in all cell lines, however, with a significant difference between BT549 where it represented 23% and MDA435 or T47D where it only represented 4%. RT–PCR experiments were performed to amplify both PRKDC isoforms (Figure 4B). The band intensities of the two isoforms were in agreement with the microarray results. The long isoform appeared more expressed in T47D and MDA435 than in MCF7 and the short isoform was barely visible on the gel for T47D and MDA435 while it appeared more abundant in BT549.
Table 3.
Fold changes and relative abundance for deregulated isoforms in MDA435, T47D or BT549 versus MCF7
| Gene | Isoforms | Experimental sample | Fold changes (Exp/MCF7) | Relative abundance | |||
|---|---|---|---|---|---|---|---|
| LF + SF (RE1) | LF (RE2) | SF (RJ) | Experimental sample | ||||
| % LF | % SF | ||||||
| FKBP1A | Wild type (SF) | MDA435 | 3.07 | 1.16 | 3.25 | 3.2 | 96.8 |
| Intron 4 retention (LF) | T47D | 1.14 | 1.13 | 1.2 | NA | NA | |
| BT549 | 1.58 | 0.98 | 1.73 | 12.4 | 87.6 | ||
| GSTZ1 | Wild type (LF) | MDA435 | 0.62 | 0.58 | 1.21 | 87.6 | 12.4 |
| Exon 5 skipping (SF) | T47D | 0.97 | 0.96 | 1.39 | 96.7 | 3.3 | |
| BT549 | 0.53 | 0.50 | 0.93 | 89.0 | 11.0 | ||
| PRKDC | Wild type (LF) | MDA435 | 1.93 | 1.97 | 1.23 | 96.3 | 3.7 |
| Exon 80 skipping (SF) | T47D | 2.19 | 2.22 | 1.66 | 95.9 | 4.1 | |
| BT549 | 1.56 | 1.60 | 1.45 | 76.8 | 23.2 | ||
| MCL1 | Wild type (LF) | MDA435 | 0.91 | 0.53 | 1.08 | 17.8 | 82.2 |
| Exon 2 skipping (SF) | T47D | 1.22 | 0.95 | 1.19 | NA | NA | |
| BT549 | 1.28 | 1.87 | 0.96 | 51.3 | 48.7 | ||
Significant deregulations are reported in bold. RE2 values correspond to the mean of fold changes calculated with each LF-specific probes (Ex2, J1-2 and J2-3). NA, not applicable (see in text the limitations for the relative abundance determination).
Figure 4.
Isoform study for PRKDC and APP genes. (A and C) Splicing events and probe location to monitor the skipping of exon 80 from PRKDC and exon 7 from APP, respectively. Expression values from probes were used to calculate the PRKDC and APP fold changes and relative abundance reported in Tables 3 and 4. Numbers in the boxes refer to the actual exon identification. (B) RT–PCR analysis of PRKDC in breast cell lines. The wild-type sequence (wt) and the short form (−80) were amplified with sets of primers located in sequences adjacent to exons 79 and 81. (D) RT–PCR analysis of APP showing the relative amount of each isoform in melanoma and brain tissues. C+ is a positive control made with equimolar amount of cloned isoforms, amplified with the same set of primers.
In another set of experiments, cRNAs from melanoma and brain tissues were co-hybridized in dye-swap assays. We observed a significant deregulation for three isoforms (fold changes >1.80 or <0.55) (Table 4). Among those, APP is a well-described alternatively spliced gene (42,45). The set of APP spotted probes was designed to monitor the splice event corresponding to the double skipping of exons 7 and 8 (APP [−7/8] or APP SF). It turns out that another isoform corresponding to the skipping of exon 8 is also expressed in the melanoma and brain tissues (APP [−8]) (Figure 4C). The APP probes enabled us to calculate the relative abundance of APP SF with respect to the two longer isoforms (APP wt and APP [−8]). The exon 6 probe is located in an exon common to the three isoforms (wt, [-8] and [-7/8]). Exon 7 is skipped in APP SF and the corresponding specific probe (E7) allows the quantification of the two longer isoforms (wt and [−8]). Probe Jct 6-9 is specific for APP SF.
Table 4.
Fold changes and relative abundance for deregulated isoforms in brain versus melanoma
| Gene | Isoform | Fold changes | Relative abundance | |||||
|---|---|---|---|---|---|---|---|---|
| Brain/Melanoma | Melanoma | Brain | ||||||
| LF+SF (RE1) | LF (RE2) | SF (RJ) | Reference | Experimental | ||||
| % LF | % SF | % LF | % SF | |||||
| PPIB | Wild type (SF) | 0.38 | 1.17 | 0.36 | 2.5 | 97.5 | 7.6 | 92.4 |
| Intron 4 retention (LF) | ||||||||
| APPa | Wild type + APP [-8] (LF) | 1.39 | 0.94 | 3.81 | 84.3 | 15.7 | 57.0 | 43.0 |
| Exon7- 8 skipping (APP [-7/8] (SF) | ||||||||
| MGC8721 | Wild type (LF) | 3.41 | 3.18 | 1.71 | NA | NA | NA | NA |
| Exon 2 skipping (SF) | ||||||||
Significant deregulations are reported in bold. RE2 values correspond to the mean of fold changes calculated with each LF-specific probes (Ex2, J1-2 and J2-3). NA, not applicable (see in text the limitations for the relative abundance determination).
aSee description of APP in Figure 4C.
In Table 4, a clear difference in the relative abundance of APP SF was observed between melanoma and brain samples. This is in agreement with the validation by RT–PCR (Figure 4D) which showed a preponderance of the two longer isoforms (APP wt and APP [−8]) in melanoma (84% versus 16% from the splice-array data) while the APP [−8] and APP [−7/8] isoforms were roughly in equal amounts in brain (57% versus 43% from the splice-array data). Our results for APP are in agreement with a recent study using another junction array platform, although no quantitative measurements were reported (42).
It should also be noted that results from the E6 probe located in the common exon (exon 6) demonstrated no significant deregulation of the APP messengers between brain and melanoma (fold change = 1.39). Hence, by using single probes (or probe sets) located in constitutive regions, one would have concluded to the absence of gene deregulation, while isoform-specific probes were able to detect and quantify a specific deregulation of the splice variants.
DISCUSSION
Microarray platforms constitute the standard tools to assess transcripts expression levels on a large scale. Alternative RNA splicing has been addressed in some reports but none displayed the type of data required to validate the quantitative nature of such platforms. This was the purpose of our study.
A first study described the expression of novel or cassette exons via the design of exon specific probes (46). Later, the same group described the concept of tiling arrays in which oligonucleotide probes are sequentially generated every 10 bp along a predefined genomic region. This allowed a more precise definition of exon/intron boundaries by searching for consensus splice sites within 20–30 bp sequences. Tiling arrays at different resolutions have also been described to monitor transcriptional activity in chromosomes 21 and 22 (9,47). Hu et al. (39) studied alternative splicing in the 3′ end regions of 1600 rat genes by designing 20 different probes per gene. Hybridizations performed with 10 normal tissues predicted that 268 (17%) genes could be regulated via alternative splicing. In another study, a global analysis of constitutive RNA splicing in yeast has been performed via the design of intron and exon/exon-junction-specific probes (40). This study introduced the usage of exon junction spanning probes interrogating sequences that are juxtaposed at the RNA level but not at the genome level. The optimization and use of junction probes to monitor exon skipping events were further validated by Castle et al. (43) on the study of two genes (RB1 and ANXA7) and by Wang et al. (41) on another set of two genes (CD44 and TPM2). Finally, an extensive analysis of exon skipping events has been carried out by Johnson et al. (42) using a microarray containing exon/exon junction probes for over 10 000 human genes that has been hybridized against 52 different biological samples. A prediction model was applied to qualitatively detect differential splicing patterns within the tested samples.
Our study describes a specific configuration for monitoring splice variant expression enabling the quantification of the absolute and relative splice variant expression levels when comparing two samples. This configuration requires a set of junction and exon probes to characterize a splice event with accuracy and sensitivity at the picomolar level. Alternate transcripts can also be generated by mechanisms not related to alternative splicing such as promoter usage and alternative polyadenylation. Such events affecting the 5′ and 3′ extremities will be monitored by fewer probes as junction probes can not be designed at these endings.
Probe design was optimized for sensitivity and specificity to include homogenous predicted melting temperature, minimal self-hybridization and minimal hairpin tendencies. In addition, we chose to use shorter oligonucleotides rather than 40mer or above to avoid the potential hybridization of half of the junction probe to a single exon. The design of exon probes rarely presents difficulties except when the exon size is limited. As with conventional microarrays, care should be taken to avoid cross-hybridization in particular with close homologs via searching for sequence similarities with mRNAs and ESTs populations. The location of the junction probes is restricted to a specific region centered on the splice junction. This positional constraint may complicate probe design, making probe composition not suitable to get the desired thermodynamic parameters. However, junction probes can be designed with a sequence up to two nucleotides off-centre, which maintained the expected specificity (data not shown). In addition, shorter probes could also be designed for problematic sequences such as GC-rich regions. It should also be noted that, in the case of the detection and quantification of the long isoform, up to three probes are designed (Ex2, J1-2 and J2-3) which allows some flexibility for the choice of the probe for quantification.
Junction probes are expected to bring the expected specificity in the case of closely related splice events For instance, it has been recently reported that NAG insertions-deletions in transcripts can occur due to splice acceptor sites displaying a NAGNAG motif (48). Such three nucleotides insertion will create a significant mismatch between the probe and the target that should guarantee the desired specificity of the junction probes.
This microarray configuration was initially validated by performing spiking experiments where known amounts of long and short recombinant RNA isoforms were co-hybridized at various concentrations. Calculated fold changes were found consistent with the expected values within ratios ranging from 0.25 to 12. Additionally, we were able to quantify relative amounts in each tested sample using simple algebra. We further demonstrated that we could quantify specific isoform expression deregulations within complex RNAs derived from breast cell lines, brain and melanoma tissue samples that were next confirmed by RT–PCR. Importantly, we had decided to evaluate the performances of the splice-array configuration by selecting the difficult but frequent setting in which isoforms generated via exon skipping are expressed at lower levels concomitantly to the wild-type isoform (cf. PRKDC). Instances in which alternative cassette exons are exclusively expressed in given tissues are indeed easy to quantify and could only rely on cassette exonic probes.
This microarray platform provides various levels of valuable information: at a very basic level, probes located in common exons will provide expression values and deregulations for given genes as observed with a regular microarray system. The addition of junction probes and probes specific for alternative events gives access to isoform deregulation between two zsamples. Finally, when two isoforms with a significant deregulation are present in the samples to be compared, we have developed a calculation method that will allow quantification of the relative ratios between the isoforms in each sample.
We are facing the same limitations associated with any microarray platform. Thus, probes with expression values below the sensitivity of the platform will not be appropriate. In addition, determination of the isoform relative abundance in each sample is dependent of the fold change values calculated for RE1, RE2 and RJ. Inaccurate relative abundance determination can occur when: (i) RE1 ≈ RE2 (the LF proportion in each sample is close to 100%), (ii) RE1 ≈ RJ (the SF proportion in each sample is close to 100%) and (iii) RE1 ≈ RE2 ≈ RJ (the SF to LF ratios are similar in each sample). Additionally, when RE1, RE2 and RJ fall within 0.6 and 1.8, we consider no significant isoform deregulation between the samples and calculation of the relative isoform abundance may be biased.
In addition, like any microarray, we detect and quantify targets that are complementary to probes displayed on the array, i.e. splice events and not full-length spliced transcripts. Any splicing event can be present in more than one isoform and one isoform can include more than one splice event. This was exemplified in the APP isoforms that were studied in the brain and melanoma samples.
Today's commercial microarray platforms proposed for global human genome expression are inappropriate to systematically detect splice variants. Probe sets are often 3′ biased and are not designed according to the types of configuration described in this study. In addition, data analysis is performed in such a way that multiple measurements are used to define a consensual behavior amongst the probes, remove outliers and produce a unique figure associated with the expression level of the gene in a given sample. The APP isoforms deregulations described in this study would have been missed by such platforms.
The new type of microarrays described here will certainly provide the necessary tools to obtain a more comprehensive and accurate picture of gene expression, help decipher the regulation networks of alternative RNA splicing and open the way to innovative and better profiled therapeutics. We are currently evaluating this microarray configuration for the detection and the quantification of splice events from the GPCR gene family.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Richard Einstein, Dr Jonathan Kearsey and Dr Aram Mangasarian for critical reading of the manuscript. They also wish to acknowledge Jerome Fouse for technical assistance. Funding to pay the Open Access publication charges for this article was provided by Exonhit Therapeutics.
REFERENCES
- 1.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Lander E.S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 5.Pospisil H., Herrmann A., Bortfeldt R.H., Reich J.G. EASED: Extended Alternatively Spliced EST Database. Nucleic Acids Res. 2004;32:D70–D74. doi: 10.1093/nar/gkh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thanaraj T.A., Stamm S., Clark F., Riethoven J.J., Le Texier V., Muilu J. ASD: the Alternative Splicing Database. Nucleic Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y.H., Chen Y.T., Lai J.J., Yang S.T., Yang U.C. PALS db: Putative Alternative Splicing database. Nucleic Acids Res. 2002;30:186–190. doi: 10.1093/nar/30.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn J.L., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., et al. The transcriptional activity of human Chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saha S., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E. Using the transcriptome to annotate the genome. Nat. Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- 11.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 13.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 14.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 15.Ladd A.N., Cooper T.A. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-reviews0008. reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 17.Stamm S. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet. 2002;11:2409–2416. doi: 10.1093/hmg/11.20.2409. [DOI] [PubMed] [Google Scholar]
- 18.Migliaccio E., Mele S., Salcini A.E., Pelicci G., Lai K.M., Superti-Furga G., Pawson T., Di Fiore P.P., Lanfrancone L., Pelicci P.G. Opposite effects of the p52shc/p46shc and p66shc splicing isoforms on the EGF receptor-MAP kinase-fos signalling pathway. EMBO J. 1997;16:706–716. doi: 10.1093/emboj/16.4.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson C.A., Payton M.N., Elliott G.S., Buaas F.W., Cajulis E.E., Grosshans D., Ramos L., Reese D.M., Slamon D.J., Calzone F.J. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene. 1997;14:1–16. doi: 10.1038/sj.onc.1200924. [DOI] [PubMed] [Google Scholar]
- 20.Wang J., Gao Q.S., Wang Y., Lafyatis R., Stamm S., Andreadis A. Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. J. Neurochem. 2004;88:1078–1090. doi: 10.1046/j.1471-4159.2003.02232.x. [DOI] [PubMed] [Google Scholar]
- 21.D'Souza I., Schellenberg G.D. Determinants of 4-repeat tau expression. Coordination between enhancing and inhibitory splicing sequences for exon 10 inclusion. J. Biol. Chem. 2000;275:17700–17709. doi: 10.1074/jbc.M909470199. [DOI] [PubMed] [Google Scholar]
- 22.Umeda Y., Taniguchi S., Arima K., Piao Y.S., Takahashi H., Iwatsubo T., Mann D., Hasegawa M. Alterations in human tau transcripts correlate with those of neurofilament in sporadic tauopathies. Neurosci. Lett. 2004;359:151–154. doi: 10.1016/j.neulet.2004.01.060. [DOI] [PubMed] [Google Scholar]
- 23.de Silva R., Lashley T., Gibb G., Hanger D., Hope A., Reid A., Bandopadhyay R., Utton M., Strand C., Jowett T., et al. Pathological inclusion bodies in tauopathies contain distinct complements of tau with three or four microtubule-binding repeat domains as demonstrated by new specific monoclonal antibodies. Neuropathol. Appl. Neurobiol. 2003;29:288–302. doi: 10.1046/j.1365-2990.2003.00463.x. [DOI] [PubMed] [Google Scholar]
- 24.Kalbfuss B., Mabon S.A., Misteli T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J. Biol. Chem. 2001;276:42986–42993. doi: 10.1074/jbc.M105113200. [DOI] [PubMed] [Google Scholar]
- 25.Feldkotter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time light Cycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada Y., Sutomo R., Sadewa A.H., Akutsu T., Takeshima Y., Wada H., Matsuo M., Nishio H. Correlation between SMN2 copy number and clinical phenotype of spinal muscular atrophy: three SMN2 copies fail to rescue some patients from the disease severity. J. Neurol. 2002;249:1211–1219. doi: 10.1007/s00415-002-0811-4. [DOI] [PubMed] [Google Scholar]
- 27.Cartegni L., Krainer A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nature Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- 28.Sumner C.J., Huynh T.N., Markowitz J.A., Perhac J.S., Hill B., Coovert D.D., Schussler K., Chen X., Jarecki J., Burghes A.H., et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 29.Skordis L.A., Dunckley M.G., Yue B., Eperon I.C., Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl Acad. Sci. USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brichta L., Hofmann Y., Hahnen E., Siebzehnrubl F.A., Raschke H., Blumcke I., Eyupoglu I.Y., Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 31.Wirth B. Spinal muscular atrophy: state-of-the-art and therapeutic perspectives. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2002;3:87–95. doi: 10.1080/146608202760196057. [DOI] [PubMed] [Google Scholar]
- 32.Darreh-Shori T., Hellstrom-Lindahl E., Flores-Flores C., Guan Z.Z., Soreq H., Nordberg A. Long-lasting acetylcholinesterase splice variations in anticholinesterase-treated Alzheimer's disease patients. J. Neurochem. 2004;88:1102–1113. doi: 10.1046/j.1471-4159.2003.02230.x. [DOI] [PubMed] [Google Scholar]
- 33.Vandenbroucke I.I., Vandesompele J., Paepe A.D., Messiaen L. Quantification of splice variants using real-time PCR. Nucleic Acids Res. 2001;29:E68. doi: 10.1093/nar/29.13.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez C., Vandesompele J., Vandenbroucke I., Holtappels G., Speleman F., Gevaert P., Van Cauwenberge P., Bachert C. Quantitative real time polymerase chain reaction for measurement of human interleukin-5 receptor alpha spliced isoforms mRNA. BMC Biotechnol. 2003;3:17. doi: 10.1186/1472-6750-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veistinen E., Liippo J., Lassila O. Quantification of human Aiolos splice variants by real-time PCR. J. Immunol. Methods. 2002;271:113–123. doi: 10.1016/s0022-1759(02)00370-8. [DOI] [PubMed] [Google Scholar]
- 36.Shulzhenko N., Smirnova A.S., Morgun A., Gerbase-DeLima M. Specificity of alternative splice form detection using RT–PCR with a primer spanning the exon junction. Biotechniques. 2003;34:1244–1249. doi: 10.2144/03346rr02. [DOI] [PubMed] [Google Scholar]
- 37.Wagner E.J., Curtis M.L., Robson N.D., Baraniak A.P., Eis P.S., Garcia-Blanco M.A. Quantification of alternatively spliced FGFR2 RNAs using the RNA invasive cleavage assay. RNA. 2003;9:1552–1561. doi: 10.1261/rna.5840803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeakley J.M., Fan J.B., Doucet D., Luo L., Wickham E., Ye Z., Chee M.S., Fu X.D. Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol. 2002;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- 39.Hu G.K., Madore S.J., Moldover B., Jatkoe T., Balaban D., Thomas J., Wang Y. Predicting splice variant from DNA chip expression data. Genome Res. 2001;11:1237–1245. doi: 10.1101/gr.165501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark T.A., Sugnet C.W., Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 41.Wang H., Hubbell E., Hu J.S., Mei G., Cline M., Lu G., Clark T., Siani-Rose M.A., Ares M., Kulp D.C., et al. Gene structure-based splice variant deconvolution using a microarry platform. Bioinformatics. 2003;19(Suppl 1):I315–I322. doi: 10.1093/bioinformatics/btg1044. [DOI] [PubMed] [Google Scholar]
- 42.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 43.Castle J., Garrett-Engele P., Armour C.D., Duenwald S.J., Loerch P.M., Meyer M.R., Schadt E.E., Stoughton R., Parrish M.L., Shoemaker D.D., et al. Optimization of oligonucleotide arrays and RNA amplification protocols for analysis of transcript structure and alternative splicing. Genome Biol. 2003;4:19. doi: 10.1186/gb-2003-4-10-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connelly M.A., Zhang H., Kieleczawa J., Anderson C.W. Alternate splice-site utilization in the gene for the catalytic subunit of the DNA-activated protein kinase, DNA-PKcs. Gene. 1996;175:271–273. doi: 10.1016/0378-1119(96)00135-7. [DOI] [PubMed] [Google Scholar]
- 45.Bergsdorf C., Paliga K., Kreger S., Masters C.L., Beyreuther K. Identification of cis-elements regulating exon 15 splicing of the amyloid precursor protein pre-mRNA. J. Biol. Chem. 2000;275:2046–2056. doi: 10.1074/jbc.275.3.2046. [DOI] [PubMed] [Google Scholar]
- 46.Shoemaker D.D., Schadt E.E., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. doi: 10.1038/35057141. [DOI] [PubMed] [Google Scholar]
- 47.Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 48.Hiller M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nature Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.