Summary
Traditional cellular and live-virus methods for detection of SARS-CoV-2 neutralizing antibodies (nAbs) are labor- and time-intensive, and thus not suited for routine use in the clinical lab to predict vaccine efficacy and natural immune protection. Here, we report the development and validation of a rapid, high throughput method for measuring SARS-CoV-2 nAbs against native-like trimeric spike proteins. This assay uses a blockade of human angiotensin converting enzyme 2 (hACE-2) binding (BoAb) approach in an automated digital immunoassay on the Quanterix HD-X platform. BoAb assays using Wuhan-WT (vaccine strain), delta (B.1.167.2), omicron BA1 and BA2 variant viral strains showed strong correlation with cell-based pseudovirus neutralization activity (PNA) and live-virus neutralization activity. Importantly, we were able to detect similar patterns of delta and omicron variant resistance to neutralization in samples with paired vaccine strain and delta variant BoAb measurements. Finally, we screened clinical samples from patients with or without evidence of SARS-CoV-2 exposure by a single-dilution screening version of our assays, finding significant nAb activity only in exposed individuals. Importantly, this completely automated assay can be performed in 4 h to measure neutralizing antibody titers for 16 samples over 8 serial dilutions or, 128 samples at a single dilution with replicates. In principle, these assays offer a rapid, robust, and scalable alternative to time-, skill-, and cost-intensive standard methods for measuring SARS-CoV-2 nAb levels.
Subject areas: Diagnostics, Virology, Biochemical assay
Graphical abstract
Highlights
-
•
This assay is significantly faster than regular neutralization assays (4 h vs. 5 days)
-
•
Detects neutralizing Abs against prefusion state of the spike trimer
-
•
Assay is automated, significantly reducing labor and cost
-
•
128 samples (in duplicate) can be completed in 4 h
Introduction
Levels of neutralizing antibodies (nAbs) against SARS-CoV-2 and other viruses predict vaccine efficacy and immune protection after natural infection.1,2,3,4,5 In addition, the degree of protection from sterilizing immunity to prevention of severe disease correlates strongly with nAb levels at any given time post-vaccination or infection.6 Thus, the ability to reliably detect and quantify SARS-CoV-2 nAbs at scale is critical in public health effort to reach population level protection in the face of waning immunity and a need for boosters.7 In addition, the emergence of viral variants that escape neutralization by vaccine-induced antibodies underscores the importance of building efficient and reliable pipelines for nAb assay development as new variants are sequenced and rise to the level of interest or concern (VOI or VOC).
SARS-CoV-2 spike (S) protein is a large homotrimeric glycoprotein, which adopts a metastable prefusion conformation before its high affinity interaction with host-membrane-associated angiotensin converting enzyme 2 (ACE-2).8,9,10 Native S protein forms two proteolytically cleaved extracellular subunits (S1 and S2), with S1 containing a specific 222 amino acid (AA) receptor binding domain (RBD) that binds to ACE-2.11,12,13 Thus, S1 promotes receptor recognition and high affinity binding. The S2 subunit, in turn, drives membrane fusion through a fusion peptide (FP), two heptad repeat regions (HR1/2), and a transmembrane domain linked to the cytoplasmic tail.14 To date, studies of neutralizing antibodies elicited by vaccination and natural infection as well as monoclonal antibody therapies have largely focused on antibodies that bind and inhibit interactions through SARS-CoV-2 RBD.15 However, studies have also identified targets of neutralizing activity in SARS-CoV-2 S protein outside of the RBD, including regions in S2 proximal to the FP and HR2.16 These findings were recently bolstered in a study by Garrett et al. using phage deep mutation scanning (Phage-DMS) to comprehensively interrogate immunodominant epitopes of antibodies in SARS-CoV-2 convalescent plasma as well as routes of antibody escape by the virus. This study independently identified non-RBD epitopes for neutralizing antibodies in FP and HR2.16 Together these findings highlight the importance of closely approximating the native structure and domain organization of spike in any robust assay for SARS-CoV-2 neutralizing antibodies.
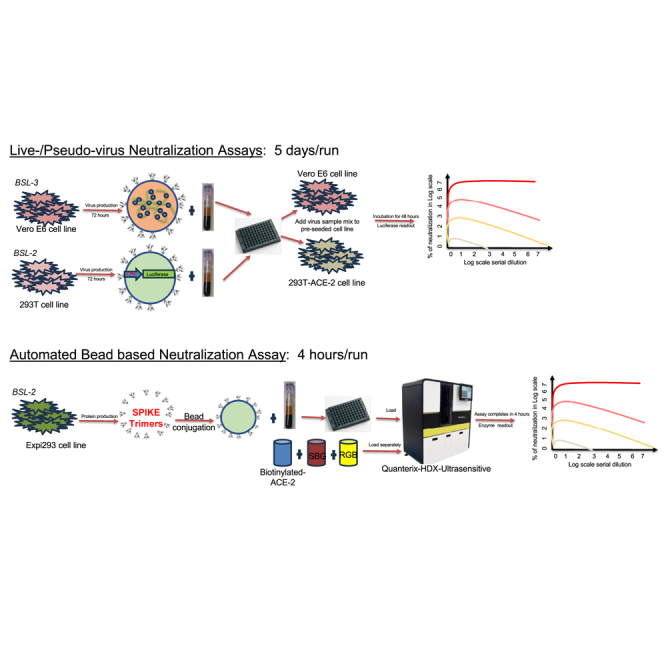
Current standard assays for measuring nAbs against SARS-CoV-2 require live, replication-competent wild virus isolates or infectious molecular clones.17,18 While these assays are important tools for research, they require a biosafety level 3 (BSL3) environment, are difficult to standardize, and are poorly suited for any scaled clinical application due to facilities, personnel, and safety requirements. A second tier of widely accepted nAb assays employs replication incompetent reporter viruses—commonly using backbones derived from either HIV or VSV—pseudotyped with SARS-CoV-2 Spike (S).19,20 These pseudovirus neutralization assays (PNAs) require only BSL2 working conditions and can be scaled for higher throughput. However, both live and pseudoviral assays require use and maintenance of living target cells, which introduces technical variability as well as regulatory complications to clinical testing operations that may seek to employ them. Furthermore, they are manual, labor-intensive assays with turn-around-times of several days. Some groups have developed ELISA- or bead-based assays to detect SARS-CoV-2 neutralizing antibodies, but these are performed manually, typically just use the RBD peptide as a target, and their sensitivity is unclear.21,22,23,24,25,26 However, the Mindray company established an automated assay to detect SARS-CoV-2 neutralizing antibodies using exclusively RBD protein as a target.27
To address these limitations, we developed and validated a rapid, high throughput, automated blockade of ACE-2 binding (BoAb) assay to quantify SARS-CoV-2 nAb activity against the native-like trimeric spike proteins of vaccine (Wuhan-1), delta (B.1.167.2), Omicron BA1, and Omicron BA2 variants. This assay is performed on the ultrasensitive Quanterix-HDX platform and is amenable to routine clinical use. We validated our BoAb by comparison to standard live virus and pseudovirus neutralization assays as well as clinically in samples from a cohort of SARS-CoV-2 exposed and vaccinated individuals collected during a serosurvey in the spring of 2021. In principle, our approach offers a rapid, adaptable, and scalable solution for detection of nAbs against any SARS-CoV-2 variant, against other viral pathogens or against emerging viruses of pathogenic potential.
Results
Detection of SARS-CoV-2 neutralizing antibodies by novel automated assay for blockade of ACE-2 binding (BoAb)
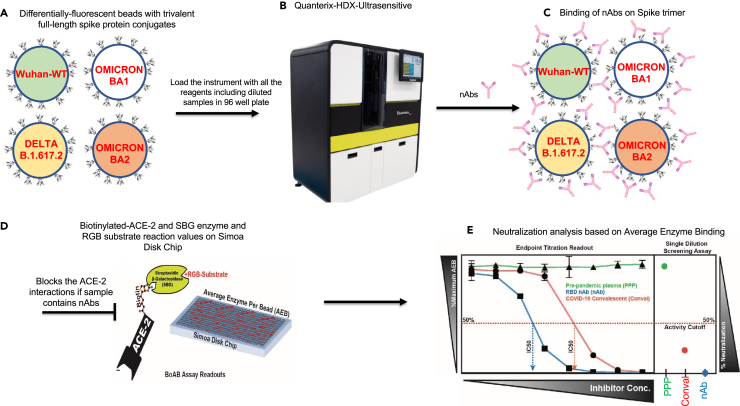
The majority of SARS-CoV-2 neutralizing antibodies prevent viral entry by inhibiting the biochemical interaction between S protein and ACE-2. We therefore designed our assay to detect inhibition of this interaction by nAbs in patient samples. The assay uses SARS-CoV-2 spike conjugated beads (Figure 1A); the beads are available tagged with different fluorophores, allowing multiplexed assays to be developed. After the beads are incubated with antibody-containing samples (Figure 1A), they are washed and incubated with a biotinylated ACE-2 detector followed by streptavidin beta-galactosidase (Figures 1B–1D). The signal was developed by addition of the ß-D-galactopyranoside substrate. To compare and quantify levels of neutralization from our BoAb assays, we engineered two primary readouts of the assay: an eight-point titration to identify the 50% inhibitory dilution or concentration (ID50 or IC50) (Figure 1E), and a single dilution readout calculated as a percentage of the maximum ACE-2 binding signal for a given spike target bead set. The latter approach was conceived as a potential screening tool for potent neutralizing antibodies while the former titering approach is appropriate for more rigorous comparisons among subjects or between candidate inhibitors.
Figure 1.
Schematic of blockade of ACE-2 binding (BoAb) assay design
The assay is automated on the Quanterix HDX platform using single molecule array (SIMOA) technology with a readout of average enzymes per bead (AEB).
(A) Magnetic beads labeled with four distinct fluorophores are conjugated with full-length trimeric spike proteins (Wuhan WT, Delta, Omicron BA1 and BA2), (B) Load the Quanterix HDX instrument with all the reagents (Diluents, an in-house biotinylated ACE-2, diluted samples in 96 well plate, RGB substrate and SBG.
(C) Spike trimer will be bound by nAbs if sample contains neutralizing antibodies (D) Blocks the spike interactions with ACE-2 and readout is based on the ACE-2 binding signal is amplified by streptavidin-beta-galactosidase and a fluorescent RGB-substrate (E) Processed data for 50% inhibitory concentration (IC50) titration. Signals from pre-pandemic samples (PPS) are compared against those from RBC neutralizing antibodies and COVID-19 convalescent samples. All the washes in between each step done by automatically.
In-house generated vaccine strain and delta variant spike proteins adopt native trimeric structures and bind with high affinity to ACE-2
In order to present authentic, native-like spike targets for neutralizing antibody detection, we utilized a soluble, stabilized prefusion spike ectodomain construct originally designed in work by Hsieh et al.28 Our plasmid DNA sequences are different for WT, Delta, Omicron BA1 and BA2 strains which were human codon optimized and synthesized at GenScript (supplementary text). This construct contains 6 stabilizing proline mutations in S2, which prevent the spontaneous and irreversible formation of a post-fusion state (Figure S1A).28 Spike targets for vaccine strain (Wuhan-Hu-1 GenBank: MN908947), delta variant (B.1.617.2) and Omicron BA1(B.1.1.529) and BA2 (B.1.1.529.2) were produced in human 293F cells and purified by affinity and size exclusion chromatography (SEC). A soluble, human IgG Fc chimera of ACE-2 was produced in a similar system before affinity and SEC. Purity of all in-house generated protein reagents was determined to be >95% using reducing sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Figure S2). We studied the structures of purified proteins using negative stain electron microscopy NS-EM (Figure S1B, S1E, and S1H) with 2-dimensional class averaging and found that the proteins are formed in trimers. Further, we confirmed that our spike proteins adopted the expected homotrimeric structures with 3-fold symmetry at the apex and an expected tapering in the S1 to S2 transition (Figures S1C, S1F, and S1I). Finally, we confirmed that our ACE-2 detector bound stably and with high affinity to both prefusion constructs using biolayer interferometry (Figure S1D, S1G, and S1J). Wuhan-Wt and delta variant spike reagents bound the ACE-2 detector at a similar steady-state level but Omicron BA1 showed high affinity to bind ACE-2 very rapidly and all variants showed stable, slow dissociation rates. Together these data confirm the authentic structure of our spike reagents and their binding activity toward the ACE-2 detector used in our BoAb assays.
Vaccine strain (Wuh-1) BoAb neutralizing activity correlates strongly with corresponding live virus and pseudovirus neutralization results
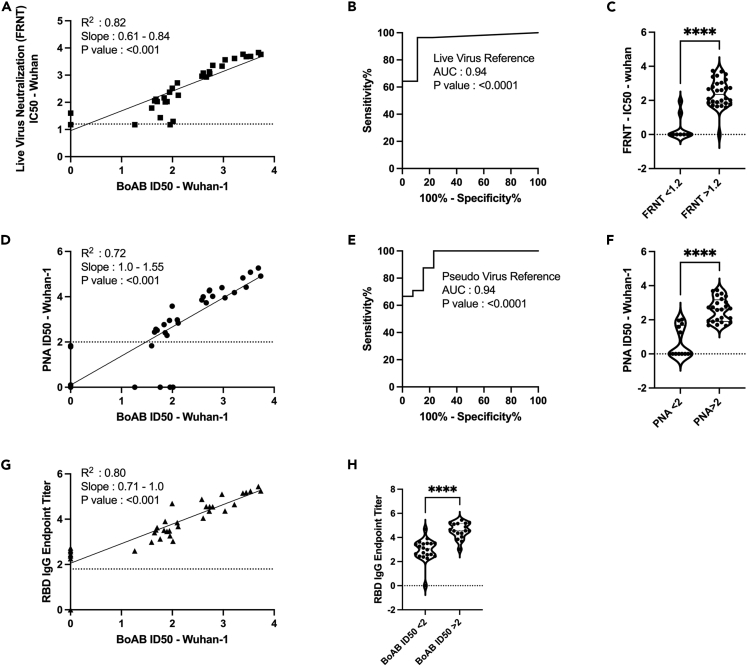
To determine the performance of our new test for SARS-CoV-2 neutralizing antibodies, we evaluated the correlation between titering results with the vaccine strain BoAb assay versus live virus and pseudovirus neutralization assays using plasma samples from patients vaccinated against COVID-19. Results from our vaccine strain assay showed strong correlation with results from a standard live virus focus reduction neutralization test (FRNT) (Figure 2A) as well as strong performance in an ROC analysis using the lowest reported log ID50 (1.17) as a cutoff for activity (AUC 0.94; p < 0.0001) (Figures 2B and 2C). Similarly, our assay correlated strongly with vaccine strain pseudovirus neutralization activity, particularly in samples above the log ID50 limit of quantification (2.0) for our pseudovirus assay (R2 = 0.72; p < 0.001) (Figure 2D). Using this pseudovirus limit of quantification (LOQ) as a cutoff for positivity, we performed a second ROC analysis comparing vaccine strain BoAb activity in samples below and above the PNA LOQ. Our assay showed robust performance (ROC AUC 0.94; p < 0.0001) with PNA results as a reference (Figures 2E and 2F). Our new assay also demonstrated strong correlation with levels of receptor binding domain (RBD) IgG, with samples containing higher levels of neutralizing antibodies as measured by BoAb showing significantly higher levels of RBD binding IgG (Figures 2G and 2H).
Figure 2.
Correlation of vaccine strain (Wuhan-Wt) BoAb IC50s with live virus and pseudovirus neutralization assays (LVN and PNA)
(A) Linear regression analysis of vaccine lineage (Wuh-1 & WA1/2020) live virus 50% focus reduction neutralization activity (FRNT50) against ID50s in the vaccine strain BoAb. The absolute value of the log dilution factor at which a sigmoidal curve (fit to duplicate eight-point dilution series for each sample) crossed 50% is plotted for the BoAb assay.
(B and C) Receiver operator characteristic (ROC) curve and categorical comparison of the vaccine strain BoAb using a live virus FRNT cutoff of 1.17 (representing a linear dilution of 1 in 15) as the reference standard for neutralizing activity.
(D) Linear regression analysis of vaccine strain (Wuh-1 & WA1/2020) pseudovirus 50% inhibitory dilution (ID50) against ID50s in the vaccine strain BoAb.
(E and F) Receiver operator characteristic (ROC) curve and categorical comparison of the vaccine strain BoAb using a pseudovirus neutralization ID50 of 2 (representing a linear dilution of 1 in 100) as the reference standard for neutralizing activity.
(G) Linear regression analysis of vaccine strain (Wuh-1 & WA1/2020) receptor binding domain (RBD) specific IgG titers against IC50s in the vaccine strain BoAb. Values are plotted as in (A) using an optical density cutoff of 0.2 to quantify levels of binding antibodies.
(H) Comparison of RBD IgG endpoint titers in samples with vaccine strain BoAb activity less than or greater than a log IC50 of 2. Statistical significance was evaluated by unpaired non-parametric t tests. ns = not significant, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. PNA, RBD endpoint titer, FRNT values are in log scale.
Levels of delta variant (B.1.167.2) BoAb neutralization correlate strongly with corresponding live and pseudovirus virus neutralization results and accurately reflect patterns of escape from neutralizing antibodies
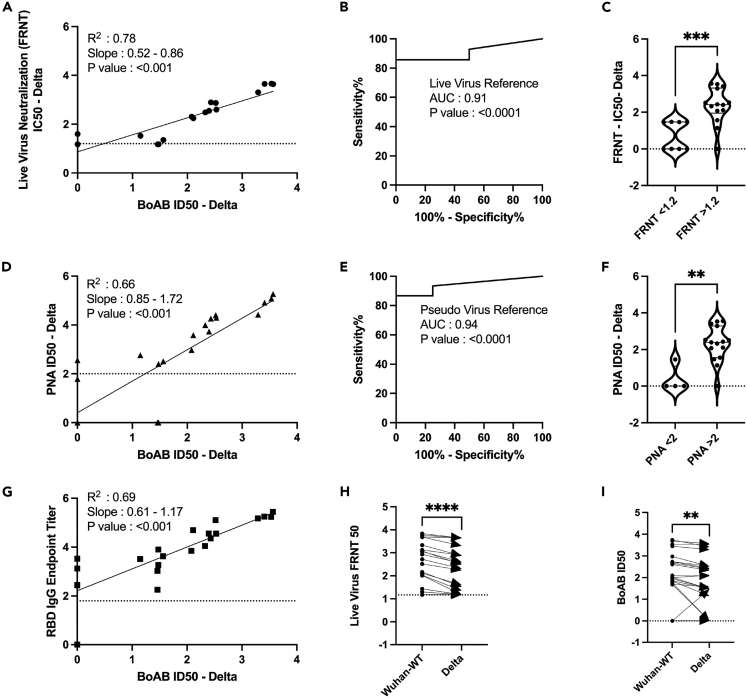
We next evaluated the performance of our assay for delta variant (B.1.167.2) neutralizing activity. We found a strong correlation and robust performance by ROC analysis for our new delta variant BoAb assay (R squared of 0.78; p < 0.001) as compared to live delta variant neutralizing activity determined by standard FRNT assay (Figures 3A–3C). ID50 results from our delta variant assay also correlated strongly with activity in our vaccine strain PNA and the corresponding live virus neutralizing antibody assay though, as expected, with a lower degree of correlation than that seen within strain (R squared of 0.66) (Figure 3D). ROC analysis revealed a similar performance of the delta variant BoAb assay to the vaccine strain PNA assay using a vaccine strain PNA log ID50 cutoff of 2 for positivity (Figures 3E and 3F). Delta variant BoAb activity also correlated with vaccine strain RBD binding titers though to a lesser extent (Figure 3G). Finally, we evaluated the decrement between vaccine strain neutralizing antibody activity and delta variant activity, observed consistently in vaccinated individuals and postulated to be, at least in part, responsible for an increased frequency of delta variant breakthrough infections among vaccinated individuals.28 Importantly, for each sample we found a similar pattern of decrement in vaccine strain and delta variant BoAb activity compared to live virus vaccine strain and delta variant FRNT results (Figures 3H and 3I). Together these data suggest that our delta variant BoAb assay correlates strongly with standard assays for neutralizing activity and may similarly detect deficits in delta variant specific activity observed among vaccinated individuals and those who experienced infection prior to the emergence of SARS-CoV-2 spike variants with the ability to escape nAbs.
Figure 3.
Correlation of delta variant BoAb ID50s with live virus and pseudovirus neutralization assays (LVN and PNA)
(A) Linear regression analysis of delta variant (B.1.617.2) live-virus FRNT 50% inhibitory concentration (ID50) against ID50s in the delta variant BoAb.
(B and C) Receiver operator characteristic (ROC) curve and categorical comparison of the delta variant BoAb using a live virus FRNT cutoff of 1.17 (representing a linear dilution of 1 in 15) as the reference standard for neutralizing activity.
(D) Linear regression analysis of vaccine strain (Wuh-1 & WA1/2020) pseudovirus 50% inhibitory concentration (ID50) against ID50s in delta variant (B.1.617.2) BoAb.
(E and F) Receiver operator characteristic (ROC) curve and categorical comparison of the delta variant BoAb using a pseudovirus neutralization ID50 of 2 (representing a linear dilution of 1 in 100) as the reference standard for neutralizing activity.
(G) Linear regression analysis of vaccine strain (Wuh-1) receptor binding domain (RBD) specific IgG titers against ID50 values in the delta variant BoAb. Values are plotted as in (A).
(H and I) Paired comparison of live virus FRNT50 and BoAb ID50 values between vaccine strain lineage (Wuhan-Hu-1 or WA1/2020) and delta variant assays. Statistical significance was evaluated by paired non-parametric t tests ns = not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001,∗∗∗∗p < 0.0001. PNA, RBD endpoint titer, FRNT values are in log scale.
Omicron BA1 and BA2 BoAb neutralization correlate strongly with corresponding pseudovirus virus neutralization results and accurately reflect patterns of BA2 escape from neutralizing antibodies
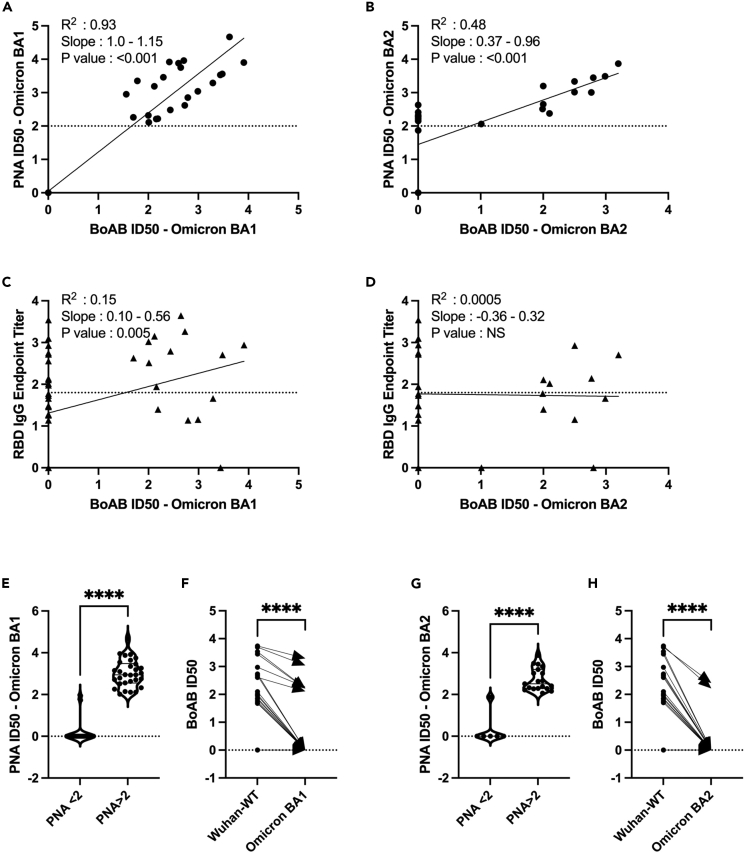
We next evaluated the performance of our assay for SARS-CoV-2 Omicron BA1 and BA2 variants neutralizing activity. We found a strong correlation for BA1 variant BoAb assay (R squared of 0.93; p < 0.001), but BA2 variant showed weak neutralization (R squared of 0.48; p < 0.001) as compared to BA1 variant neutralizing activity (Figures 4A and 4B). Omicron BA1 BoAb activity correlated with activity in RBD-specific endpoint titers but not BA2 (Figures 4C and 4D). ROC analysis revealed a similar performance of the Omicron BA1 and BA2 variant BoAb assay to the vaccine strain PNA assay using a vaccine strain (Wuhan-Wt) PNA log ID50 cutoff of 2 for positivity (Figures 4E and 4F). Finally, we evaluated the decrement between vaccine strain neutralizing antibody activity and Omicron BA1 and BA2 variants activity, observed consistently in vaccinated individuals and postulated to be, at least in part, responsible for a decreased frequency of omicron variant breakthrough infections among vaccinated individuals.29 Importantly, for each sample we found a similar pattern of decrement in vaccine strain and Omicron variant BoAb activity (Figures 4G and 4H). Together these data suggest that Omicron variant BoAb assay correlates strongly with standard assays for neutralizing activity and may similarly detect deficits in omicron variant specific activity observed among vaccinated individuals and those who experienced infection prior to the emergence of SARS-CoV-2 spike variants with the ability to escape nAbs.
Figure 4.
Correlation of Omicron variant BA1 and BA2 BoAb ID50s with pseudovirus neutralization assays (PNA)
(A and B) Linear regression analysis of Omicron variant BA1 and BA2 pseudovirus 50% inhibitory concentration (ID50) against ID50s in the BA1 and BA2 variant BoAb, respectively.
(C and D) Linear regression analysis of vaccine strain (Wuh-1) receptor binding domain (RBD) specific IgG titers against ID50 values in the BA1 and BA2 variants respectively, BoAb values are plotted as in (A and B).
(E‒H) Paired comparison of BoAb ID50 values between Wuhan strain lineage (Wuhan-Hu-1 or WA1/2020) and Omicron BA1 and BA2 variant assays. Statistical significance was evaluated by paired non-parametric t tests ns = not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001,∗∗∗∗p < 0.0001. NS: No Significance. PNA, RBD Endpoint Titer values are in log scale.
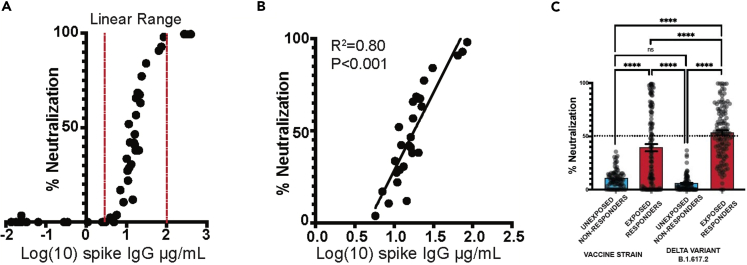
Screening for neutralizing antibody activity by single dilution BoAb among SARS-CoV-2 exposed patients
An ideal clinical screening test for SARS-CoV-2 neutralizing activity, in addition to being automated and well correlated with accepted standard assays, should not require limiting-dilution analysis which carries significant costs associated with skilled labor and resources. We therefore generated a single dilution screening test at a sample dilution that was well correlated with live-virus neutralizing activity (1:50) (Figure 5). Next, we evaluated the correlation between single-dilution blockade of binding at 1:50 with quantitative spike IgG serology, also performed on the Quanterix platform using an EUA serology assay in samples from vaccinated individuals at various times after vaccination. We found a strong linear correlation between blockade of binding and levels of spike IgG in samples with spike-specific IgG levels between 5 and 100 μg/mL. At higher concentrations, blockade of binding was saturated at 100% inhibition. Significant blockade was not detected in samples with less than 5 μg/mL of spike-specific IgG (Figures 5A and 5B). Finally, the percentage neutralization at a 1:50 dilution was evaluated in a subset of samples from a serosurvey cohort collected in the Emory Healthcare system between January and March of 2021 among inpatients and outpatients who received a blood draw during the relevant encounter. Using available SARS-CoV-2 PCR testing data and serological results, we categorized patients into individuals more likely to have neutralizing activity at the time of sampling (exposed responders) and those unlikely to have nAb activity (unexposed, non-responders). Among 278 patients tested, we identified 115 who were serologically positive with evidence of SARS-CoV-2 exposure. Eighty-five patients were serologically negative without evidence of SARS-CoV-2 exposure at the time of the blood draw. All individuals who screened positive for significant neutralizing activity (>50% inhibition at a 1:50 dilution) in vaccine strain and delta variant single dilutions assays fell into the exposed responder category or had an unknown exposure status at the time of blood draw. Significantly more neutralizing activity was detected against the delta variant in this cohort, perhaps due to the fact that the circulating strain at the time (B1.167-Alpha)29,30 carries many of the same spike mutations as the delta variant (Figure 5C) and BoAB activity not performed for omicron variants BA1 and BA2 due to insufficient volume of these samples. Together these data provide proof of concept for use and further validation of our multi-variant BoAb tests as screening tools in patients with evidence of SARS-CoV-2 exposure or vaccination.
Figure 5.
Correlation between BoAb single dilution testing and quantitative SARS-CoV-2 IgG serology
(A and B) Correlation of quantitative anti-spike IgG levels with 1:50 single dilution BoAb in all tested vaccinated individuals. Additionally, the correlation was plotted with a separate linear regression that was limited to samples with inhibition in the linear range (C). Comparison of single dilution delta and vaccine strain neutralizing antibody activity in patients with or without evidence of SARS- CoV-2 exposure and seroconversion from a serosurvey conducted in the spring of 2021 at Emory University Hospital Midtown. Statistical significance was evaluated by unpaired non-parametric t tests ns = not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001,∗∗∗∗p < 0.0001.
Discussion
We report the development and validation of blockade of binding assays for the detection of nAbs against multiple SARS-CoV-2 variants. Results from our assays correlate strongly with established methods for nAb detection including live virus FRNT. Unlike these standard methods, our approach is completely automated and rapid and does not require cell culture, BSL3 facilities, or extensive manual liquid handling. In addition, we employ spike antigens with native trimeric structure in our assays to capture the breadth of epitopes bound by vaccine and infection induced antibody responses. This latter point is particularly important with the roll out of boosters, which purportedly broaden the antibody response. An enhancement in neutralizing activity mediated by breadth of epitope specificity would be difficult to detect using subdomain and non-native spike targets.
Most viral infections are controlled by functional antibodies, like nAb that block interactions between viral antigens and host receptors.1,30,31,32,33 In the case of SARS-CoV-2, spike protein interacts with the ACE-2 host cell receptor to enter host cells. Before interacting with host receptor, the spike antigen exists in a prefusion state. Conformational changes occur in the spike protein once it binds to ACE-2 receptor. Based on this finding, most assays were developed to detect nAbs in blood samples by incubating plasma or serum with pseudo-typed viruses or live viruses prior to co-culture with target cells.34,35 It is well understood that viruses maintain the prefusion state of viral antigens (spike for SARS-CoV-2 and Env for HIV-1 and HIV-2) before interacting with host receptor, and most nAbs target the prefusion state of viral antigen.36,37 Based on this model, we established an assay for the identification of nAbs against the prefusion state of SARS-CoV-2 spike protein that blocks interactions with human ACE-2 receptor. We have observed that the purified version of spike protein from Wuh-1 (WT), Delta, Omicron BA1 and BA2 used in these studies-maintained structures with one RBD up (opened confirmation) and two RBD down (closed confirmation) which was previously observed by Z Ke et al., when they studied spike structures on intact virion by cryo-EM images.9 Thus, the purified spike proteins used in this assay maintain the prerequisite conformational state for the RBD-ACE2 interaction. Further, we observed that the spike proteins showed strong hACE-2 interaction by BLI.
For the initial analysis of BoAb assay, we used samples from Nooka et al.,38 who studied the neutralization determinants in myeloma patients. With this cohort, we observed that results in the BoAb assay strongly correlated with both live virus neutralization (Wuhan R2 = 0.82 and p ≤ 0.001; Delta R2 = 0.78 and p ≤ 0.001) and pseudovirus neutralization results (Wuhan R2 = 0.72 and p ≤ 0.001; Delta R2 = 0.66 and p ≤ 0.001). Live virus neutralization was not performed against Omicron BA1 and BA2, but pseudovirus neutralization against BA1 correlated strongly (BA1 R2 = 0.93) Interestingly, we also identified significant correlation of BoAb results with RBD endpoint titers for Wuhan-WT (p ≤ 0.001), Delta strains (p = 0.002), Omicron BA1 (0.005), but BA2 did not show significant correlation. These results agree with other studies in which RBD titers are strongly correlated with neutralization titers and Omicron lineages are strongly escaped from antibodies.39,40,41 Further, we have observed that infected and vaccinated individuals showed strong neutralizing antibody response against Wuhan-WT, Delta, and Omicron-BA1 strains when compared with un-infected and vaccinated individuals (Figure S5), and our observation is lined with the studies which are showed strong neutralizing antibody responses in infected vaccinated individuals than un-infected vaccinated candidates.42,43,44
We evaluated the utility of our BoAb assay in a second cohort of samples from a serological survey of patients requiring a blood draw in the Emory Healthcare system from January to March of 2021. We used available COVID-19 PCR testing results and combined serological status to categorize patients who were likely to have been exposed to SARS-CoV-2 and to have developed a humoral immune response against the virus. We hypothesized that this group of exposed responders had the highest pre-test probability in the cohort of harboring SARS-CoV-2 neutralizing activity and screened them using a 1:50 single dilution version of our BoAb assay. In agreement with our hypothesis, we observed strong neutralization against SARS-CoV-2 spike that was consistent with the patients’ exposure status. Therefore, our data are strongly aligned with correlations of antibody responses vs. neutralization responses against SARS-CoV-2.45,46,47 The rapid, automated, high-throughput BoAb assay described here may be useful for large-scale quantification of nAbs against SARS-CoV-2 variants for both clinical and investigational uses.
The BoAb assay developed for the Quanterix HDX platform provides five significant advantages over other neutralizing antibody assays. First, as compared to live virus and pseudovirus neutralization methods, the BoAb assay is significantly faster to complete, offers much higher throughput, and does not require special biocontainment facilities. Second, as compared to other non-culture-based neutralization assays, which are usually run in a standard ELISA format, the HDX platform provides ultrasensitive detection capabilities. Third, the assay is fully automated. Fourth, the Quanterix format allows the test to be multiplexed so that antibody blockade of multiple spike targets can be investigated simultaneously. Additionally, the fifth novel aspect of our assay is the use of the full-length trimeric spike protein as the antibody target. The other available assays either use just the RBD peptide or an S1 subunit monomer including RBD. We believe that trimeric spike provides a more physiological binding environment since antibodies that interfere with ACE-2 binding by targeting non-RBD epitopes of spike could be detected with our test. For example, neutralizing antibodies have been shown to bind outside of the RBD, including regions in S2 proximal to the FP and HR2.16
Limitations of the study
It should be noted that our study was limited by availability of standard live-virus neutralizing antibody data and a need to directly correlate activity measured in our assay with known correlates of nAb activity in standard cell-based assays. While our data suggest that biochemical neutralization as measured by BoAb correlates well with results from these more established tests, additional work is needed to evaluate the implications of this association for vaccine efficacy and protection after natural infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS-CoV-2 (COVID-19) Spike RBD antibody, (Clone: HL1003-HU) | GeneTex | Cat# GTX635866; RRID:AB_2888584 |
| SARS-CoV-2 (COVID-19) Spike RBD antibody, (Clone: HL1003) | GeneTex | Cat# GTX635792; RRID:AB_2888574 |
| SARS-CoV-2 (COVID-19) Spike antibody, (Clonality: Polyclonal) | GeneTex | Cat# GTX135356; RRID:AB_2887482 |
| Biological samples | ||
| Serum or Plasma from SARS-CoV-2 vaccinated or Un-vaccinated | Emory Hospital, Atlanta, GA, USA. | N/A |
| Chemicals and Recombinant proteins | ||
| Dulbecco’s Modified Eagle Medium/Ham F-12 50/50 Mix (DMEM F12) | Corning | Cat#10-090-CV |
| TrypLE™ Express Enzyme (1X), phenol red | Gibco | Cat#12605010 |
| Pen-strep (5000 IU/mL, 5 mg/mL), 100 mL | MPBiologicals | Cat#1670049 |
| DPBS | Gibco | Cat#14190-144 |
| 2-Mercaptoethanol | Gibco | Cat#21985023 |
| Bovine Serum Albumin (BSA) | Fisher Bioreagents | Cat#BP1600-100 |
| Sodium Chloride | Sigma | Cat#S3014 |
| Sodium phosphate monobasic monohydrate | Sigma | Cat#S3522 |
| Imidazole | Sigma | Cat#5513 |
| Mini-PROTEAN® TGX™ Precast Gels | Bio Rad | Cat#4561084 |
| Amicon® Ultra-15 Centrifugal Filter Unit | Millipore | Cat#UFC905008 |
| Amicon®Ultra-4 Centrifugal Filter Unit | Millipore | Cat#UFC805008 |
| Amicon® Ultra-0.5Centrifugal Filter Unit | Millipore | Cat#UFC503024 |
| Amicon® Ultra-15 Centrifugal Filter Unit | Millipore | Cat#UFC903024 |
| NativeMark™ Unstained Protein Standard | Thermo Fisher Scientific | Cat#LC0725 |
| Corning® 500 mL Polycarbonate Erlenmeyer Flask with Vent Cap | Corning | Cat#431145 |
| ExpiFectamine™ 293 Transfection Kit | Thermo Fisher Scientific | Cat#A14524 |
| Expi293™ Expression Medium | Thermo Fisher Scientific | Cat#A1435102 |
| HisPur Ni-NTA Resin | Thermo Fisher Scientific | Cat#88221 |
| Opti-MEM™ I Reduced Serum Medium | Thermo Fisher Scientific | Cat#31985070 |
| Pierce™ Protein G Agarose | Thermo Fisher Scientific | Cat#20399 |
| Stericup filters | Thermo Fisher Scientific | Cat#290-4520 |
| Slide-A-lyzer Dialysis Cassette | Thermo Fisher Scientific | Cat#66030 |
| 50x TAE (Tris/Acetic Acid/EDTA) Buffer, 5 L | BioRad | Cat#1610773 |
| Transfection Reagent - BioT (1 mL) | Bioland Scientific | Cat#BioT1 |
| Pierce™ Protein G IgG Binding Buffer | Thermo Fisher Scientific | Cat#21011 |
| Pierce™ IgG Elution Buffer | Thermo Fisher Scientific | Cat#21009 |
| TWEEN® 20 | Sigma | Cat#P9416-100 mL |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma | Cat#11836170001 |
| Bio-Safe™ Coomassie Stain | BioRad | Cat#1610786EDU |
| Falcon® 96-well White/Clear Flat Bottom TC-treated Microplate, with Lid, Sterile, | Corning | Cat#353377 |
| White 96-well Microplate with Clear Bottom, Sterile and Tissue Culture Treated, Lid Included | PerkinElmer | Cat#6005181 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| Anti-His Biosensors | ForteBio | Cat#18-5114 |
| ProA Biosensors | Sartorius | Cat#18-5010 |
| EDAC, 1-Ethyl-3-(3-Dimethylaminopropyl)carbodiimide, Hydrochloride | Thermo Fisher Scientific | Cat#E2247 |
| Streptavidin-β-galactosidase (SBG) | Quanterix | Cat# 103798 |
| Resorufin ß-D-galactopyranoside (RGP) | Quanterix | Cat#103159 |
| Homebrew beads-750 | Quanterix | Cat#103529 |
| Homebrew beads-488 | Quanterix | Cat#103526 |
| Bead Conjugation buffer | Quanterix | Cat#101357 |
| Biotinylation Reaction buffer | Quanterix | Cat#101358 |
| Blocking buffer | Quanterix | Cat#101356 |
| Detector/Sample Diluent buffer | Quanterix | Cat#101359 |
| Bead Diluent | Quanterix | Cat#100458 |
| SARS-CoV-2 lentiviral kit | BEI-Resources | Cat# NR-53816 |
| Bright-Glo kit | Promega | Cat#E2610 |
| SARS-CoV-2 Spike (pre-fusion) trimer-Wuhan-WT | This study | jroback@emory.edu |
| SARS-CoV-2 Spike (pre-fusion) trimer-Delta | This study | jroback@emory.edu |
| SARS-CoV-2 Spike (pre-fusion) trimer-Omicron BA1 | This study | jroback@emory.edu |
| SARS-CoV-2 Spike (pre-fusion) trimer-Omicron-BA2 | This study | jroback@emory.edu |
| Experimental models: Cell lines | ||
| HEK-293T | ATCC | Cat#CRL-3216 |
| HEK-293T-ACE-2 | BEI-Resources | Cat#NR-52511 |
| Expi-293 | Thermo Fisher Scientific | Cat#A14527 |
| Software and algorithms | ||
| Prism 10 | GraphPad | https://www.graphpad.com/ |
| PyMoL | Schrödinger | https://pymol.org/2/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact: John D Roback; jroback@emory.edu.
Method details
Samples
Samples were sourced from various studies with IRB 00001663 at Emory University after obtaining the approval and consent from Institutional Review Board. Samples (total n = 300) were tested on the Quanterix HD-X instrument, which uses single molecule arrays (SIMOAS) of femtoliter-sized reaction chambers etched into a disk to detect single enzyme labeled proteins. The HD-X is comparable in cost to clinical immunoassay instruments and has similar preventative maintenance requirements. As the instrument is completely automated, the increased costs of purchase and maintenance are offset by reduced operator time. The automated steps of the BoAB assay begin with an initial incubation of antibody-containing sample with spike conjugated, fluorescently coated magnetic bead reagents. The beads are then separated magnetically and washed. Next, a biotinylated ACE2 detector is added and incubated, and a second round of magnetic separation and wash are performed. Following incubation with streptavidin beta-galactosidase, a third wash is performed followed by addition of the ß-D-galactopyranoside substrate. Spike antigens that are bound by the ACE2 detector can form the SBG/RGP fluorescent product that is detected and counted digitally; if the samples contain anti-spike antibodies, the ACE2 detector is prevented from binding, reducing the signal. The instrument can handle up to 288 replicates at a time.
Pseudovirus neutralization assay (PNA)
Neutralization activities against SARS-CoV-2 WT (Wuh-1), Delta (B.1.617.2), Omicron BA1 and BA2 strains were measured in a single-round-of-infection assay with pseudo viruses, as previously described.38 Briefly, to produce SARS-CoV-2 WT, Delta, Omicron BA1 and BA2 pseudoviruses, an expression plasmid bearing codon optimized SARS-CoV-2 full-length S plasmid (Parental sequence Wuhan-1, GenBank #: MN908947.3) was co-transfected into HEK293T cells (ATCC#CRL-3216) with plasmids encoding non-surface proteins for lentivirus production and a lentiviral backbone plasmid expressing a Luciferase-IRES-ZsGreen reporter, HIV-1 Tat and Rev packing plasmids (BEI Resources; cat# NR-53816) and pseudoviruses harvested after 48 h of post transfection and performed titration. Pseudoviruses were mixed with serial dilutions of plasma or antibodies and then added to monolayers of ACE-2-overexpressing 293T cells (BEI Resources; cat#52511), in duplicate. 24 h after infection, cells were lysed, luciferase was activated with the Luciferase Assay System (Promega), and relative light units (RLU) were measured on a synergy Biotek reader. We have tested and compared pre-pandemic samples in the pseudovirus neutralization assay and BoAB assay (Figure S4).
Protein expression and purification
Trimeric SARS-CoV-2 Spike (Wuh-1, Delta B.1.617.2, Omicron BA1 and BA2) as well as Angiotensin Converting Enzyme-2 (ACE-2)-IgFC chimera proteins were produced by transfection in FreeStyle 293-F cells using plasmids (MN908947 for Wuhan-1 and Delta strains, and NM_021804.3 for hACE-2). Briefly, FreeStyle 293F cells were seeded at a density of 2E6 cells/mL in Expi293 expression media and incubated with shaking on at 37°C and 127 rpm with 8% CO2 overnight. The following day, 2.5E6 cells/mL were transfected using ExpiFectamine 293 transfection reagent (ThermoFisher, cat. no. A14524) according to the manufacturer protocol. Transfected cells were then incubated with orbital shaking for 4–5 days at 37°C, 127 rpm, 8% CO2. Supernatants containing secreted trimeric ectodomains were collected by centrifugation at 4,000xg for 20 min at 4°C. Clarified supernatants were then filtered using a 0.22 μm stericup filter (ThermoFisher, cat.no. 290–4520) and loaded onto pre-equilibrated affinity columns for protein purification. The SARS-CoV-2 Spike trimer and ACE-2 proteins were purified using His-Pur Ni-NTA resin (ThermoFisher, cat.no. 88221) and Protein G Agarose (ThermoFisher, cat.no. 20399) respectively. Briefly, His-Pur Ni-NTA resin was washed twice with PBS by centrifugation at 2000xg for 10 min. The resin was resuspended with the spike-trimer supernatant and incubated for 2 h on a shaker at RT. Gravity flow columns were then loaded with supernatant-resin mixture and washed (25mM Imidazole, 6.7 mM NaH2PO4.H2O and 300 mM NaCl in PBS) four times, after which the protein was eluted in elution buffer (235 mM Imidazole, 6.7 mM NaH2PO4.H2O and 300 mM NaCl in PBS). Eluted protein was dialyzed against PBS using Slide-A-lyzer Dialysis Cassette (ThermoScientific, Cat# 66030) and concentrated using 100 kDa Amicon Centrifugal Filter Unit, at 2000 g at 4°C. The concentrated protein eluate was then run and fractionated on a Sepharose 600 (GE Healthcare) column on an AktaPure (GE Healthcare). Fractions corresponding to the molecular weight of each protein were pooled and concentrated as described above. Proteins were quantified by BCA Protein Assay Kit (Pierce) and quality was confirmed by SDS-PAGE (Figure S2). The spike proteins were tested against antibodies which are targeting S1 subunit, RBD and prefusion state of spike proteins (Figure S3).
ACE-2 protein expression and purification
The soluble ACE-2 IgFC chimera was expressed as described above. Clarified supernatants were diluted 1:1 with binding buffer before loading on a protein G gravity flow column, pre-equilibrated with 10 mL of binding buffer (Pierce cat.no.21011). Columns were washed with 20 mL of binding buffer, and the protein was eluted in 40 mL of elution buffer. Following elution, samples were first neutralized to pH 7.5 using 1 M Tris, pH 9.0. Eluted protein was dialyzed against 50mM Tris (pH7.5), 150mM NaCl using a Slide-A-lyzer Dialysis Cassette (ThermoScientific, Cat# 66030) and concentrated using 50 kDa Amicon Centrifugal Filter Unit, at 2000 g at 4°C. Size exclusion chromatography and quality control was performed on the concentrated protein as described above.
Assessment of spike-ACE-2 binding by biolayer interferometry
6x His-tagged spike was diluted to 50 μg/mL in PBS before immobilization on nickel NTA biosensors (fortebio). Association of ACE-2 was monitored using and OctetRED96e instrument (fortebio) in 2-fold dilutions series starting at 100 μg/mL for 600s followed by dissociation in PBS for 500s. Tips were regenerated using 10mM glycine and regenerated in 10 mM NiCl2 before re-loading with equivalent concentrations of spike. Performed affinity studies on generated spike trimers (Figure S1).
Generation of detector and conjugated beads
ACE-2 detector biotinylation and spike bead conjugation were performed per the Quanterix Homebrew Detection Antibody Biotinylation and Bead Conjugation Protocols.
ACE-2 Biotinylation
Briefly, ACE-2 was buffer exchanged using Amicon filtration into Quanterix biotinylation reaction buffer prior to mixing at 1 mg/mL with a 40x challenge ratio of NHS-PEG4-biotin for 30 min at room temperature. Cleanup of the biotinylated detection reagent was achieved by a further round of amicon filtration following recovery in biotinylation reaction buffer and determination of protein concentration. A final detector concentration of 0.5μg/mL was used in the assay.
Spike conjugation with magnetic beads
Paramagenetic beads were activated after washing with bead conjugation buffer using 9ug EDC (10 mg/mL) in a final bead volume of 300 μL containing 4.2E8 beads for 30 min at 4°C with rocking. Following activation, beads were washed with Bead Conjugation Buffer and 300 μL cold spike at 0.2 mg/mL to the beads followed by incubation at 4°C with rocking for 2 h. Beads were then washed and blocked for 45 min at room temperature, followed by a final wash and resuspension in 300 μL bead diluent. Spike capture beads were stored at 4°C until use in the assay.
Negative stain sample preparation, data collection and data analysis
Spike protein was diluted to 0.05 mg/mL in PBS prior to grid preparation. A 3 μL drop of diluted protein applied to previously glow-discharged, carbon coated grids for ∼60 s, blotted and washed twice with water, stained with 0.75% uranyl formate, blotted and air dried. Between 25 and 35 images were collected on Talos L120C microscope (Thermo Fisher, Waltham, MA) at 73,000 magnification and 1.97 Å pixel size. Relion-3.1 was used for particle picking, 2D classification and 3D reconstruction [PMID: 23000701].
Quantification and statistical analysis
GraphPad PRISM version 9 was used to perform the statistical analysis. Correlation between the assays was performed by Pearson r correlation method and linear regression analysis. All statistical tests were two-sided, unless otherwise noted, and statistical significance was assessed at the ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 and further details are provided in the figure legend where analysis was performed.
Acknowledgments
We would like to thank Lee Cato from Eric Ortlund’s lab for help in processing SEC for protein purification, and we also acknowledge Amit Joshi, Todd Glynn, and Danielle Svancara from Quanterix team for their tremendous support while developing this assay for use on the HDX instrument. We acknowledge Biodefense and Emerging Infections (BEI) Resources for providing plasmid (NR-52309) and 293T-ACE-2 cell line (NR-52511). The Robert P. Apkarian Integrated Electron Microscopy Core (IEMC) at Emory University is subsidized by the School of Medicine and Emory College of Arts and Sciences. Additional support is provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health under award number UL1TR000454. We would like to acknowledge Connie Arthur and Cheryl Meier for their fabulous support with serosurvey specimen processing. A.S.N. and J.D.R received grants from NIH/NCI (1 U54 CA260563-01: Immune regulation of COVID-19 infection in cancer and autoimmunity) and the Emory CURE Foundation.
Author contributions
N.C., H.V., A.S.N., and J.D.R. contributed to the acquisition, analysis, and interpretation of the data, S.P., K.B.M., W.H.Os., S.C., T.J.B., and F.H. interpretation of data, A.P. and F.F. involved in the interpretation of the NS-EM images, G.D., H.W., J.B., J.G., S.A., S.S., and M.V.D provided samples from Emory Healthcare, D.N.A., E.O., W.L., S.R.S., A.S.N., and J.D.R. contributed to the data analysis.
Declaration of interests
N.C., H.V., S.P., K.B.M., W.H.Os., H.W., A.S.N., and J.D.R. are co-inventors of BoAb assay technology. Emory University filed a patent on this technology.
Published: October 18, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108256.
Contributor Information
Sean R. Stowell, Email: srstowell@bwh.harvard.edu.
Andrew S. Neish, Email: aneish@emory.edu.
John D. Roback, Email: jroback@emory.edu.
Supplemental information
References
- 1.Tea F., Ospina Stella A., Aggarwal A., Ross Darley D., Pilli D., Vitale D., Merheb V., Lee F.X.Z., Cunningham P., Walker G.J., et al. SARS-CoV-2 neutralizing antibodies: Longevity, breadth, and evasion by emerging viral variants. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 3.Overbaugh J., Morris L. The Antibody Response against HIV-1. Cold Spring Harb. Perspect. Med. 2012;2:a007039. doi: 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan C.-W., Chia W.-N., Young B.E., Zhu F., Lim B.L., Sia W.R., Thein T.L., Chen M.I.C., Leo Y.S., Lye D.C., Wang L.F. Pan-Sarbecovirus Neutralizing Antibodies in BNT162b2-Immunized SARS-CoV-1 Survivors. N. Engl. J. Med. 2021;385:1401–1406. doi: 10.1056/NEJMoa2108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milne G., Hames T., Scotton C., Gent N., Johnsen A., Anderson R.M., Ward T. Does infection with or vaccination against SARS-CoV-2 lead to lasting immunity? Lancet Respir. Med. 2021;9:1450–1466. doi: 10.1016/S2213-2600(21)00407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine-Tiefenbrun M., Yelin I., Alapi H., Katz R., Herzel E., Kuint J., Chodick G., Gazit S., Patalon T., Kishony R. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat. Med. 2021;27:2108–2110. doi: 10.1038/s41591-021-01575-4. [DOI] [PubMed] [Google Scholar]
- 8.Wang M.Y., Zhao R., Gao L.J., Gao X.F., Wang D.P., Cao J.M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seyran M., Takayama K., Uversky V.N., Lundstrom K., Palù G., Sherchan S.P., Attrish D., Rezaei N., Aljabali A.A.A., Ghosh S., et al. The structural basis of accelerated host cell entry by SARS-CoV-2. FEBS J. 2021;288:5010–5020. doi: 10.1111/febs.15651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomassetti F., Nuccetelli M., Sarubbi S., Gisone F., Ciotti M., Spinazzola F., Ricotta C., Cagnoli M., Borgatti M., Iannetta M., et al. Evaluation of S-RBD and high specificity ACE-2-binding antibodies on SARS-CoV-2 patients after six months from infection. Int. Immunopharmacol. 2021;99 doi: 10.1016/j.intimp.2021.108013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 14.Xia X. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses. 2021;13:109. doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett M.E., Galloway J., Chu H.Y., Itell H.L., Stoddard C.I., Wolf C.R., Logue J.K., McDonald D., Weight H., Matsen F.A., 4th, Overbaugh J. High-resolution profiling of pathways of escape for SARS-CoV-2 spike-binding antibodies. Cell. 2021;184:2927–2938.e11. doi: 10.1016/j.cell.2021.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bewley K.R., Coombes N.S., Gagnon L., McInroy L., Baker N., Shaik I., St-Jean J.R., St-Amant N., Buttigieg K.R., Humphries H.E., et al. Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat. Protoc. 2021;16:3114–3140. doi: 10.1038/s41596-021-00536-y. [DOI] [PubMed] [Google Scholar]
- 18.Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., Vanderheiden A., Nyhoff L., Davis C.W., Adekunle O., et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong H.L., Wu Y.T., Cao J.L., Yang R., Liu Y.X., Ma J., Qiao X.Y., Yao X.Y., Zhang B.H., Zhang Y.L., et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg. Microbes Infect. 2020;9:2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.https://www.bio-rad.com/en-us/product/bio-plex-multiplex-sars-cov-2-neutralization-antibody-assay-kits-reagents?ID=01588ca5-0c9b-967d-140e-6e8dbb425c7b
- 22.https://www.mblbio.com/bio/g/product/sars-cov-2/pickup/SARS-CoV-2-neutralizing-antibody.html
- 23.https://www.genscript.com/covid-19-detection-svnt.html
- 24.https://www.thermofisher.com/elisa/product/SARS-CoV-2-Neutralizing-Ab-ELISA-Kit/BMS2326
- 25.https://thenativeantigencompany.com/products/sars-cov-2-neutralization-assay-development-kit-rbd-ace2/
- 26.https://www.caymanchem.com/product/502070/sars-cov-2-neutralizing-antibody-detection-elisa-kit
- 27.Zedan H.T., Yassine H.M., Al-Sadeq D.W., Liu N., Qotba H., Nicolai E., Pieri M., Bernardini S., Abu-Raddad L.J., Nasrallah G.K. Evaluation of commercially available fully automated and ELISA-based assays for detecting anti-SARS-CoV-2 neutralizing antibodies. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-21317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheedarla N., Hanna L.E. In: Recent Developments in Applied Microbiology and Biochemistry. Buddolla V., editor. Academic Press; 2019. Chapter 7-Functional and Protective Role of Neutralizing Antibodies (NAbs) Against Viral Infections; pp. 83–93. [DOI] [Google Scholar]
- 30.Murin C.D., Wilson I.A., Ward A.B. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat. Microbiol. 2019;4:734–747. doi: 10.1038/s41564-019-0392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020;58:e02107–e02120. doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padilla-Quirarte H.O., Lopez-Guerrero D.V., Gutierrez-Xicotencatl L., Esquivel-Guadarrama F. Protective Antibodies Against Influenza Proteins. Front. Immunol. 2019;10:1677. doi: 10.3389/fimmu.2019.01677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montefiori D.C., Roederer M., Morris L., Seaman M.S. Neutralization tiers of HIV-1. Curr. Opin. HIV AIDS. 2018;13:128–136. doi: 10.1097/COH.0000000000000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 36.Pancera M., Zhou T., Druz A., Georgiev I.S., Soto C., Gorman J., Huang J., Acharya P., Chuang G.Y., Ofek G., et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutten L., Lai Y.T., Blokland S., Truan D., Bisschop I.J.M., Strokappe N.M., Koornneef A., van Manen D., Chuang G.Y., Farney S.K., et al. A Universal Approach to Optimize the Folding and Stability of Prefusion-Closed HIV-1 Envelope Trimers. Cell Rep. 2018;23:584–595. doi: 10.1016/j.celrep.2018.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nooka A.K., Shanmugasundaram U., Cheedarla N., Verkerke H., Edara V.V., Valanparambil R., Kaufman J.L., Hofmeister C.C., Joseph N.S., Lonial S., et al. Determinants of Neutralizing Antibody Response After SARS CoV-2 Vaccination in Patients With Myeloma. J. Clin. Oncol. 2022;40:3057–3064. doi: 10.1200/JCO.21.02257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ripperger T.J., Uhrlaub J.L., Watanabe M., Wong R., Castaneda Y., Pizzato H.A., Thompson M.R., Bradshaw C., Weinkauf C.C., Bime C., et al. Orthogonal SARS-CoV-2 Serological Assays Enable Surveillance of Low-Prevalence Communities and Reveal Durable Humoral Immunity. Immunity. 2020;53:925–933.e4. doi: 10.1016/j.immuni.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein S.L., Pekosz A., Park H.S., Ursin R.L., Shapiro J.R., Benner S.E., Littlefield K., Kumar S., Naik H.M., Betenbaugh M.J., et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J. Clin. Invest. 2020;130:6141–6150. doi: 10.1172/JCI142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 42.Servellita V., Syed A.M., Morris M.K., Brazer N., Saldhi P., Garcia-Knight M., Sreekumar B., Khalid M.M., Ciling A., Chen P.Y., et al. Neutralizing immunity in vaccine breakthrough infections from the SARS-CoV-2 Omicron and Delta variants. Cell. 2022;185:1539–1548.e5. doi: 10.1016/j.cell.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans J.P., Zeng C., Carlin C., Lozanski G., Saif L.J., Oltz E.M., Gumina R.J., Liu S.L. Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muena N.A., García-Salum T., Pardo-Roa C., Avendaño M.J., Serrano E.F., Levican J., Almonacid L.I., Valenzuela G., Poblete E., Strohmeier S., et al. Induction of SARS-CoV-2 neutralizing antibodies by CoronaVac and BNT162b2 vaccines in naïve and previously infected individuals. EBioMedicine. 2022;78 doi: 10.1016/j.ebiom.2022.103972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z., Xu W., Xia S., Gu C., Wang X., Wang Q., Zhou J., Wu Y., Cai X., Qu D., et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct. Target. Ther. 2020;5:282. doi: 10.1038/s41392-020-00402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.