Abstract
Downregulating Angiotensin Converting Enzyme2 (ACE2) expression may be a shared mechanism for RNA viruses.
Aim
Evaluate the expressions of ACE2 effectors: the long non-coding RNA ‘MALAT-1’, the micro-RNA ‘miR-200c-3p’ and the histone deacetylase ‘SIRT1’ in SARS-COV-2 patients and correlate to disease severity. Sera samples from 98 SARS-COV-2 patients and 30 healthy control participants were collected. qRT-PCR was used for MALAT-1 and miR-200c-3p expression. SIRT1 was measured using ELISA.
Results
In sera of COVID-19 patients, gene expression of miR-200c-3p is increased while MALAT-1 is decreased. SIRT1 protein level is decreased (P value < 0.001). Findings are accentuated with increased disease severity. Serum MALAT-1, miR-200c-3p and SIRT1 could be used as diagnostic markers at cut off values of 0.04 (95.9 % sensitivity), 5.59 (94.9 % sensitivity, 99 % specificity), and 7.4 (98 % sensitivity) respectively. A novel MALAT-1-miR-200c-3p-SIRT1 pathway may be involved in the regulation of SARS-COV-2 severity.
Keywords: SARS-COV-2, MALAT-1, SIRT1, miR-200c-3p, ACE2, COVID-19
Highlights
-
•
Downregulating ACE2 by miR-200c-3p may be a shared mechanism for RNA viruses.
-
•
MALAT-1 sponges miR-200c-3p, suggesting a protective role.
-
•
A novel MALAT-1-miR-200c-3p-SIRT-1 pathway may regulate COVID-19 severity.
-
•
Decreased MALAT-1 relieves miR-200c-3p inhibition, thus downregulating SIRT-1 expression.
1. Introduction
The coronavirus family has been known since 1965 to cause upper-respiratory tract infection in humans [1]. Symptoms were usually mild to moderate; however, Severe pandemics have been caused by three well known viruses of the family: SARS coronavirus (SARS-CoV) in 2002; MERS coronavirus (MERS-CoV) in 2012; and SARS-CoV-2 (COVID-19) in 2019. These three viruses caused fatal lower-respiratory tract infections and heavily impacted healthcare systems due to pneumonia and other complications. This illustrated the need of quick modalities for early diagnosis and follow up of complicated cases [2].
Previous research on SARS-CoV and MERS-CoV allowed physicians to identify early markers of disease complications. In addition, scientists quickly developed COVID-19 therapeutics and vaccines. Conduction of basic research on COVID-19 is still mandatory to identify viral biomarkers, host defense mechanisms, and possible therapeutics [3].
SARS-COV-2 entry into human cells occurs by clathrin mediated endocytosis after viral attachment to the extracellular domain of the angiotensin converting enzyme 2 receptors (ACE2R) found on the surface of many human cells, including lung cells [4]. This attachment is immunologically responded to by downregulating ACE2R in an attempt to decrease viral entry. Eventually, Angiotensin II levels increase causing inflammation and thrombosis, which further contributes to the acute lung injury seen in COVID-19. It is thus logical to investigate the factors that affect ACE2 levels, such as non-coding RNAs [5].
MicroRNAs (MiRNAs) are tiny noncoding RNAs (ncRNAs) around 22 nucleotides long that bind to regions on target transcripts known as miRNA response elements (MREs), causing either transcript destruction or translational inhibition [6]. miR-200c-3p is a member of the miR-200 family. miR-200c-3p was one of the first to be linked to viral infections and cell-mediated antiviral responses. Overexpression of miR-200c-3p has a critical role in the development of post viral lung injury and ARDS as it downregulates the expression of the mRNA that transcribes ACE2 [7]. This increases levels of angiotensin II leading to lung injury [8].
Long noncoding RNAs (LncRNAs) are transcripts with a length of more than 200 nucleotides, that are transcribed by RNA polymerase II but are not translated into proteins [9]. LncRNAs have now been demonstrated to play critical epigenetic regulatory functions [10]. MALAT-1 (metastasis-associated lung adenocarcinoma transcript 1) is a > 8000 nucleotide long intergenic non-coding RNA (lincRNA) found on chromosome 11q13 [11], that contains several putative miRNA binding sites, one of these is the miR-200c-3p. Sponging of miR-200c-3p by MALAT-1 suppresses it [12,13].
Sirtuins are a family of nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylase that are involved in cellular stress response and organism longevity [14]. Sirtuins also provide defense against viral infections [15]. SIRT1 can suppress NF-κB [16], as well as bind to ACE2 promotor enhancing its expression [17,18].
Considering the above-mentioned data, a MALAT-1/miR-200c-3p/Sirt1 axis may serve as a regulatory mechanism involved in the progression and/or the prevention of COVID-19 [19]. Our aim was to investigate the possible role of MALAT-1 in SARS-COV-2 patients and its potential effect on miR-200c-3p and SIRT1. The Expression of MALAT-1 and miR-200c-3p in addition to SIRT1 protein levels in serum of SARS-COV-2 patients was also correlated to the severity of lung affection of patients.
1.1. Study participants and methodology
Ninety-eight COVID-19 positive patients were recruited in this study from the ward and the ICU of Kasr-Alainy Internal Medicine Isolation Hospital, Cairo University, from April 2021 till December 2021. The study has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) and approved by the Research Ethics Committee of Kasralainy School of Medicine [code: MS-604-2021]. Written consents were taken from participants. Both males and females between 18 and 70 years old were included. Those who were suffering from malignancy, autoimmune, chronic severe lung diseases, and chronic renal diseases were excluded as well as pregnant females.
Study participants were divided into: Group 1 included 30 controls of age and sex matched to group 2, which included 98 COVID-19 patients further subdivided into: group (2a): 19 non-diabetic normotensive COVID-19 patients, group (2b): 20 type II DM normotensive COVID-19 patients and group (2c): 59 type II DM with idiopathic hypertension COVID-19 patients.
Group (2b) participants were previously diagnosed as diabetics according to the WHO diagnostic criteria for diabetes: Fasting Plasma Glucose (FPG) ≥ 125 mg/dL or 2-h post-load plasma glucose ≥200 mg/dL or HbA1c ≥ 6.5 mg/dL. Group (2c) participants were previously diagnosed as hypertensives according to the International Society of Hypertension (ISH) diagnostic criteria for HTN: Systolic/diastolic Blood Pressure (BP) ≥ 140/90 mmHg following standard protocols for measurement, including repeat measurements.
All patients were subjected to thorough history taking and clinical examination including age, gender, BMI, and comorbid diseases (DMII, HTN).
Five ml blood was taken from each participant after getting a written informed consent. Two ml were taken on EDTA. The remaining 3 ml were centrifuged to separate serum that was used for the determination of the relative expression of miR-200c-3p and MALAT-1 by RT-PCR, in addition to the detection of serum protein level of SIRT1 using ELISA.
CBC, HbA1c, kidney function tests (serum creatinine, urea, and albumin), liver function tests (alanine aminotransferase [ALT] and aspartate aminotransferase [AST]), lipid profile, FBG, serum ferritin, D-dimer, LDH, and CRP levels were investigated.
RNA was extracted from serum by (Qiagen, Valencia, CA, USA). Briefly, one mL QIAzol lysis reagent was added to 200 μL serum and incubated for 5 min at room temperature, followed by chloroform phase separation. The upper watery phase was mixed with 100 % ethanol and placed in RNeasy Mini spin columns in collection tubes and centrifuged at 8000 g at room temperature for 15 s eventually the RNA was eluted, quantified, and assessed for purity using the NanoDrop® (ND)-1000 spectrophotometer (NanoDrop Technologies, Inc. Wilmington, USA).
Reverse transcription of extracted RNA in a final volume of 20 μL RT reactions using the miScript II RT kit (Qiagen, Valencia, CA, USA) was done.
Quantitative Real-time PCR (qPCR) technique, using miScript SYBR green PCR kit and miScript primer assays [hsa-miR-200c-3p: (YP0020448, Thermofisher)], was used for the Detection of miR-200c-3p relative expression levels in sera of participants. qPCR for Detection of lncRNA MALAT-1 was done using Quantitect SYBR green PCR master mix and MALAT-1 primer assay [Human MALAT-1: (LPH18065A, Thermofisher)] in a 25 μl per well reaction volume. The real-time cycler was programmed on 95 °C for 15 min, followed by a three-step cycling of 15 s on 94 °C, then 30 s on 55 °C and finally 30 s on 70 °C. After 40 cycles, analysis of melting curves was done to validate specific production of the expected PCR product. No control miRNA is known in serum, so as an endogenous control, SNORD 68 was used for miR-200c-3p, while GAPDH was used for MALAT-1. The expression levels were evaluated using the ΔCt method, where ΔCt was calculated by subtracting the Ct values of SNORD 68/GAPDH from those of target micro-RNAs. ΔΔCt was calculated by subtracting the ΔCt of group 1 samples from the ΔCt of group 2 samples. The fold change in miR-200c-3p and MALAT-1 expression levels were calculated by equation 2–ΔΔCt.
Quantitation of Human Sirtuin 1 (SIRT1) in serum was done by an ELISA kit from Bioassay Technology Laboratory (Cat. No E2557Hu), Zhejiang, China.
The Chest X-ray score was calculated according to Brixia's scoring system. The lungs were divided into six areas by two lines, the first at the level of the right inferior pulmonary vein and the second at the level of the inferior wall of the aortic arch. According to the degree of the lesion, radiologists assigned a score from 0 to 3 to each region. Scores range from 0 for no lung abnormalities to 1 for interstitial infiltrates, 2 for interstitial and alveolar infiltrates (interstitial dominating), and 3 for interstitial and alveolar infiltrates (alveolar dominant). The total score should range between 0 and 18. The Brixia scoring system excluded other symptoms including pleural effusion and pulmonary vascular enlargements [20]. A Brixia score < 8 accounts for mild lung affection, while a Brixia score ≥ 8 accounts for a more severe affection [21].
CT chest was done. Total severity score (TSS) was calculated. TSS is a quantitative score assessing the inflammatory abnormalities in each of the five lobes of both lungs, including the presence of GGOs, consolidation, or mixed GGOs. Each lobe received a score between 0 and 4 points depending on how much of it was affected: (0) = 0 %, (1) = 1–25 %, (2) = 26–50 %, (3) = 51–75 %, or (4) = 76–100 %. The overall score, which varied from 0 to 20, was the sum of the points from each lobe [22]. A total score from 0 to 5 is classified as mild, 6 to 10 as moderate, and 11 to 20 as severe [23].
1.2. Statistical analysis
Statistical analysis was performed with IBM1® SPSS2® Statistics Version 25. Numerical data were presented as mean, standard deviation (SD). Data were explored for normality by checking the data distribution using Kolmogorov-Smirnov and Shapiro-Wilk tests. Data were analyzed using independent t-test for normally distributed data and Mann Whitney test for non-normally distributed data. Categorical data was analyzed by chi2 test. ROC analysis was done to detect the diagnostic value of studied markers. Correlation between quantitative variables was done by pearsons correlation. The significance level was set at P ≤ 0.05 within all tests.
2. Results
2.1. Most study participants were admitted to the ICU, with hypoxia, acidosis and altered biochemical investigations
Out of the 98 COVID-19 patients enrolled in this study, 47 were females (48 %) and 51 were males (52 %). 75 % of study participants required admission to the Intensive care unit (ICU) (Table 1).
Table 1.
Sex of the studied COVID-19 group and percentage of ICU admission.
| Frequency | Percent % | ||
|---|---|---|---|
| Sex | Female | 47 | 48 % |
| Male | 51 | 52 % | |
| Place of admission | Ward | 24 | 24.5 % |
| ICU | 74 | 75.5 % |
As shown in Table 2, COVID-19 group participants had lower mean levels of PH, PO2, HCO3 and O2RA than the normal average ranges, while the mean levels of FBS, neutrophils, ALT, LDH and CRP were higher than the normal average ranges.
Table 2.
Biochemical data of COVID-19 group.
| Minimum | Maximum | Mean | Normal values | |
|---|---|---|---|---|
| PH | 6.65 | 7.55 | 7.32 ± 0.19 | 7.35–7.45 |
| PCO2 | 20.6 | 184 | 43.64 ± 21.55 | 35–45 mmHg |
| PO2 | 11.9 | 136 | 53.35 ± 22.47 | 75–100 mmHg |
| HCO3 | 4.3 | 35 | 21.04 ± 6.36 | 22-26 mEq/L |
| O2RA | 50 | 97 | 82.44 ± 9.44 | 97–100 % |
| FBS | 100 | 873 | 308.87 ± 152.76 | 70–99 mg/dl Up to 130 in diabetics |
| Platelet count | 45 | 420 | 228.78 ± 87.73 | 150–450 × 103/uL |
| Neutrophil | 0.7 | 29.99 | 8.26 ± 6.21 | 2.6–6 × 103/uL |
| Lymphocyte | 0.2 | 7.1S | 1.15 ± 0.88 | 1–4.8 × 103/uL |
| ALT | 10 | 578 | 67.57 ± 104.03 | 7–55 U/L |
| LDH | 153 | 1229 | 484.2 ± 259.47 | 105–333 IU/L |
| CRP | 6 | 96 | 47.64 ± 32.3 | <10 mg/L |
O2RA: O2 at room air, FBS: Random blood sugar, ALT: alanine transaminase, LDH: lactate dehydrogenase, CRP: C-Reactive Protein.
2.2. Decreased expression of lncRNA MALAT-1 in COVID-19 group along with elevated expression of miR-200c-3p
Fig. 1 shows LncRNA MALAT-1 and miR-200c-3p relative gene expression in control and in COVID-19 groups. The relative gene expression of lncRNA MALAT-1 in COVID-19 patients was significantly lower than in control group (P value < 0.001), while that of miR-200c-3p in COVID-19 patients was significantly higher than in control group (P value < 0.001).
Figure 1.
LncRNA MALAT-1 and miR-200c-3p relative gene expression in control and in COVID-19 groups.
2.3. A significant decrease of the anti-inflammatory, anti-fibrotic SIRT1 in COVID-19 group sera
Fig. 2 shows that the protein level of SIRT1 in COVID-19 patients was significantly lower than in control group (P value < 0.001).
Figure 2.
SIRT1 serum protein levels in control and COVID-19 groups.
2.4. Classification of COVID-19 group according to the total severity score (TSS) scoring system
To detect the variations in the levels of relative gene expression of MALAT-1 and miR-200c-3p, and in the serum protein levels of SIRT1 between different degrees of COVID-19 severity, COVID-19 group patients were divided according to the degree of severity in computed tomography (CT) lung scans. Each patient was given a score according to the TSS scoring system [23]. After which, patients were divided into three groups. Patients with scores (0–5) were classified as mild, patients with scores (6–10) were classified as moderate, and patients with scores (11–20) were classified as severe. Then, we compared the expression levels of MALAT-1 and miR-200c-3p, along with SIRT1 protein serum levels among the three groups.
2.5. Increased disease severity correlates with further decreased expression of MALAT-1 and SIRT1, along with increased miR-200c-3p expression and GGO percentage
As shown in Table 3, with increased disease severity among COVID-19 patients, the expression level of MALAT-1 and the protein level of SIRT1 become significantly lower. On the other hand, the expression level of miR-200c-3p is significantly higher (P value < 0.001). In addition, there is a significant difference in the percentage of GGO between mild, moderate, and severe COVID-19 patients (P value < 0.001).
Table 3.
Comparison of MALAT-1 and miR-200c-3p gene expressions, SIRT1 protein levels and GGO according to disease severity (TSS).
| Mild N = 19 |
Moderate N = 48 |
Severe N = 31 |
P value | |
|---|---|---|---|---|
| MALAT-1 | 1.3 ± 0.9 | 0.31 ± 0.3 | 0.06 ± 0.09 | <0.001 |
| miR-200c-3p | 7.05 ± 6.56 | 14.15 ± 2.6 | 19.04 ± 3.19 | <0.001 |
| SIRT1 | 24.06 ± 10.56 | 14.64 ± 2.62 | 10.67 ± 2.42 | <0.001 |
| GGO percentage | 18.72 ± 8.54 | 42.23 ± 11.01 | 64.41 ± 10.89 | <0.001 |
2.6. Classification of COVID-19 group according to the presence of diabetes mellitus (DM) and/or hypertension (HTN)
COVID-19 group participants (N = 98) were divided according to the presence of DM and HTN into three groups: (a) nondiabetic non hypertensive (N = 19) (b) diabetic normotensive (N = 20) and (c) diabetic and hypertensive (N = 59), with a total of 79 diabetic patients and 59 hypertensive patients.
We compared MALAT-1 and miR-200c-3p relative gene expressions, and SIRT1 sera protein levels among diabetics and hypertensives in comparison to nondiabetics and non-hypertensive patients respectively.
2.7. DM and HTN may influence COVID-19 severity as shown by increased TSS score in DM and HTN patients
In addition, we detected statistically significant difference in the TSS between diabetics and nondiabetics, and between hypertensive and non-hypertensive COVID-19 patients (Table 4) in the three categories: mild, moderate, and severe COVID-19 patients (P value < 0.001).
Table 4.
TSS in diabetic and hypertensive versus non-diabetic, non-hypertensive COVID-19 patients.
| Diabetic (N = 79) | Non-Diabetic (N = 19) | P value | Hypertensive (N = 59) | Non- Hypertensive (N = 39) | P value | |||
|---|---|---|---|---|---|---|---|---|
| TSS | Mild | N | 6 | 14 | <0.001 | 0 | 20 | <0.001 |
| % | 7.60 % | 73.70 % | 0.00 % | 51.30 % | ||||
| Moderate | N | 42 | 5 | 29 | 18 | |||
| % | 53.20 % | 26.30 % | 49.20 % | 46.20 % | ||||
| severe | N | 31 | 0 | 30 | 1 | |||
| % | 39.20 % | 0.00 % | 50.80 % | 2.60 % |
TSS: Total severity score.
The percentage of mild cases was higher in non-diabetics (73.7 %). A higher percentage of moderate (53.2 %) and severe cases (39.2 %) is seen in diabetic COVID-19 patients, indicating the influence of DM on COVID-19 disease course.
Similarly, the percentage of severe cases was higher among hypertensive patients (50.8 %) and the percentage of mild cases was higher in non-hypertensive patients (51.3 %), denoting increased severity of COVID-19 in the presence of HTN.
2.8. COVID-19 participants previously diagnosed with DM and/or HTN show more profound upregulation of miR-200c-3p, and downregulation of MALAT-1 and SIRT1
Table 5 shows that the expression level of MALAT-1 and the protein level of SIRT1 are significantly decreased while the expression level of miR-200c-3p is significantly increased in diabetic and in hypertensive COVID-19 patients when compared to non-diabetic and non-hypertensive COVID-19 patients respectively. This denotes the influence of DM and HTN on the pathogenesis of COVID-19.
Table 5.
Comparison of MALAT-1 and miR-200c-3p expression levels, and SIRT1 protein level between diabetic and non-diabetic COVID-19 patients, and between hypertensive and non-hypertensive COVID-19 patients.
| Diabetic (N = 79) | Non-Diabetic (N = 19) | P value | Hypertensive (N = 59) | Non-Hypertensive (N = 39) | P value | |
|---|---|---|---|---|---|---|
| MALAT-1 | 0.19 ± 0.2 | 1.45 ± 0.83 | <0.001 | 0.09 ± 0.08 | 0.96 ± 0.75 | <0.001 |
| miR-200c-3p | 16.22 ± 3.42 | 6.04 ± 6.17 | <0.001 | 17.54 ± 2.92 | 9.27 ± 5.34 | <0.001 |
| SIRT1 | 12.97 ± 2.93 | 25.02 ± 10.36 | <0.001 | 11.7 ± 2.19 | 20.76 ± 8.3 | <0.001 |
2.9. O2 levels correlate positively to MALAT-1 and SIRT1, while GGO, TSS, and CXR score correlate positively to miR-200c-3p
Fig. 3 shows a significant positive correlation between MALAT-1 and O2 (r = 0.49, P value < 0.01), and a significant negative correlation between MALAT-1 and GGO, TSS, and CXR score with the following values respectively (r = 0.697, P value < 0.01), (r = 0.619, P value < 0.01) (r = 0.676, P value < 0.01).
Figure 3.
Correlations of MALAT-1 with O2 saturation, GGO, TSS and CXR score. TSS (Total severity score), GGO (ground glass opacity), CXR: chest x-ray, O2 on RA (Oxygen at room air).
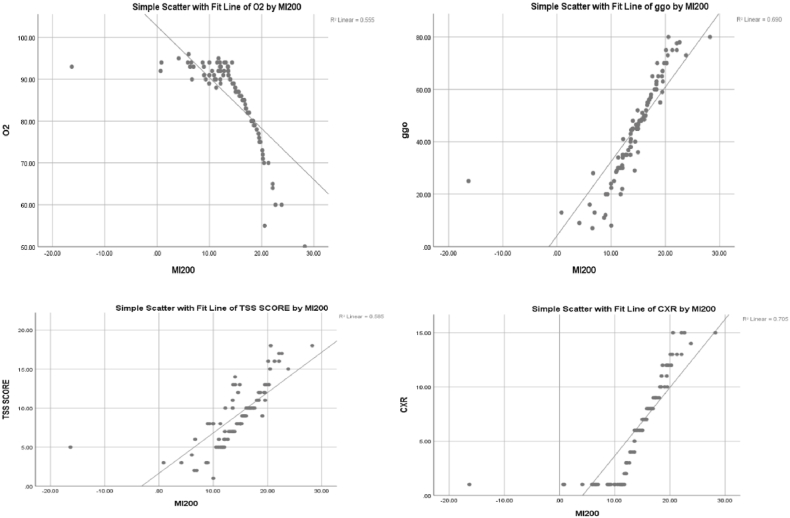
Fig. 4 shows a significant negative correlation of miR-200c-3p with O2 (r = 0.745, P value < 0.01), and a significant positive correlation of miR-200c-3p with GGO, TSS and CXR score with the following values respectively (r = 0.831, P value < 0.01), (r = 0.765, P value < 0.01), (r = 0.84, P value < 0.01).
Figure 4.
Correlations of miR-200c-3p with O2 saturation, GGO, TSS and CXR score. TSS (Total severity score), GGO (ground glass opacity), CXR: chest x-ray, O2 on RA (Oxygen at room air).
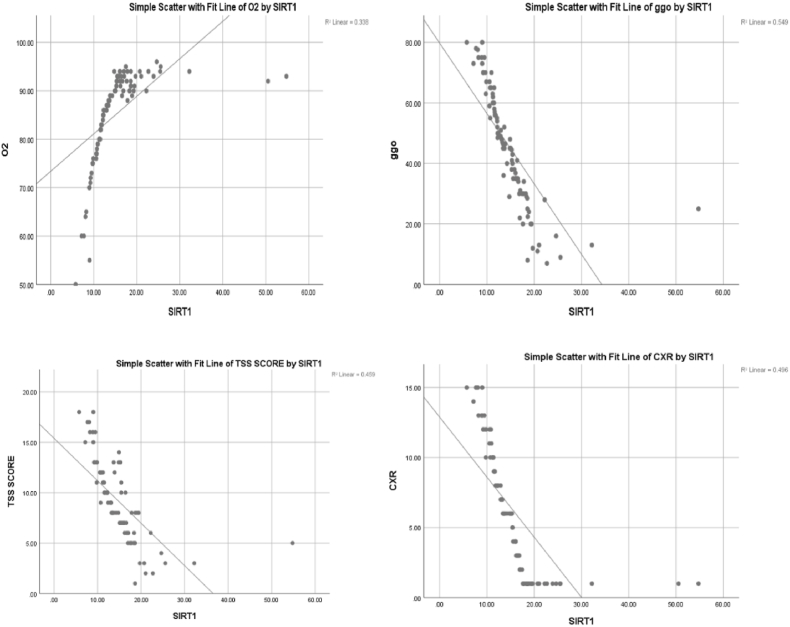
Fig. 5 shows a significant positive correlation between SIRT1 and O2 (r = 0.581, P value < 0.01), and a significant negative correlation between SIRT1 and GGO, TSS and CXR score with the following values respectively (r = 0.741, P value < 0.01), (r = 0.677, P value < 0.01) (r = 0.704, P value < 0.01).
Figure 5.
Correlation of SIRT1 with O2 saturation, GGO, TSS and CXR score. TSS (Total severity score), GGO (ground glass opacity), CXR: chest x-ray, O2 on RA (Oxygen at room air).
2.10. The correlation of MALAT-1 and SIRT1 is significantly positive, while miR-200c-3p correlates negatively to MALAT-1. Moreover miR-200c-3p negatively correlates to SIRT1
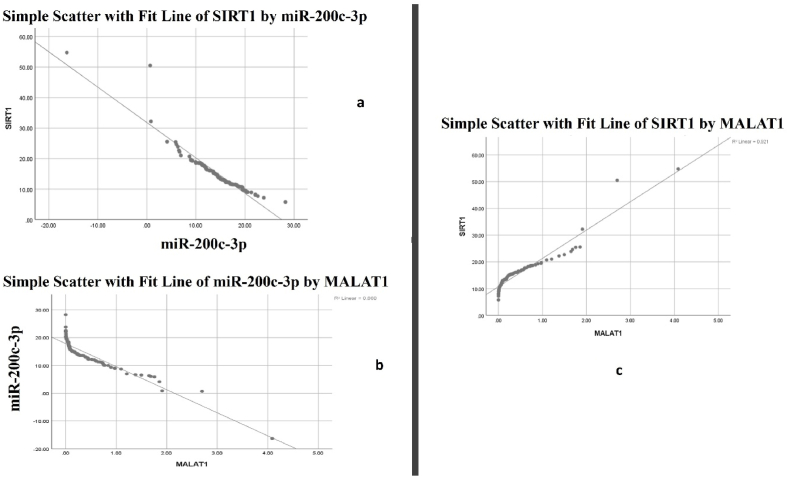
Fig. 6a shows a significant negative correlation between SIRT1 and miR-200c-3p (r = 0.941, P value < 0.01). Fig. 6b shows a significant negative correlation between miR-200c-3p and MALAT-1 (r = 0.927, P value < 0.01). Fig. 6c shows a significant positive correlation between SIRT1 and MALAT-1 (r = 0.96, P value < 0.01).
Figure 6.
a: Correlation analysis of SIRT1 and miR-200c-3p in COVID-19 patients. b: Correlation analysis of miR-200c-3p and MALAT-1 in COVID-19 patients. C: Correlation analysis of SIRT1 and MALAT-1 in COVID-19 patients.
2.11. The application of MALAT-1, SIRT1, and miRNA-200c-3p as diagnostic markers
Using ROC curve (Fig. 7), we found that serum MALAT-1 expression and Serum protein levels of SIRT1 could be used as diagnostic markers for COVID-19 at cut off values of 0.04 (95.9 % sensitivity) and 7.4 (98 % sensitivity) respectively, yet both show low specificity. Serum expression of miR-200c-3p could be used as a diagnostic marker for COVID-19 at a cut off value of 5.59, with sensitivity 94.9 %, and specificity 99 %
Figure 7.
ROC curve for MALAT-1, SIRT1 and miRNA-200c-3p.
3. Discussion
As with previous pandemics caused by coronaviruses, the underlying pathophysiology of COVID-19 is still being investigated to identify prognostic biomarkers and therapeutic opportunities.
Ninety-eight COVID-19 study participants were divided according to two parameters (i) the presence of previously diagnosed diabetes mellitus (DM) and/or hypertension (HTN) (ii) the severity of lung affection. Both genders are included almost equally, which coincides with previous studies that show no difference in the prevalence of COVID-19 infection between males and females [24].
To assess the severity of COVID-19, we calculated the Total Severity Score (TSS) in COVID-19 study group. TSS is a score that illustrates the degree of lung affection as seen in computed tomography (CT) lung scan. Each lobe is given a score, after which lobar scores are added to give a final TSS. TSS from 0 to 5 was classified as mild, 6 to 10 as moderate, and 11 to 20 as severe [23].
In this study, the TSS score was significantly higher among diabetic and hypertensive when compared to non-diabetic and non-hypertensive COVID-19 patients respectively. In fact, two meta-analysis studies previously declared an up to a 2.5-fold and a 2-fold higher risk of severe and fatal COVID-19 when associated with pre-existing hypertension and diabetes respectively [25,26].
Studying human genome expression is an important angle that could explain how individual genetic variations may impact COVID-19 severity and prognosis. Despite being extensively transcribed, only 2 % of RNAs code for proteins. Noncoding RNAs are important epigenetic regulators. They include microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). LncRNAs are long transcripts (>200 nucleotide), that are involved in viral infection, immunomodulation, and inflammatory response [27,28]. MiRNAs are short (∼22-nucleotide) noncoding RNAs that bind to the 3′-UTRs of their target mRNAs, leading to posttranscriptional silencing, translational repression, or RNA degradation. It was noticed that miRNAs levels are subjected to changes and play critical roles during viral infections as previously reported in H5N1 Influenza virus, Ebola virus, and HIV-1 [29,30].
miR-200c-3p is a noncoding miRNA known for its ability to regulate angiotensin converting enzyme II (ACE2) by binding to the 3’ untranslated region (UTR) region of its mRNA. ACE2 receptor (ACE2R) is an identified receptor for SARS-COV-2 [[31], [32], [33], [34]], previously reported that ACE2 mRNA expression cannot be relied on to display protein levels in a tissue as mRNA and protein quantities of ACE2 hugely vary within and across tissues. Wang et al. also exhibited low ACE2 protein levels in blood cells. It is thus valid to detect the expression levels of miRNAs regulating ACE2 during SARS-COV-2 infection [35,36].
ACE2 has many physiological roles in the lung tissue, among which, it physiologically hydrolyzes angiotensin II (AngII) to Ang-(1–7) resulting in anti-inflammatory and antifibrotic effects [[37], [38], [39]]. Comprehensively, it is hypothesized that the upregulation of miR-200c-3p acts as a defensive mechanism against viral infection, by downregulating ACE2R, consequently decreasing SARS-COV-2 host cell entry. Moreover, as a consequence of decreased ACE2 by miR-200c-3p, the profibrotic AngII increases [40]. AngII acts in a pro-inflammatory and a pro-oxidant manner inducing vasoconstriction, enhanced inflammation, thrombosis, and increased collagen synthesis in lung fibroblasts [41,74] causing acute lung injury, alveolar edema, and increased incidence of secondary lung fibrosis [[42], [43], [44], [45]]. Interestingly, ACE2 can counter-regulate the wound healing response [40]. The forementioned mechanism is thought to participate in the pathogenesis of acute respiratory distress syndrome in H5N1 virus infection, in severe pneumonia, and in interstitial lung disease [8,46,47]. Indeed, Fibrosis is a well-known untreatable long-term complication of covid19 that may be precipitated by the same mechanism.
In the present study, miR-200c-3p expression levels are significantly higher in COVID-19 patients. Moreover, the expression level of miR-200c-3p is significantly increased with the increase in severity among COVID-19 patients. Furthermore, we found a significant negative correlation of miR-200c-3p with O2 saturation, and a significant positive correlation of miR-200c-3p with GGO, TSS score and CXR score. This mimics results of studies conducted by [48,77], who found greater expression of miR-200c-3p in severe COVID-19 patients who were on oxygen therapy (P < 0.0001).
Another study compared miR-200c-3p at times of admission and discharge of COVID-19 patients. They revealed higher expression levels at the time of discharge, along with a negatively correlated IL-6 expression, which may imply a defensive response by the body, where higher miR-200c-3p expression occurs to decrease viral entry to host cell [49].
Previous research using bioinformatics analysis and luciferase reporter assays suggested that lncRNA Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT-1) directly binds to and downregulates miR-200c-3p. In addition, miR-200c-3p could bind to 3′UTR of Silent Information Regulator T-1(SIRT1), inhibiting it – an effect reversed by the overexpression of MALAT-1 [[50], [51], [52]]. SIRT1 is a NAD + -dependent histone deacetylase that targets several transcriptional factors that mediate inflammation, stress, mitochondrial biogenesis, insulin secretion, and aging [53,76]. SIRT1 can suppress the transcription of several cytokine genes by directly regulating the NF-κB pathway [54]. Additionally, SIRT- 1 can inhibit A Disintegrin and Metalloproteinase Domain 17 (ADAM17) activation, consequently decreasing proinflammatory cytokines TNF-α and IL-6 [15]. Moreover, SIRT1 directly decreases viral replication [55]. It was also established that SIRT1 can bind to the ACE2 promoter increasing its expression [17].
In this study our aim was to evaluate the effect of a possible interaction of lncRNA MALAT-1, miR-200c-3p and SIRT1 in COVID-19 patients, and to correlate the severity of clinical symptoms and signs to their serum expression levels.
MALAT-1 is a lncRNA that regulates genes involved in immunomodulation and cell cycle regulation [27]. The relative gene expression of MALAT-1 in COVID-19 patients in this study is significantly lower than in control group (p value < 0.001). Moreover, the expression level of MALAT-1 is significantly decreased with the increase in severity among COVID-19 patients. This agrees with [56] who reported lower MALAT-1 expression in monocytes and macrophages from bronchoalveolar lavage (BAL) samples of mild COVID-19 patients. In another study on CD4+ T cells from BAL of COVID-19 patients, MALAT-1 was upregulated in Mild cases and downregulated in Severe cases [57].
A significant positive correlation is detected between MALAT-1 and O2 saturation. On the other hand, MALAT-1 significantly negatively correlates to Ground glass opacity (GGO), TSS score, and CXR score. Previous data demonstrate that MALAT-1 downregulation is involved in pulmonary profibrotic pathogenesis due to atypical macrophage activation favoring M2 upregulation [56]. Furthermore, previous studies on MALAT-1-knockout mice reported lower expression of the immunosuppressant IL-10 in Th1 and Th2 cells, accentuating the immune responses to infection [58], which imitates exaggerated immune responses that occur in SARS-COV-2 infection. MALAT- 1 inhibition was also found to direct CD4+ T cells away from the T regulatory cell (Tregs) subtype, which adds more evidence to the protective immunomodulatory anti-inflammatory role of MALAT-1, as Studies have shown that Tregs are able to repress cytokine secretion and play a crucial role in preventing autoimmunity [59].
In this study, SIRT1 serum protein levels are significantly lower in COVID-19 patients than in control group. Moreover, SIRT1 expression is significantly decreased along with an increase in severity among COVID-19 patients. In addition, a significant positive correlation between SIRT1 and O2 saturation is detected, along with a significant negative correlation with GGO, TSS score and CXR score, providing proof of the possible protective effect of SIRT1.
This is in accordance with [60] who detected significantly decreased SIRT1 expression in COVID-19 patients, which was negatively correlated with p53, IL1-β, and TNF. [16] showed that SIRT1 can reduce inflammation in COVID-19 patients. [75] found that SIRT1 inactivation was linked to COVID-19 severity and its low level served as a predictor of COVID-19 that was not under control.
Statistical correlations in our study show a highly significant positive correlation between MALAT-1 and SIRT1, while highly significant negative correlations were found between MALAT-1 with miR-200c-3p and miR-200c-3p with SIRT1. Previous studies elaborated the possible regulatory role of MALAT-1 on miR-200c-3p and SIRT1, through interactive sites, where MALAT-1 could directly bind miR-200c-3p, downregulating it. Similarly, in brain microvascular endothelial cells (BMECs), miR-200c-3p can modulate SIRT1 expression by binding to SIRT1 3′UTR. Furthermore, MALAT-1 was able to reverse the inhibitory effect of miR-200c-3p on SIRT1 [32], suggesting a novel feedback loop.
The comparison of MALAT-1 and SIRT1 expression between hypertensive and non-hypertensive COVID-19 patients reveales significantly decreased expression levels in hypertensives. This may be explained by the protective antihypertensive effect of MALAT-1 and SIRT1 as mentioned by [61,62] respectively.
A similar observation was seen in diabetic COVID-19 patients, who had significantly decreased expression levels of MALAT-1 and SIRT1 when compared to non-diabetics. This agrees with [63] who described downregulated SIRT1 in trigeminal sensory neurons of diabetic mice and with [73], but may be not in line with other studies in which MALAT-1 was upregulated in diabetes [64,65]. This may be explained with variance in expression of MALAT-1 in COVID-19 patients [66].
Another important finding is that the expression level of miR-200c-3p is significantly increased in diabetic and in hypertensive COVID-19 patients [48]. mentioned an increased level of miRNA200 with hypertension, and [67] stated that the expression of the miRNA200 family is strongly induced in islets of diabetic mice and that beta cell specific overexpression of miR-200c-3p in mice is sufficient to induce beta cell apoptosis and lethal type 2 diabetes. This could explain the higher liability for type II DM post-COVID-19 infection.
Receiver operating characteristic (ROC) curves compare sensitivity versus specificity across a range of values, enabling the detection of the best cut-off value that could be used clinically as a diagnostic biomarker. High sensitivity indicates a high negative predictive value, an ideal “rule-out” test. High specificity indicates a high positive predictive value, an ideal “rule-in” test [68]. Using ROC curve, we find that serum MALAT-1 expression and Serum protein levels of SIRT1 could be used as diagnostic markers for COVID-19 at cut off values of 0.04 (95.9 % sensitivity) and 7.4 (98 % sensitivity) respectively, yet both show low specificity. Serum expression of miR-200c-3p could be used as a diagnostic marker for COVID-19 at a cut off value of 5.59, with sensitivity 94.9 %, and specificity 99 %.
As regards the clinical picture, 75.5 % of COVID-19 study participants were admitted to the intensive care unit (ICU), most of them suffered from metabolic acidosis and hypoxia [69]. illustrated the vicious cycle between the development of hypoxia and acidosis in COVID-19 patients. Inflammation, difficulty with gas exchange in the lungs and thrombosis contribute to the onset of acidosis. According to the Verigo-Bohr effect, a drop in blood pH decreases oxygen saturation, which exacerbates acidosis followed by deterioration of the patient's condition. Decreased pH can also change the conformation of the SARS-COV-2 S-protein, which reduces the affinity and the avidity of the protective antibodies. Furthermore, Hypoxia and acidosis cause immune system dysregulation and multidirectional pro- and anti-inflammatory reactions, resulting in the formation of a cytokine storm, a major indication for ICU admission.
The mean random Blood Glucose (RBG) of COVID-19 group is high. Previous research suggested that diabetic patients were more susceptive to SARS-COV-2 infection and complications. In addition, high blood glucose level in previously non-diabetic patients is indicative of poorer outcomes [70,71].
Furthermore, laboratory findings show higher-than-normal mean levels of neutrophils, neutrophil to lymphocyte ratio (NLR), alanine aminotransferase (ALT), lactate dehydrogenase (LDH), and C-reactive protein (CRP) in COVID-19 patients. These findings are in accordance with [72] who compared these laboratory data between the non-severe COVID-19 patients and those with the severe course. They reported increased neutrophils, NLR, ALT, LDH, CRP and blood glucose levels in the severe group.
Collectively, we hypothesize a possible protective role of MALAT-1 and SIRT1 in SARS-COV-2 infection, possibly by suppressing miR-200c-3p, which decreases inflammation and fibrosis, improving diseases outcomes. Furthermore, MALAT-1 can be used as a biomarker for the severity of the disease.
4. Conclusion
We found that the expression of lncRNA MALAT-1 and the serum protein level of SIRT1 in COVID-19 patients are significantly lower than in control group (P value < 0.001). Moreover, with increased disease severity and with the presence of DM or HTN, lower results are recorded. The expression of miR-200c-3p in COVID-19 patients is significantly higher than in control group (P value < 0.001), with accentuated results seen in severe cases and in DM or HTN cases when compared to moderate and mild cases, non-diabetics, and non-hypertensives.
Ground glass appearance, chest x-ray score and total severity score of CT lung scans positively correlate to miR-200c-3p, and negatively correlate to MALAT-1 and SIRT1. On the other hand, Oxygen saturation positively correlates to MALAT-1 and SIRT1, and negatively correlates to miR-200c-3p.
SIRT1 positively correlates to MALAT-1, while it negatively correlates to miR-200c-3p. MALAT-1 and miR-200c-3p serum relative gene expression levels could be used as diagnostic markers for COVID-19, at cut off values of 0.04 (95.9 % sensitivity) and 7.4 (98 % sensitivity) respectively, yet both show low specificity. Serum expression of miR-200c-3p could be used as a diagnostic marker for COVID-19 at a cut off value of 5.59, with sensitivity 94.9 %, and specificity 99 %.
5. Recommendations
We recommend that more studies should be done involving a larger sample size to confirm our results. Indeed, further exploration of molecular pathways affected by MALAT-1, miR-300c-3p and SIRT1 in viral infections could open pathways for new prognostic and/or therapeutic markers targeting fatal pandemics affecting the world every now and then.
Ethics approval and consent to participate
Consents from included study participants were taken and are available upon request. This study has been conducted in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. The Research Ethics Committee of Kasralainy School of Medicine has approved this study accordingly with code: MS-604-2021.
Funding
This study was self-funded by the authors.
Authors' contributions
OS and MEA performed the practical work. HE researched the study point and wrote the manuscript. YAE collected and revised study participants data. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Contributor Information
Olfat Shaker, Email: Olfat.shaker@kasralainy.edu.eg.
Monica El Amir, Email: monicaelamirmina@cu.edu.eg.
Yasmine Abd Elfatah, Email: yasmineabdelfatah@kasralainy.edu.eg.
Heba M. Elwi, Email: Heba.elwi@kasralainy.edu.eg.
Abbreviations
- ACE2
Angiotensin converting enzyme 2
- ADAM17
A Disintegrin and Metalloproteinase Domain 17
- ALT
Alanine aminotransferase
- ARDS
Acute respiratory distress syndrome
- BAL
Bronchoalveolar lavage
- CBC
Complete blood count
- COVID-19
Coronavirus Disease 2019
- CRP
C- reactive protein
- CXR
Chest x-ray
- ELISA
Enzyme-linked immunosorbent assay
- ESR
Erythrocyte sedimentation rate
- GGO
Ground glass opacity
- ICU
Intensive care unit
- IL1B
Interleukin 1 beta
- LDH
Lactate dehydrogenase
- lincRNAs
Long intergenic noncoding ribonucleic acid
- LncRNAs
Long noncoding ribonucleic acid
- MALAT-1
Metastasis associated lung adenocarcinoma transcript 1
- miRNA
Micro ribonucleic acid
- MRES
MiRNA response elements
- mRNA
Messenger ribonucleic acid
- ncRNAs
Non-coding ribonucleic acid
- NF-kB
Nuclear factor kappa B
- NLR
Neutrophil to lymphocyte ratio
- qRT PCR
Quantitative real time polymerase chain reaction
- RAS
Renin angiotensin system
- RBG
Random blood glucose
- SARS-COV
Severe acute respiratory syndrome coronavirus
- SIRT1
Silent information regulator 1
- TGF
Transforming growth factor
- TH1
Type 1 helper cells
- TH2
Type 2 helper cells
- TNF
Tumor necrosis factor
- Tregs
Regulatory T cells
- TSS
Total severity score
- UTR
Untranslated region
- WHO
World Health Organization
Data availability
Data will be made available on request.
References
- 1.Tyrrell D.A., Bynoe M.L. Cultivation of viruses from a high proportion of patients with colds. Lancet. 1966;(1):76–77. doi: 10.1016/S0140-6736(66)92364-6. 1966. [DOI] [PubMed] [Google Scholar]
- 2.Sampath S., Khedr A., Qamar S., et al. Pandemics throughout the history. Cureus. 2021;13(9) doi: 10.7759/cureus.18136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M., Wang H., Tian L., et al. COVID-19 vaccine development: milestones, lessons and prospects. Sig. Transduct. Target Ther. 2022;7:146. doi: 10.1038/s41392-022-00996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boopathi S., Poma A., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2020;39(9):1–10. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta O., Bhandari P., Raut A., et al. Coronavirus disease (COVID-19): comprehensive review of clinical presentation. Front. Public Health. 2021;8 doi: 10.3389/fpubh.2020.582932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J., Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int. J. Mol. Sci. 2018;19(5):1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelhamid A., El Deeb M., Zaafan M. The protective effect of xanthenone against LPS-induced COVID-19 acute respiratory distress syndrome (ARDS) by modulating the ACE2/Ang-1-7 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2022;26(14):5285–5296. doi: 10.26355/eurrev_202207_29320. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q., Du J., Yu X., et al. miRNA-200c-3p is crucial in acute respiratory distress syndrome. J. Cell Disc. 2017;3(1):1–17. doi: 10.1038/celldisc.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattick J., Makunin I. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 10.Wang C., Wang L., Ding Y., et al. LncRNA structural characteristics in epigenetic regulation. Int. J. Mol. Sci. 2017;18(12):2659. doi: 10.3390/ijms18122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao M., Wang S., Li Q., et al. MALAT-1: a long non-coding RNA highly associated with human cancers. Oncol. Lett. 2018;16(1):19–26. doi: 10.3892/ol.2018.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q., Zhang C., Chen R., et al. Disrupting MALAT-1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Canc. Lett. 2016;383(1):28–40. doi: 10.1016/j.canlet.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y., Ma L. New insights into long non-coding RNA MALAT-1 in cancer and metastasis. Cancers. 2019;11(2):216. doi: 10.3390/cancers11020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C., Zhou M., Ge Y., et al. SIRT1 and aging related signaling pathways. Mech. Ageing Dev. 2020;187 doi: 10.1016/j.mad.2020.111215. [DOI] [PubMed] [Google Scholar]
- 15.Miller R., Wentzel A., Richards G. COVID-19: NAD+ deficiency may predispose the aged, obese and type2 diabetics to mortality through its effect on SIRT1 activity. Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.110044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chattree V., Singh K., Singh K., et al. A comprehensive review on modulation of SIRT1 signaling pathways in the immune system of COVID-19 patients by phytotherapeutic melatonin and epigallocatechin-3-gallate. J. Food Biochem. 2022;1:15. doi: 10.1111/jfbc.14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke N., Belyaev N., Lambert D., et al. Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin. Sci. 2014;126(7):507–516. doi: 10.1042/CS20130291. [DOI] [PubMed] [Google Scholar]
- 18.McLachlan C. The angiotensin-converting enzyme 2 (ACE2) receptor in the prevention and treatment of COVID-19 are distinctly different paradigms. Clin. Hypert. 2020;26(1):1–3. doi: 10.1186/s40885-020-00147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barangi S., Hayes A.W., Karimi G. The role of lncRNAs/miRNAs/Sirt1 axis in myocardial and cerebral injury. Cell Cycle. 2023;22(9):1062–1073. doi: 10.1080/15384101.2023.2172265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Setiawati R., Widyoningroem A., Handarini T., et al. Modified chest X-ray scoring system in evaluating severity of COVID-19 patient in Dr. Soetomo General Hospital Surabaya, Indonesia. Int. J. Gen. Med. 2021;14:2407. doi: 10.2147/IJGM.S310577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balbi M., Caroli A., Corsi A., et al. Chest X-ray for predicting mortality and the need for ventilatory support in COVID-19 patients presenting to the emergency department. Eur. Radiol. 2021;31(4):1999–2012. doi: 10.1007/s00330-020-07270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elmokadem A., Mounir A., Ramadan Z., et al. Comparison of chest CT severity scoring systems for COVID-19. Eur. Radiol. 2022;32(5):3501–3512. doi: 10.1007/s00330-021-08432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagci A., Sarinoglu R., Bilgin H., et al. Relationship of the cycle threshold values of SARS-CoV-2 polymerase chain reaction and total severity score of computerized tomography in patients with COVID 19. Int. J. Infect. Dis. 2020;101:160–166. doi: 10.1016/j.ijid.2020.09.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin J.M., Bai P., He W., Wu F., Liu X.F., Han D.M., et al. Gender differences in patients with COVID-19: focus on severity and mortality. Front. Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippi G., Wong J., Henry B.M. Hypertension in patients with coronavirus disease 2019 (COVID-19): a pooled analysis. Pol. Arch. Intern. Med. 2020;130(4):304–309. doi: 10.20452/pamw.15272. [DOI] [PubMed] [Google Scholar]
- 26.Mantovani A., Byrne C., Zheng M., et al. Diabetes as a risk factor for greater COVID-19 severity and in-hospital death: a meta-analysis of observational studies. Nutr. Metabol. Cardiovasc. Dis. 2020;30(8):1236–1248. doi: 10.1016/j.numecd.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S., Zhang E., Senapati P., et al. A novel angiotensin II–induced long noncoding RNA giver regulates oxidative stress, inflammation, and proliferation in vascular smooth muscle cells. Circ. Res. 2018;(123):12. doi: 10.1161/CIRCRESAHA.118.313207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahaa S., Sahab C., Dutta S., et al. In silico analysis of altered expression of long non-coding RNA in SARS-CoV-2 infected cells and their possible regulation by STAT1, STAT3 and interferon regulatory factors. Heliyon. 2021;7(3) doi: 10.1016/j.heliyon.2021.e06395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deogharia M., Gurha P. The “guiding” principles of noncoding RNA function. Wiley Inter. Rev. RNA. 2021;13(4) doi: 10.1002/wrna.1704. [DOI] [PubMed] [Google Scholar]
- 30.Kianmehr A., Faraoni I., Kucuk O., et al. Epigenetic alterations and genetic variations of angiotensin-converting enzyme 2 (ACE2) as a functional receptor for SARS-CoV-2: potential clinical implications. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40(8):1587–1598. doi: 10.1007/s10096-021-04264-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trojanowicz B., Imdahl T., Ulrich C., et al. Circulating miR-421 targeting leucocytic angiotensin converting enzyme 2 is elevated in patients with chronic kidney disease. Nephron. 2019;141(1):61–74. doi: 10.1159/000493805. [DOI] [PubMed] [Google Scholar]
- 32.Wang S., Han X., Mao Z., et al. MALAT lncRNA induces autophagy and protects brain microvascular endothelial cells against oxygen–glucose deprivation by binding to miR-200c-3p and upregulating SIRT1 expression. Neuroscience. 2019;397:116–126. doi: 10.1016/j.neuroscience.2018.11.024. [DOI] [PubMed] [Google Scholar]
- 33.Papannarao J., Schwenke D., Manning P., et al. medRxiv; 2021. Upregulated miR-200c May Increase the Risk of Obese Individuals to Severe COVID-19. [DOI] [Google Scholar]
- 34.Wang Y., Wang Y., Luo W., et al. A comprehensive investigation of the mRNA and protein level of ACE2, the putative receptor of SARS-CoV-2, in human tissues and blood cells. Int. J. Med. Sci. 2020;17(11):1522. doi: 10.7150/ijms.46695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chauhan N., Jaggi M., Chauhan S., et al. COVID-19: fighting the invisible enemy with microRNAs. Expert Rev. Anti-Infect. Ther. 2021;19(2):137–145. doi: 10.1080/14787210.2020.1812385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hum C., Loiselle J., Ahmed N., et al. MicroRNA mimics or inhibitors as antiviral therapeutic approaches against COVID-19. Drugs. 2021;81(5):517–531. doi: 10.1007/s40265-021-01474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lelis D., Freitas D., Machado A., et al. Angiotensin-(1-7), adipokines and inflammation. Metabolism. 2019;95:36–45. doi: 10.1016/j.metabol.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Dalan R., Bornstein S., El-Armouche A., et al. The ACE-2 in COVID-19: foe or friend? Horm. Metab. Res. 2020;52:257–263. doi: 10.1055/a-1155-0501. Google Scholar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng Y., Yu C., Li W., et al. Angiotensin-converting enzyme 2/angiotensin-(1-7)/Mas axis protects against lung fibrosis by inhibiting the MAPK/NF-kappaB pathway. Am. J. Respir. Cell Mol. Biol. 2014;50:723–736. doi: 10.1165/rcmb.2012-0451OC. [DOI] [PubMed] [Google Scholar]
- 40.Uhal B., Dang M., Dang V., et al. Cell cycle dependence of ACE-2 explains downregulation in idiopathic pulmonary fibrosis. Eur. Respir. J. 2013;42:198–210. doi: 10.1183/09031936.00015612. [Google Scholar] [DOI] [PubMed] [Google Scholar]
- 41.Verdecchia P., Cavallini C., Spanevello A., et al. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo W., Zhao X., Chen Y. In Molecular Biology of the SARS-Coronavirus. 2010. SARS coronavirus and lung fibrosis; pp. 247–258. [DOI] [Google Scholar]
- 43.Bourgonje A., Abdulle A., Timens W., et al. Angiotensin‐converting enzyme 2 (ACE2), SARS‐CoV‐2 and the pathophysiology of coronavirus disease 2019 (COVID‐19) J. Pathol. 2020;251(3):228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giovanni A., Rossi 1, Sacco1 O., et al. Can resveratrol-inhaled formulations Be considered potential adjunct treatments for COVID-19. J. Front. Immunol. 2021;(12):1591. doi: 10.3389/fimmu.2021.670955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Liu C., Hsu T., et al. Upregulation of ACE2 and TMPRSS2 by particulate matter and idiopathic pulmonary fibrosis: a potential role in severe COVID-19. Part. Fibre Toxicol. 2021;18:11. doi: 10.1186/s12989-021-00404-3. [Google Scholar] [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Z., Tao J., Zuo T., et al. The correlation between miR-200c and the severity of interstitial lung disease associated with different connective tissue diseases. Scand. J. Rheumatol. 2017;46(2):122–129. doi: 10.3109/03009742.2016.1167950. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Q., Wang Q., Zhao B., et al. Identification of a SARS-CoV-2 virus-encoded small non-coding RNA in association with the neurological disorders in COVID-19 patients. Signal Transduct. Targeted Ther. 2022;7(1):1–3. doi: 10.1038/s41392-022-00969-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pimenta R., Viana N., dos Santos G., et al. MiR-200c-3p expression may be associated with worsening of the clinical course of patients with COVID-19. Mol. Biol. Res. Commun. 2021;10(3):141–147. doi: 10.22099/mbrc.2021.40555.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdolahi S., Hosseini M., Rezaei R., et al. Evaluation of miR-200c-3p and miR-421-5p levels during immune responses in the admitted and recovered COVID-19 subjects. J. Mol. Epidemiol. Evol. Genet. Infect. Dise. 2022;98 doi: 10.1016/j.meegid.2022.105207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuo Y., Chen L., He X., Ye Z., Li L., Liu Z., et al. Atorvastatin regulates MALAT-1/miR-200c/NRF2 activity to protect against podocyte pyroptosis induced by high glucose. Diabetes, Metab. Syndrome Obes. Targets Ther. 2021;14:1631–1645. doi: 10.2147/DMSO.S298950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shan W., Xu H., Zhengchun M., et al. MALAT-1 lncRNA induces autophagy and protects brain microvascular endothelial cells against oxygen–glucose deprivation by binding to miR-200c-3p and upregulating SIRT1 expression. Neuroscience. 2019;397:116–126. doi: 10.1016/j.neuroscience.2018.11.024. [DOI] [PubMed] [Google Scholar]
- 52.Xiao H., Tang K., Liu P., et al. LncRNA MALAT-1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6(35):38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitada M., Ogura Y., Monno I., et al. Sirtuins and type 2 diabetes: role in inflammation, oxidative stress, and mitochondrial function. Front. Endocrinol. 2019;10:187. doi: 10.3389/fendo.2019.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajendrasozhan S., Yang S., Kinnula V., et al. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2008;177(8):861–870. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Budayeva H., Rowland E., Cristea I. Intricate roles of mammalian sirtuins in defense against viral pathogens. J. Virol. 2016;90(1):5–8. doi: 10.1128/JVI.03220-14. https://journals.asm.org/doi/full/10.1128/JVI.03220-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui H., Banerjee S., Guo S., et al. Long noncoding RNA MALAT-1 regulates differential activation of macrophages and response to lung injury. JCI Insight. 2019;4(4) doi: 10.1172/jci.insight.124522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang K., Wang C., Vagts C., et al. Long non-coding RNAs (lncRNAs) NEAT1 and MALAT-1 are differentially expressed in severe COVID-19 patients: an integrated single cell analysis. PLoS One. 2021;17(1) doi: 10.1371/journal.pone.0261242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hewitson J., West K., James K., et al. MALAT-1 suppresses immunity to infection through promoting expression of Maf and IL-10 in Th cells. J. Immunol. 2020;204(11):2949–2960. doi: 10.4049/jimmunol.1900940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balandya E., Wieland‐Alter W., Sanders K., et al. Human seminal plasma fosters CD 4+ regulatory T‐cell phenotype and transforming growth factor‐β1 expression. Am. J. Reprod. Immunol. 2012;68(4):322–330. doi: 10.1111/j.1600-0897.2012.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bordoni V., Tartaglia E., Sacchi A., et al. The unbalanced p53/SIRT1 axis may impact lymphocyte homeostasis in COVID-19 patients. Int. J. Infect. Dis. 2021;105:49–53. doi: 10.1016/j.ijid.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G., Li Y., Peng Y., et al. Association of polymorphisms in MALAT-1 with risk of coronary atherosclerotic heart disease in a Chinese population. Lipids Health Dis. 2018;17(1):1–7. doi: 10.1186/s12944-018-0728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren C., Wu Z., Wang W., et al. SIRT1 exerts anti-hypertensive effect via FOXO1 activation in the rostral ventrolateral medulla. Free Radic. Biol. Med. 2022;188:1–13. doi: 10.1016/j.freeradbiomed.2022.06.003. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y., Zhao X., Wu X., et al. microRNA-182 mediates Sirt1-induced diabetic corneal nerve regeneration. Diabetes. 2016;65(7):2020–2031. doi: 10.2337/db15-1283. [DOI] [PubMed] [Google Scholar]
- 64.Liu J., Yao J., Li X., et al. Pathogenic role of lncRNA-MALAT-1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5(10):e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puthanveetil P., Chen S., Feng B., et al. Long non‐coding RNA MALAT 1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell Mol. Med. 2015;19(6):1418–1425. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plowman T., Lagos D. Non‐coding rnas in covid‐19: Emerging insights and current questions. Non-Coding RNA. 2021;7(3) doi: 10.3390/ncrna7030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belgardt B., Ahmed K., Spranger M., et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat. Med. 2015;21(6):619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 68.Florkowski C. Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: communicating the performance of diagnostic tests. Clin. Biochem. Rev. 2008;29(Suppl 1):S83. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2556590/ [PMC free article] [PubMed] [Google Scholar]
- 69.Nechipurenko Y., Semyonov D., Lavrinenko I., et al. The role of acidosis in the pathogenesis of severe forms of COVID-19. Biology. 2021;10(9):852. doi: 10.3390/biology10090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu J., Huang J., Zhu G., et al. Elevation of blood glucose level predicts worse outcomes in hospitalized patients with COVID-19: a retrospective cohort study. BMJ Open Diab. Res. Care. 2020;8(1) doi: 10.1136/bmjdrc-2020-001476. https://drc.bmj.com/content/8/1/e001476.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Norris T., Razieh C., Yates T., et al. Admission blood glucose level and its association with cardiovascular and renal complications in patients hospitalized with COVID-19. Diabetes Care. 2022;45(5):1132–1140. doi: 10.2337/dc21-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ghahramani S., Tabrizi R., Lankarani K., et al. Laboratory features of severe vs. non-severe COVID-19 patients in Asian populations: a systematic review and meta-analysis. Eur. J. Med. Res. 2020;25(1):1–10. doi: 10.1186/s40001-020-00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Donmez G., Outeiro T. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol. Med. 2013;5(3):344–352. doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lambert D., Hooper N., Turner A. Angiotensin-converting enzyme 2 and new insights into the renin-angiotensin system. Biochem. Pharmacol. 2021;75:781–786. doi: 10.1016/j.bcp.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nemeth Z., Kiss E., Takacs I. The role of epigenetic regulator SIRT1 in balancing the homeostasis and preventing the formation of specific “soil” of metabolic disorders and related cancers. Front. Biosci.-Landmark. 2022;27(9):253. doi: 10.31083/j.fbl2709253. [DOI] [PubMed] [Google Scholar]
- 76.Poulose N., Raju R. Sirtuin regulation in aging and injury. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2015;1852(11):2442–2455. doi: 10.1016/j.bbadis.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roustai Geraylow K., Hemmati R., Kadkhoda S., et al. miRNA expression in COVID-19. Gene Rep. 2022;28 doi: 10.1016/j.genrep.2022.101641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.