Abstract
Unlike other lactic acid bacteria, Lactococcus lactis subsp. lactis NCDO 2118 was able to grow in a medium lacking glutamate and the amino acids of the glutamate family. Growth in such a medium proceeded after a lag phase of about 2 days and with a reduced growth rate (0.11 h−1) compared to that in the reference medium containing glutamate (0.16 h−1). The enzymatic studies showed that a phosphoenolpyruvate carboxylase activity was present, while the malic enzyme and the enzymes of the glyoxylic shunt were not detected. As in most anaerobic bacteria, no α-ketoglutarate dehydrogenase activity could be detected, and the citric acid cycle was restricted to a reductive pathway leading to succinate formation and an oxidative branch enabling the synthesis of α-ketoglutarate. The metabolic bottleneck responsible for the limited growth rate was located in this latter pathway. As regards the synthesis of glutamate from α-ketoglutarate, no glutamate dehydrogenase was detected. While the glutamate synthase-glutamine synthetase system was detected at a low level, high transaminase activity was measured. The conversion of α-ketoglutarate to glutamate by the transaminase, the reverse of the normal physiological direction, operated with different amino acids as nitrogen donor. All of the enzymes assayed were shown to be constitutive.
Lactic acid bacteria (LAB) are characterized by their numerous nutritional requirements, and in particular their incapacity to grow at the expense of mineral nitrogen in the absence of exogenous amino acids. Because of this requirement for organic nitrogen substrates, LAB are frequently grown on complex (MRS or M17) media. Despite the importance of LAB in the food industry, relatively few studies have been devoted to the complete determination of the nutritional requirements of LAB. The single-omission technique has been used to identify the amino acid requirements of Lactococcus (5, 20), Lactobacillus (14, 16, 17), and Enterococcus and Pediococcus strains (7). Certain nutritional requirements appear to be strain dependent, while the requirements for certain amino acids are more widespread, e.g., those for glutamic acid, histidine, and branched-chain amino acids. Ledesma et al. (14) proposed the requirement for glutamic acid, valine, and leucine as a taxonomic criterion for the Lactobacillus genus. The most complete studies on amino acid requirements in LAB were presented by Morishita and colleagues, who reported the systematic isolation and characterization of mutants that had lost their requirements for specific amino acids, with four Lactobacillus strains (16, 17) and later with one Enterococcus faecium strain, one Pediococcus acidilactici strain, and Lactococcus lactis ATCC 19435 (7). Such mutants were obtained for each of the species and for the majority of amino acids tested, indicating that the capacity to synthesize these amino acids was acquired by reversion of simple point mutations. However, for glutamate, such mutants were not obtained for any of the species studied, suggesting that the glutamate biosynthetic pathway is extensively impaired and that the citric acid cycle is probably inactive in all species of LAB.
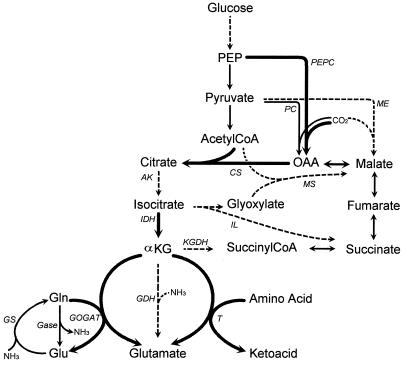
More recently, Morishita and Yajima (18) demonstrated that certain enzymes of the oxidative branch of the tricarboxylic acid (TCA) cycle leading to α-ketoglutarate are present in lactobacilli. These authors demonstrated that citrate synthase and aconitase activities were present in all strains investigated, while both isocitrate dehydrogenase and α-ketoglutarate dehydrogenase could not be detected in any of the strains. The lack of isocitrate dehydrogenase activity was used to explain the requirement for glutamate in lactobacilli. However, growth could be restored by addition of α-ketoglutarate, and glutamate dehydrogenase activity was detected in some of the strains tested. No attempts were made to detect other enzymes potentially involved in glutamate synthesis from α-ketoglutarate, despite the frequent reports that both glutamate synthase (GOGAT) (3, 6, 12, 23) and transaminases (10, 22) are known to catalyze this reaction in other microorganisms (Fig. 1).
FIG. 1.
Metabolic pathways potentially involved in the synthesis of glutamic acid from the glycolytic intermediates. Thin arrows, enzymes not assayed in this study; thick arrows, enzymes detected in L. lactis NCDO 2118; dashed arrows, enzymes not detected in L. lactis NCDO 2118; 1, enzyme not detected but considered to be present. Abbreviations: OAA, oxaloacetic acid; αKG, α-ketoglutaric acid; Glu, glutamic acid; Gln, glutamine; PEPC, PEP carboxylase; PC, pyruvate carboxylase; ME, malic enzyme; CS, citrate synthase; AK, aconitase; IDH, isocitrate dehydrogenase; KGDH, α-ketoglutarate dehydrogenase; IL, isocitrate lyase; MS, malate synthase; GDH, glutamate dehydrogenase; T, transaminase (or aminotransferase); GS, glutamine synthetase; Gase, glutaminase.
Since L. lactis NCDO 2118 grows in synthetic media lacking glutamic acid and amino acids of the glutamate family (5), some capacity to synthesize glutamate from metabolites of central metabolism must exist. In light of these preliminary indications of TCA cycle activity leading to glutamate synthesis, a more comprehensive study of the enzymes and carbon flux enabling growth of L. lactis in the absence of glutamate was undertaken.
MATERIALS AND METHODS
Organism and culture media.
Lactococcus lactis subsp. lactis NCDO 2118, obtained from the collection held at the Institut National de la Recherche Agronomique (Jouy-en-Josas, France), was used throughout this study. The medium used for the growth of the inoculum was the synthetic MS10 medium described by Cocaign-Bousquet et al. (5). The medium used for growth in tube cultures was the MS14 medium (19), with the following composition: glucose, 10 g/liter; KH2PO4, 9 g/liter; K2HPO4, 7.5 g/liter; MgCl2·6H2O, 0.2 g/liter; biotin, 10 mg/liter;nicotinic acid, 1 mg/liter; Ca-pantothenate, 1 mg/liter; riboflavin, 1 mg/liter; pyridoxamine, 5 mg/liter; glutamic acid, 0.3 g/liter; isoleucine, 0.16 g/liter; leucine, 0.33 g/liter; methionine, 0.06 g/liter; serine, 1.04 g/liter; and valine, 0.16 g/liter. The MS14 medium used for growth of cultures in a fermentor contained glucose (20 g/liter), KH2PO4 (3 g/liter), K2HPO4 (2.5 g/liter), (NH4)2SO4 (0.6 g/liter), and twofold-higher concentrations of each amino acid, while other components were not modified. Addition of other components or removal of glutamate from MS14 medium was performed as stated elsewhere in the text. These media were prepared from concentrated stock solutions stored at 4°C after filtration through cellulose nitrate membranes (0.22-μm pore size). The media (pH 6.6) were sterilized by filtration through cellulose nitrate membranes (0.22-μm pore size; Sartorius) directly into the sterilized (20 min at 121°C) culture vessel.
Culture conditions.
Cultures were grown in butyl-rubber-stoppered tubes or in a 2-liter fermentor (Sétric Génie Industriel, Toulouse, France), at a temperature of 30°C and an agitation speed of 250 rpm. The bacteria were grown under a controlled gas environment by flushing both the tubes and the medium with nitrogen. The medium used in the fermentor was aseptically gassed immediately before inoculation and maintained throughout under an N2 atmosphere at a positive pressure of 1 kPa. Cultures in the fermentor were maintained at pH 6.6 by automatic addition of 5 N KOH.
Inoculation (2% [vol/vol]) was with late-exponential-phase cells from cultures grown on MS10 medium, which were washed twice with sterile phosphate buffer (100 mM, pH 6.6) and resuspended in this same buffer to avoid carryover of amino acids. Tubes were directly inoculated with these cells. All growth experiments in tubes were performed in triplicate. For cultures grown in the fermentor with various modified MS14 media, prolonged lag phases were avoided by serial transfer of exponentially growing cells through two transfers in a butyl-stoppered shaken flask containing the modified MS14 medium.
Analytical methods.
Bacterial growth was monitored by spectrophotometric measurements at 580 nm and calibrated against cell dry weight measurements. Cells were harvested by filtration on 0.45-μm-pore-size nylon membranes, washed with 2 volumes of deionized water, and dried to a constant weight at 60°C under partial vacuum (200 mm Hg [26.7 kPa]). A change of 1 U of optical density was shown to be equivalent to 0.31 g of dry matter per liter. The biomass formula used to convert cell dry weights into molar cell carbon concentrations was determined to be C5.05H9.20O2.77N1.0 by elemental analysis at Ecole Nationale Supérieure de Chimie (Toulouse, France), with a molar mass of 140 g mol−1.
Concentrations of glucose and products (lactate, acetate, formate, ethanol, and succinate) in the fermentation broth were measured with high-pressure liquid chromatography with a Bio-Rad HPX87H+ column and the following conditions: a temperature of 48°C, solvent H2SO4 (5 mM), a flow rate of 0.5 ml min−1, and double detection in series (refractometer and UV).
Concentrations of amino acids in the medium or in the transaminase assays were determined with an AminoQuant 1090 high-pressure liquid chromatography (Hewlett-Packard) after derivatization by orthophthalaldehyde in the presence of 3-mercaptopropionic acid, separation with a C18 column, and spectrophotometric detection at 338 nm. Ammonia concentrations were determined in filtered culture samples with an ammonia-selective electrode (Orion).
The α-ketoglutaric acid concentration in the supernatant was determined by enzymatic analysis with glutamate dehydrogenase, as described by Bergmeyer and Bernt (2).
Crude cell extract preparation.
A volume of culture corresponding to 90 mg (dry weight) of cells was centrifuged (4°C, 10 min at 10,000 × g) and washed twice with KCl (0.2%). For the determination of transaminase activity, the cell pellet was resuspended in 5 ml of buffer of the following composition: phosphate buffer (400 mM, pH 7.2), 30 ml; glycerol, 10 ml; MgCl2 (50 mM), 4 ml; and dithiothreitol (300 mM), 150 μl. For the other enzymes, the phosphate buffer was replaced by a Tris (45 mM)-carballylate (15 mM) buffer (pH 7.2). The cell suspension was rapidly frozen and stored at −20°C until extraction. No detrimental effect on enzyme activity was observed for frozen cells over a period of at least 15 days compared with freshly harvested cells.
Cell extracts were prepared by disrupting the bacteria by sonication. The resulting crude extract was centrifuged (4°C, 15 min at 10,000 × g) to remove cell debris, and the supernatant was used for enzyme assays. The protein content of the extracts was determined by the method of Lowry et al. (15) with bovine serum albumin as the protein standard.
Determination of enzyme activities.
All assays were carried out at 30°C in 2-ml cuvettes containing 1 ml of the appropriate enzyme mixture at pH 7.2. The volume of extract was chosen to ensure linearity between activity and protein concentration and varied from 20 to 200 μl. Enzymes were assayed by coupling appropriate enzyme reactions to the spectrophotometric determination of NAD(P)(H) at 340 nm, except for citrate synthase, aconitase, and transaminase.
Phosphoenolpyruvate (PEP) carboxylase was assayed with Tris-HCl buffer (pH 7.2, 100 mM), MnSO4 (5 mM), PEP (5 mM), NADH (0.6 mM), malate dehydrogenase (5 U/ml), and extract, and the reaction was started with KHCO3 (10 mM).
Malic enzyme catalyzing the carboxylation of pyruvate in malate was measured with phosphate buffer (pH 7.2, 100 mM), MgCl2 (5 mM), NaHCO3 (10 mM), NADPH (0.3 mM), pyruvate (20 mM), and extract to initiate the reaction.
Isocitrate lyase was assayed at 324 nm with Tris-HCl buffer (pH 7.2, 100 mM), EDTA (0.45 mM), MgCl2 (5 mM), phenylhydrazine (0.58 g/liter), and extract, and the reaction was started with isocitrate (1 mM).
Malate synthase was assayed at 232 nm with Tris-HCl buffer (pH 7.2, 100 mM), glyoxylate (1 mM), and extract, and the reaction was started with acetyl coenzyme A (acetyl-CoA) (0.1 mM).
Citrate synthase was measured with Tris buffer (100 mM), acetyl-CoA (0.1 mM), MgCl2 (5 mM), dithionitrobenzoate (0.5 mM), and extract, and the reaction was started with oxaloacetate (0.15 mM). The CoA liberated by the citrate synthase reacted with dithionitrobenzoate to form a product measured spectrophotometrically at 412 nm.
Aconitase was assayed at 240 nm with Tris buffer (100 mM), NaCl (100 mM), cis-aconitate (2 mM), and extract to initiate the reaction.
Isocitrate dehydrogenase was measured with Tris (100 mM), NAD or NADP (0.6 mM), MnSO4 (5 mM), and extract, and the reaction was started with isocitrate (10 mM).
α-Ketoglutarate dehydrogenase was measured with Tris (100 mM), MgCl2 or MnSO4 (5 mM), CoA (0.2 mM), thiamine pyrophosphate (0.3 mM), dithiothreitol (5 mM), NADP (0.6 mM), extract, and α-ketoglutarate (5 mM) to initiate the reaction.
Glutamate dehydrogenase was measured with Tris (100 mM), NH4Cl (20 mM), NAD(P)H (0.6 mM), extract, and α-ketoglutarate (10 mM) to initiate the reaction.
GOGAT was assayed with Tris (100 mM), α-ketoglutarate (10 mM), NAD(P)H (0.6 mM), MgCl2 or MnSO4 (5 mM), and extract, and the reaction was started with glutamine (10 mM).
Transaminase catalyzing the transfer of an ammonium group from an amino acid to a ketoacid was assayed in the direction of glutamate synthesis, by measuring glutamate production and donor amino acid consumption with an AminoQuant analysis. The reaction mixture containing phosphate buffer (pH 7.2, 20 mM), pyridoxalphosphate (0.05 mM), amino acid (3 mM), and the extract was incubated for 15 min at 30°C. The reaction was started by addition of α-ketoglutarate (pH 7, 10 mM). Samples collected just before the α-ketoglutarate addition (time zero) and after 2, 5, and 10 min of incubation were immediately supplemented with sulfosalicylic acid (3%) to stop the reaction and chilled in ice. The samples were then centrifuged (4°C, 10 min at 10,000 × g) to remove precipitation and stored at −20°C. Concentrations of amino acids were determined by AminoQuant analysis after dilution in methanol and centrifugation of the samples. The donor amino acids tested for the transaminase reaction were isoleucine, leucine, valine, serine, and methionine (all included in the MS14 medium); alanine; and aspartate.
All the biochemical reagents were obtained from Sigma-Aldrich (San Quentin Fallavier, France).
RESULTS
Lag phase and growth rate.
The growth response of L. lactis NCDO 2118 was examined in culture tubes containing the minimal medium (MS14) in which glutamic acid was replaced by other amino acids, inorganic nitrogen, or citric acid cycle intermediates and compared with growth in minimal medium. The growth in MS14 medium proceeded without lag phase at a maximal growth rate of 0.16 h−1 (Table 1). When glutamic acid was replaced by glutamine, the growth behavior was identical to that in the reference medium. The removal of glutamic acid from the MS14 medium led to a reduced growth rate and a prolonged lag phase. This very long lag phase was observed only during the first transfer from the MS14 medium to the medium lacking glutamate. The growth in the second transfer in the same medium began immediately after inoculation. When the strain was cultivated again in MS14 medium and transferred to the medium lacking glutamate, a similar lag phase was observed before growth began, indicating that this long lag phase was not attributable to the selection of a variant population.
TABLE 1.
Lag phase and maximal specific growth rate (μmax) observed during growth of L. lactis subsp. lactis NCDO 2118 in MS14 medium of modified compositiona
| Medium | Lag phase (h) | μmax (h−1) |
|---|---|---|
| MS14 | 0 | 0.16 ± 0.01 |
| MS14 − Glu + Gln | 0 | 0.16 ± 0.01 |
| MS14 − Glu | 40–48b | 0.11 ± 0.01 |
| MS14 − Glu + NH3 | 40–48b | 0.11 ± 0.01 |
| MS14 − Glu + α-KGd | —c | 0.15 ± 0.01 |
| MS14 − Glu + α-KGd + NH3 | —c | 0.15 ± 0.01 |
| MS14 − Glu + K2CO3 | 40–48b | 0.10 ± 0.01 |
| MS14 − Glu + citrate | 40–48b | 0.10 ± 0.01 |
| MS14 − Glu + isocitrate | 40–48b | 0.11 ± 0.01 |
| MS14 − Glu + Ala | 40–48b | 0.10 ± 0.01 |
| MS14 − Glu + Asp | 40–48b | 0.11 ± 0.01 |
The values represent the means of about 10 measurements for each condition.
Lag phase comprised between 40 and 48 h.
Variable with the α-ketoglutarate concentration (see Fig. 2).
α-KG, α-ketoglutaric acid.
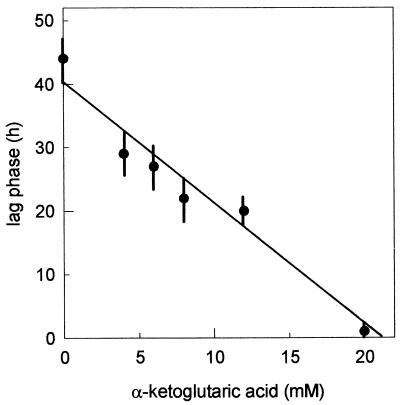
Addition of α-ketoglutarate to the MS14 medium lacking glutamate restored the growth rate to the reference value (Table 1). Moreover, the duration of the lag phase was a function of the concentration of α-ketoglutarate added to the medium (Fig. 2). No lag phase was observed in the presence of 20 mM α-ketoglutarate. Inorganic nitrogen had no effect on either growth rate or lag phase when added to MS14 medium lacking glutamate, with or without addition of α-ketoglutarate. Finally, addition of carbonate, or intermediates of the citric acid cycle, citric or isocitric acids, or alanine or aspartic acid, to the MS14 medium lacking glutamate had no effect on growth of the strain.
FIG. 2.
Effect of the α-ketoglutaric acid concentration on the lag phase observed during culture of L. lactis NCDO 2118 in MS14 medium lacking glutamic acid after a transfer from MS14 medium.
Growth and metabolic behavior.
The growth of L. lactis NCDO 2118 in the MS14 medium proceeded with the following stoichiometry (in millimolar concentrations): 100 glucose + 2.5 Glu + 1.5 Val + 3.3 Leu + 1.8 Ile + 0.6 Met + 18.7 Ser→9.4 biomass + 14.3 NH3 + 178 lactate + 20.3 formate + 13.7 acetate + 3.2 succinate + 0.4 α-ketoglutarate. Though the fermentation was homolactic whichever the medium used, formate and acetate were formed as classical minor products while ethanol was never produced, due to reducing equivalent equilibrium associated with the high conversion of serine to lactate, as previously shown (19). The stoichiometry was similar on the various media tested, except for the following points: (i) glutamate, α-ketoglutarate, or glutamine was consumed when present in the medium; (ii) glutamate was produced when α-ketoglutarate or glutamine was present; (iii) succinate was produced in a twofold-higher concentration when α-ketoglutarate was present in the medium; (iv) ammonia was produced in a higher amount when glutamine was consumed; and (v) α-ketoglutarate was produced even in the MS14 medium lacking glutamate.
Activities of anaplerotic enzymes and of enzymes catalyzing oxidative reactions of the citric acid cycle.
To examine the biosynthetic pathway leading from glycolytic intermediates to α-ketoglutarate, the activities of anaplerotic enzymes, of enzymes of the glyoxylic shunt, and of the oxidative sequence of the citric acid cycle were measured with cell extracts prepared from cells of L. lactis NCDO 2118 grown in the media used in the fermentor, i.e., MS14, MS14 without glutamate, and the latter medium supplemented with either glutamine or α-ketoglutarate.
Medium composition had no effect on the measured activity of any of the enzymes tested (shown in Fig. 1), and average values obtained from measurements performed with cells growing in these four media are presented in Table 2. A small PEP carboxylase activity, which catalyzes the carboxylation of PEP to oxaloacetate, was demonstrated. On the other hand, neither malic enzyme, catalyzing the carboxylation of pyruvate in malate, nor the glyoxylate shunt enzymes isocitrate lyase and malate synthase, mediating the synthesis of one succinate and one malate from isocitrate and acetyl-CoA (Fig. 1), were detected. Citrate synthase activity was demonstrated, as was isocitrate dehydrogenase activity when NAD was used as cofactor, while the latter enzyme had an eightfold-lower activity with NADP as cofactor. Surprisingly, aconitase activity was not detected whatever the direction used for the measurement. No α-ketoglutarate dehydrogenase activity was detected.
TABLE 2.
Activities of anaplerotic enzymes, enzymes of the glyoxylic shunt, and enzymes mediating oxidative reactions of the citric acid cycle in extracts of L. lactis subsp. lactis NCDO 2118
| Enzyme | Sp acta |
|---|---|
| Anaplerotic and glyoxylic shunt enzymes | |
| PEP carboxylaseb | 3 |
| Malic enzymec | ND |
| Isocitrate lyaseb | ND |
| Malate synthaseb | ND |
| Citric acid cycle enzymes | |
| Citrate synthaseb | 22 |
| Aconitasec | ND |
| NAD-dependent isocitrate dehydrogenaseb | 23 |
| NADP-dependent isocitrate dehydrogenaseb | 3 |
| α-Ketoglutarate dehydrogenaseb | ND |
Specific activity was expressed as nanomoles per minute per milligram of protein. The values were average values obtained during measurements with cell extracts from the four synthetic media used in fermentor cultures. ND, not detected.
Activity was assayed in the physiological direction.
Activity was assayed in both directions.
Activities of enzymes mediating the synthesis of glutamate from α-ketoglutarate.
To gain further understanding of the metabolic pathway mediating glutamate synthesis, the activities of putative enzymes converting α-ketoglutarate to glutamate were studied (Table 3). As previously noted for the other enzymes assayed (see above), no differences were observed in specific activity on the four media tested. No glutamate dehydrogenase activity was found in any of the nutritional conditions. A low activity of GOGAT was demonstrated irrespective of the NH3 concentration present in the medium. Amino acid-ketoglutarate transaminase activity was demonstrated in each medium. This activity was particularly high when isoleucine or leucine was the amino group donor, while activity was also found with valine, methionine, aspartate, and, to a lower extent, alanine. No transaminase activity was detected for serine.
TABLE 3.
Activities of enzymes mediating the synthesis of glutamate from α-ketoglutarate in extracts of L. lactis subsp. lactis NCDO 2118
| Enzyme | Sp acta |
|---|---|
| Glutamate dehydrogenasec | ND |
| GOGATb | 4 |
| Amino acid-ketoglutarate transaminase | |
| Nitrogen donor amino acids tested | |
| Isoleucine | 142 |
| Leucine | 137 |
| Valine | 69 |
| Methionine | 32 |
| Aspartate | 22 |
| Alanine | 6 |
| Serine | ND |
Specific activity was expressed as nanomoles per minute per milligram of protein. The values were average values obtained during measurements with cell extracts from the four synthetic media used in fermentor cultures. ND, not detected.
Activity was assayed in the physiological direction.
Activity was assayed in both directions.
DISCUSSION
Removing the glutamate and related amino acids, i.e., glutamine, arginine, and proline, from the MS14 medium resulted in a very long lag phase before growth began and subsequently a decreased growth rate, as was the case in other synthetic media (5). Nevertheless, it was clearly established that the strain of lactic acid bacteria used in this study was able to synthesize glutamic acid, indicating that the pathway leading from the glycolytic intermediates to glutamic acid was operative.
The enzymatic study showed the presence of a PEP carboxylase activity, providing the necessary link between glycolysis and the amphibolic TCA cycle, though this activity was very low. This study is the first demonstration of an anaplerotic activity in a lactic acid bacterium. Among the three enzymes mediating the oxidative conversion of oxaloacetate and acetyl-CoA into α-ketoglutarate, activities were measured for citrate synthase and isocitrate dehydrogenase. This latter enzyme was not detected in any strain of Lactobacillus tested by Morishita and Yajima (18), and they concluded that its absence was responsible for the glutamate auxotrophy in LAB. This is clearly not true for L. lactis NCDO 2118, the strain used in our study. Moreover, the activity was observed with both NAD and NADP as cosubstrates, though it was eightfold higher with NAD than with NADP. On the other hand, aconitase activity was not detected by us, while it was observed, though at low levels, in four Lactobacillus strains tested by Morishita and Yajima (18). Since the pathway leading from oxaloacetate and acetyl-CoA to α-ketoglutarate was shown to be operative, the apparent lack of activity of this enzyme in our strain probably indicates a value too low to be determined rather than a true absence of activity. A similar observation was reported by Ruklish et al. (21) for Brevibacterium flavum, in which the aconitase activity was difficult to measure, with a value 10 to 20 times lower than the activities of the other citric acid cycle enzymes, leading the authors to conclude that this enzyme was probably exerting a controlling influence on TCA cycle activity.
As expected for an anaerobic bacterium, no α-ketoglutarate dehydrogenase activity was observed, in agreement with the results reported by Amarasingham and Davis (1) with Escherichia coli or by Morishita and Yajima (18) with lactobacilli. Moreover, none of the enzymes of the glyoxylate shunt was detected, indicating that succinate could not be produced from isocitrate. These observations, together with the observed accumulation of both α-ketoglutarate and succinate as overflow metabolites, indicate that both the oxidative and the reductive branches of the citric acid cycle were operative. The operation of the reductive pathway leading to synthesis of succinate from citrate, fumarate, or malate was previously proposed for lactobacilli (11), though this compound is not normally associated with the metabolism of sugars by L. lactis.
As regards the synthesis of glutamic acid from α-ketoglutaric acid, no glutamate dehydrogenase activity was demonstrated despite the presence of ammonia in the medium, though this enzyme was shown to be the main glutamate-forming enzyme in a majority of microorganisms (4, 8, 9, 13) and even in lactobacilli despite the incapacity to synthesize glutamic acid (18). On the other hand, glutamine synthetase-GOGAT activity, converting α-ketoglutarate and glutamine into 2 mol of glutamate, was shown to be present in our strain, but only at a very low level compared with the transaminase activity. The activity of the transaminase varied greatly with the nitrogen donor amino acid, isoleucine and leucine being the preferred substrates for the amination of α-ketoglutarate. Among the five amino acids present in every medium tested, only serine was unable to mediate a transamination reaction with α-ketoglutarate. The stoichiometric coefficients for amino acid consumption were not significantly different in media with or without glutamate, indicating that the synthesis of glutamate from α-ketoglutarate in a medium lacking glutamate was not dependent on the consumption of one particular amino acid as the nitrogen donor but was most probably related to the consumption of several amino acids. While aminotransferases have been studied for a variety of microorganisms, Yvon et al. (24) recently purified an aminotransferase from an L. lactis strain. This enzyme exhibited activity with the aromatic amino acids, leucine and methionine, but not with isoleucine or valine. These results suggested that the transaminase activity measured in our strain was probably due not to a single enzyme, but rather to at least two different enzymes with different affinities for the amino acids.
In view of our kinetic and enzymatic results, the crucial problem of reduced growth rate and very long lag phases in media lacking glutamate can be better explained. As regards the decreased growth rate, one of the metabolic steps involved in glutamic acid synthesis from the central metabolism, i.e., the glycolytic intermediates, was responsible for a metabolic bottleneck. The addition of α-ketoglutarate to the culture medium created culture conditions in which the synthesis of α-ketoglutarate was no longer necessary. Since this addition restored the growth rate to the value obtained in the MS14 medium containing glutamate, the limiting step was located in the part of the pathway leading to α-ketoglutarate. Further attempts to identify more precisely the location of the metabolic bottleneck were unsuccessful, because the addition of citrate or isocitrate was without effect on growth, probably because the strain was unable to use these compounds as substrates due to the lack of plasmid-encoded uptake systems. Moreover, supplementing the culture medium lacking glutamate with other compounds, including those directly involved in the conversion of α-ketoglutarate into glutamate, i.e., ammonia, alanine, and aspartate, which are considered to be nitrogen donors in the transamination reaction leading to glutamate (10, 22), was without effect on the growth characteristics. Taking into account the enzymatic results, the metabolic bottleneck could be at the level of the aconitase.
It is interesting to note that all of the enzymes assayed in our study were constitutive because they exhibited similar levels of activity whatever the medium used. This is somewhat surprising because some of them should not be necessary for growth in certain media. Growth in MS14 medium should not necessitate the enzymes of the oxidative part of the citric acid cycle mediating the synthesis of α-ketoglutarate from oxaloacetate and acetyl-CoA, and it seems obvious that, in this medium, the excretion of α-ketoglutarate was the result of the deamination of glutamate in transamination reactions. Therefore, the question remains as to why a lag phase was observed when the synthesis of glutamate became necessary. While the lag phase is frequently attributed to the time necessary for the synthesis of enzymes required for the growth in new medium, this induction process is not usually so long. Moreover, in our case, all of the required enzymes were present in all media, indicating that the lag phase was not related to this phenomenon. Since a high α-ketoglutarate concentration enabled the lag phase to be shortened, the lag phase was clearly related to the metabolic step between α-ketoglutarate and glutamate catalyzed by the transaminase. Because the substrate profile of transaminase activity was identical in all media, the possibility of the synthesis of isoenzymes specific to this particular reaction can be excluded. It must be remembered that the physiological function of the transaminases is to form amino acids from ketoacids, by transferring the amino group of glutamate, mainly, which is then deaminated to α-ketoglutarate. In the particular case where glutamate was lacking, the reverse direction might operate at the expense of other amino acids present in the medium. It can be postulated that, since the α-ketoglutarate synthesis is rate limiting, the long lag phase corresponds to the time necessary for the metabolism to accumulate an α-ketoglutarate concentration high enough to enable glutamate synthesis and subsequently to allow growth to proceed. Hence, the apparently curious excretion of α-ketoglutarate despite its rate-limited synthesis was certainly related to its necessary accumulation in the cell. Unfortunately, little is known about the transaminases in lactic acid bacteria, the number of enzymes present, their specificities, and their affinities, and this is an important task in better understanding the nitrogen metabolism of LAB.
ACKNOWLEDGMENTS
We thank Monique Suderie for technical assistance and Nic Lindley for valuable discussions.
REFERENCES
- 1.Amarasingham C R, Davis B D. Regulation of α-ketoglutarate dehydrogenase formation in Escherichia coli. J Biol Chem. 1965;240:3664–3667. [PubMed] [Google Scholar]
- 2.Bergmeyer H U, Bernt E. 2-Oxoglutarate UV spectrophotometric determination. In: Bergmeyer H U, editor. Methods of enzymatic analysis. New York, N.Y: Academic Press; 1963. pp. 1577–1580. [Google Scholar]
- 3.Börman-El Kholy E R, Eikmanns B J, Gutmann M, Sahm H. Glutamate dehydrogenase is not essential for glutamate formation by Corynebacterium glutamicum. Appl Environ Microbiol. 1993;59:2329–2331. doi: 10.1128/aem.59.7.2329-2331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burchall J J, Niederman R A, Wolin M J. Amino group formation and glutamate synthesis in Streptococcus bovis. J Bacteriol. 1964;88:1038–1044. doi: 10.1128/jb.88.4.1038-1044.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocaign-Bousquet M, Garrigues C, Novak L, Lindley N D, Loubiere P. Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J Appl Bacteriol. 1995;79:108–116. [Google Scholar]
- 6.Dainty R H. Glutamate biosynthesis in Clostridium pasteurianum and its significance in nitrogen metabolism. Biochem J. 1972;126:1055–1056. doi: 10.1042/bj1261055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deguchi Y, Morishita T. Nutritional requirements in multiple auxotrophic lactic acid bacteria: genetic lesions affecting amino acid biosynthetic pathways in Lactococcus lactis, Enterococcus faecium and Pediococcus acidilactici. Biosci Biotechnol Biochem. 1992;56:913–918. doi: 10.1271/bbb.56.913. [DOI] [PubMed] [Google Scholar]
- 8.Griffith C J, Carlsson J. Mechanism of ammonia assimilation in Streptococci. J Gen Microbiol. 1974;82:253–260. doi: 10.1099/00221287-82-2-253. [DOI] [PubMed] [Google Scholar]
- 9.Halpern Y S, Umbarger H E. Conversion of ammonia to amino groups in Escherichia coli. J Bacteriol. 1960;80:285–288. doi: 10.1128/jb.80.3.285-288.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong M M, Shen S C, Braunstein A E. Distribution of L-alanine dehydrogenase and L-glutamate dehydrogenase in Bacilli. Biochim Biophys Acta. 1959;36:288–289. doi: 10.1016/0006-3002(59)90111-8. [DOI] [PubMed] [Google Scholar]
- 11.Kaneuchi M, Seki M, Komagata K. Production of succinic acid from citric acid and related acids by Lactobacillus strains. Appl Environ Microbiol. 1988;54:3053–3056. doi: 10.1128/aem.54.12.3053-3056.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kusnan M B, Berger M G, Fock H P. The involvement of glutamine synthase/glutamate synthase in ammonia assimilation by Aspergillus nidulans. J Gen Microbiol. 1987;133:1235–1242. doi: 10.1099/00221287-133-5-1235. [DOI] [PubMed] [Google Scholar]
- 13.Kusnan M B, Klug K. Ammonia assimilation by Aspergillus nidulans: 15N-ammonia study. J Gen Microbiol. 1989;135:729–738. doi: 10.1099/00221287-135-4-729. [DOI] [PubMed] [Google Scholar]
- 14.Ledesma O V, de Ruiz Holgado A, Oliver G, de Giori G S, Raibaud P, Galpin J V. A synthetic medium for comparative nutritional studies of Lactobacilli. J Appl Bacteriol. 1977;42:123–133. doi: 10.1111/j.1365-2672.1977.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 15.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 16.Morishita T, Fukada T, Shirota M, Yura T. Genetic basis of nutritional requirements in Lactobacillus casei. J Bacteriol. 1974;120:1078–1084. doi: 10.1128/jb.120.3.1078-1084.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morishita T, Deguchi Y, Yajima M, Sakurai T, Yura T. Multiple nutritional requirements of lactobacilli: genetic lesions affecting amino acid biosynthetic pathways. J Bacteriol. 1981;148:64–71. doi: 10.1128/jb.148.1.64-71.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morishita T, Yajima M. Incomplete operation of biosynthetic and bioenergetic functions of the citric acid cycle in multiple auxotrophic Lactobacilli. Biosci Biotechnol Biochem. 1995;59:251–255. [Google Scholar]
- 19.Novak L, Cocaign-Bousquet M, Lindley N D, Loubiere P. Metabolism and energetics of Lactococcus lactis during growth in complex or synthetic media. Appl Environ Microbiol. 1997;63:2665–2670. doi: 10.1128/aem.63.7.2665-2670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiter B, Oram J D. Nutritional studies on cheese starters. I. Vitamin and amino acid requirements of single strain starters. J Dairy Res. 1962;29:63–77. [Google Scholar]
- 21.Ruklish M P, Duntse M E, Levane V A, Kalninya R V. Characteristics of the operation of the tricarboxylic acid cycle in Brevibacterium flavum and Micrococcus glutamicus. Mikrobiologiya. 1987;56:759–763. [Google Scholar]
- 22.Shen S C, Hong M M, Braunstein A E. The main path of nitrogen assimilation in Bacillus subtilis. Biochim Biophys Acta. 1959;36:290–291. doi: 10.1016/0006-3002(59)90112-x. [DOI] [PubMed] [Google Scholar]
- 23.Tempest D W, Meers J L. Synthesis of glutamate in Aerobacter aerogenes by a hitherto unknown route. Biochem J. 1970;117:405–407. doi: 10.1042/bj1170405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yvon M, Thirouin S, Rijnen L, Fromentier D, Gripon J-L. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl Environ Microbiol. 1997;63:414–419. doi: 10.1128/aem.63.2.414-419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]