Abstract
Mis-folding of the prion protein (PrP) is known to cause neurodegenerative disease; however, the native function of this protein remains poorly defined. PrP has been linked with many cellular functions, including cellular proliferation and senescence. It is also known to influence epidermal growth factor receptor (EGFR) signaling, a pathway that is itself linked with both cell growth and senescence. Adult neural stem cells (NSCs) persist at low levels in the brain throughout life and retain the ability to proliferate and differentiate into new neural lineage cells. KO of PrP has previously been shown to reduce NSC proliferative capacity. We used PrP KO and WT NSCs from adult mouse brain to examine the influence of PrP on cellular senescence, EGFR signaling, and the downstream cellular processes. PrP KO NSCs showed decreased cell proliferation and increased senescence in in vitro cultures. Expression of EGFR was decreased in PrP KO NSCs compared with WT NSCs and additional supplementation of EGF was sufficient to reduce senescence. RNA-seq analysis confirmed that significant changes were occurring at the mRNA level within the EGFR signaling pathway and these were associated with reduced expression of mitochondrial components and correspondingly reduced mitochondrial function. Metabolomic analysis of cellular energy pathways showed that blockages were occurring at critical sites for production of energy and biomass, including catabolism of pyruvate. We conclude that, in the absence of PrP, NSC growth pathways are downregulated as a consequence of insufficient energy and growth intermediates.
Keywords: prion, PrP, senescence, epidermal growth factor, EGFR, mitochondria, glycolysis
The prion protein (PrP) is most widely known for its causative role in the transmissible spongiform encephalopathies or prion diseases. Prion diseases encompass a family of neurodegenerative conditions that affect humans and animals. During disease PrP becomes misfolded into conformers that can recruit and convert more PrP, thus resulting in propagation through the brain (1, 2). While this change from normal PrP into its disease-associated conformers is deleterious in all prion diseases, much remains to be determined about the normal function of PrP within the cell.
PrP KO mice show limited consequences of the complete loss of PrP (3, 4) and, therefore, the importance of PrP was considered questionable. However, despite developing normally and having a normal life expectancy, PrP KO mice do have more subtle phenotypes, some of which only become apparent under stress conditions. In an unchallenged environment, PrP KO differences from WT mice include defects in sleep wake cycles/circadian rhythms (5) and peripheral myelination (6). When PrP KO mice are stressed, further phenotypes are discovered; for example, with increasing exercise stress KO mice demonstrated less muscle capacity of the heart and skeletal system (7), seizure inducing agents showed PrP KO mice have greater susceptibility to seizure (8) and when challenged with a severe brain insult such as cerebral occlusion, lesions are more extensive in PrP KO mice compared with WT (9, 10). The increased infarct size may be related to a function of PrP in protection against oxidative stress (11, 12, 13, 14) and may also be influenced by a lesser capacity to repopulate lost neurons from neural stem cell (NSC) reserves in PrP KO mice.
Adult NSCs are found in the subventricular zone and the subgranular zone of the hippocampus in mouse brain. They continue to grow throughout life with a limited capacity to repopulate neurons. PrP KO NSCs demonstrate reduced proliferation capacity compared with WT NSCs (15, 16). Increased concentrations of soluble full-length PrP have been shown to maintain embryonic stem cells in a highly proliferative state (17), however, cleavage fragments from the N terminus of PrP induce quiescence, a recoverable nonproliferative state (18). Together this indicates that the presence of PrP or soluble fragments of PrP have dynamic effects on NSC growth. Cell growth can also be reduced by an increased propensity to become senescent. Senescence occurs when normally proliferative cells irrecoverably exit the cell cycle. PrP has additionally been linked with a protective role against senescence (19, 20). Therefore, the impaired growth in PrP KO NSCs might be dually due to loss of proliferation and increased senescence.
The epidermal growth factor receptor (EGFR) signaling pathway has been linked with both proliferation and senescence. Overexpression or mis-regulation of EGFR has long been associated with aberrant cell growth in cancer. Absence of epidermal growth factor (EGF) not only impairs proliferation but directs cells into a senescent state (21). Furthermore, EGFR signaling has been connected with PrP function in NSCs through control of the Notch pathway, where PrP was shown to control expression of EGFR (22). In dental pulp-derived stem cells, PrP coimmunoprecipitated with EGFR and siRNA silencing of PrP resulted in inhibition of signaling stimulated by EGF (23). Therefore, PrP may be a major player in the control of EGFR-associated proliferation and senescence.
We hypothesized that the reduction in PrP KO NSC proliferation might also correlate with increased senescence mediated by alterations in the EGFR signaling pathway. Our data indicate that reduced EGFR signaling in the PrP KO NSCs increases senescence through changed cellular energy pathways.
Results
PrP KO NSCs are less proliferative and more senescent than WT NSCs
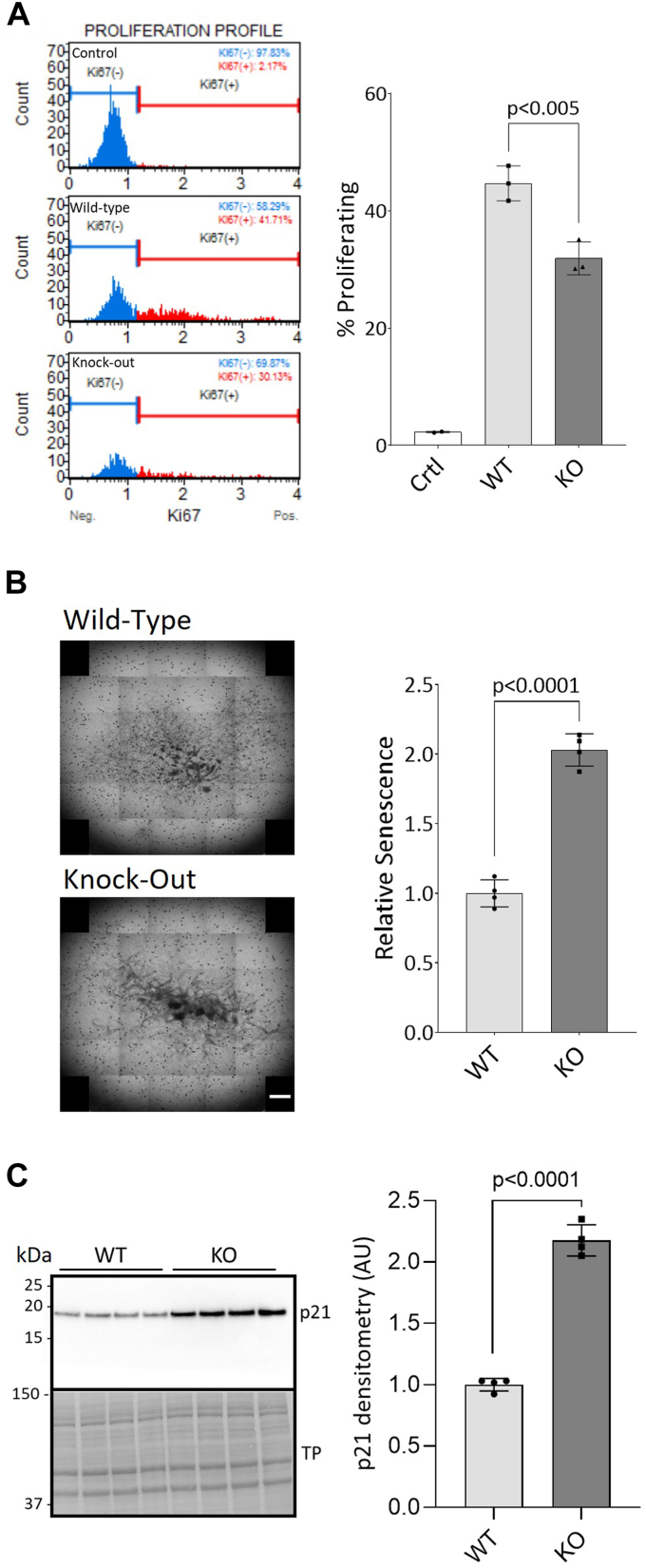
PrP KO NSCs have previously been shown to grow slower than WT NSCs (15) and less cells are found in the S and G2/M proliferative phases of the cell cycle (16). We first validated that the PrP KO NSCs were less proliferative by measuring the number of cells expressing Ki67, a marker of proliferation. WT NSC cultures contained approximately 10% more Ki67-positive cells than the PrP KO NSCs (Fig. 1A). To determine if the reduced number of proliferating cells corresponded with higher senescence in the PrP KO NSC cultures, cells were stained for beta-galactosidase activity, a marker of senescence, and the intensity of staining quantified (Fig. 1B). A 2-fold increase in staining was observed in the KO NSCs, confirming that the PrP KO NSCs were more senescent than WT NSCs. In agreement with previous studies showing soluble PrP stimulated NSC growth (17), we found that secreted anchorless PrP produced by NSCs from Tg44 GPI−/− mice was able to partially rescue the senescence phenotype (Supporting information 1). The PrP KO NSCs also had increased protein levels of p21, a cell cycle inhibitor that induces senescence and prevents apoptosis (Fig. 1C). Thus, our results confirm that the PrP KO NSCs are less proliferative and more senescent than WT NSCs.
Figure 1.
Cellular proliferation is reduced and senescence is increased in PrP KO NSCs.A, flow cytometry profiles showing the cell count of Ki67-negative (blue) and Ki67-positive (red) WT and KO NSCs with quantification (right). B, beta-galactosidase staining of senescent cells. Well scanning showing representative staining intensity of a single well of WT and KO NSCs from a 24-well plate (left) and quantification (right). The scale bar indicates 1000 μm. C, Western blotting for the marker of senescence, p21 (left) and quantification (right). WT and KO results were compared by two-tailed unpaired student’s t test with Welch’s correction (p-values are indicated on graphs), n = 3 (A) and n = 4 (B and C). Graphs are displayed relative to control (A; “Ctrl”: no antibody) or the average of the WT (B and C). TP, total protein; NSC, neural stem cell; PrP, prion protein.
KO NSC senescence is linked with less EGFR
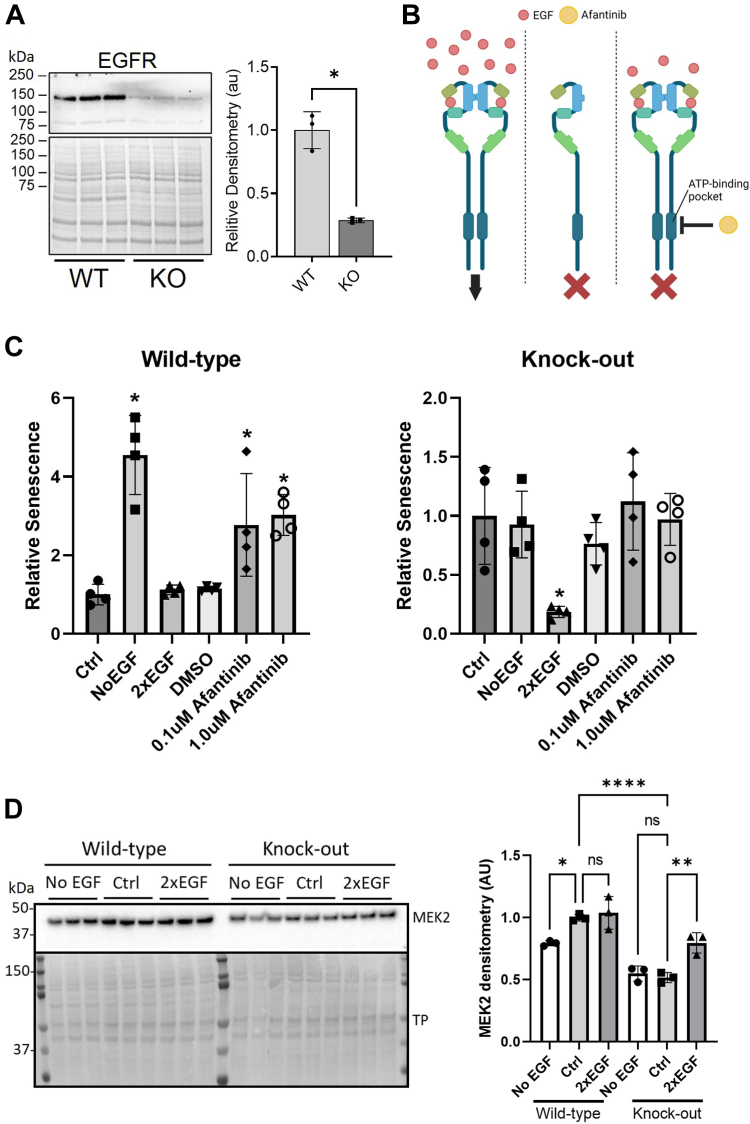
EGFR is a well-known target for anticancer drugs, which act by blocking its signaling and thereby slowing cell proliferation. We therefore investigated EGFR signaling and its link with senescence and PrP. In similarity with previous observations (22), we found significantly decreased protein levels of EGFR in the PrP KO NSCs (Fig. 2A). Conversely, depriving WT NSCs of EGF for 24 h resulted in an increase in PrP expression (Supporting information 2 and (22)). Manipulation of EGF media concentrations has also been shown to influence cell proliferation (21). In WT NSCs removal of EGF from the media or inhibition of EGFR by afatinib for 24 h increased senescence (Fig. 2, B and C and Supporting information 3). Conversely removal of EGF or blockage of EGFR signaling had no effect on the PrP KO NSCs but increasing EGF media concentrations significantly reduced their senescence staining, suggesting fewer growing cells were irreversibly exiting the cell cycle (Fig. 2, B and C and Supporting information 3). Western blotting for protein levels of MEK2, a downstream intermediate in EGFR signaling, showed changes in its levels at 24 h that correlated with the EGF-associated detection of senescence (Fig. 2D). This indicates that the increased senescence in the PrP KO NSCs is driven by a deficiency in the EGFR signaling pathway that is linked with PrP.
Figure 2.
PrP KO–induced cellular senescence is regulated by EGFR.A, representative Western blotting of EGFR in WT and KO NSCs (left) and quantification (right). B, schematic showing media and inhibitor manipulations of EGFR signaling carried out for 24 h in C. Image created with BioRender.com. C, relative senescence staining (beta-galactosidase) of WT (left) and PrP KO (right) NSCs following removal of EGF (NoEGF) or addition of the EGFR inhibitor afatinib to block EGFR signaling or doubling of the EGF concentration in the growth to increase EGFR signaling. D, Western blotting (left) and densitometry (right) of MEK2 detection, after 24 h in media with normal, 0, or double EGF concentrations. Results were compared by two-tailed unpaired student’s t test with Welch’s correction (A; n = 4) or by one-way ANOVA compared to control (C, n = 4, and D, n = 3) ∗p < 0.05. Graphs are displayed relative to the average of the WT (A), normal 1× EGF media controls (“Ctrl”; C), or WT Ctrl (D). EGFR, epidermal growth factor receptor; NSC, neural stem cell; PrP, prion protein.
KO NSC senescence is associated with transcriptome changes in senescence, cell cycle, and EGFR-associated signaling pathway genes
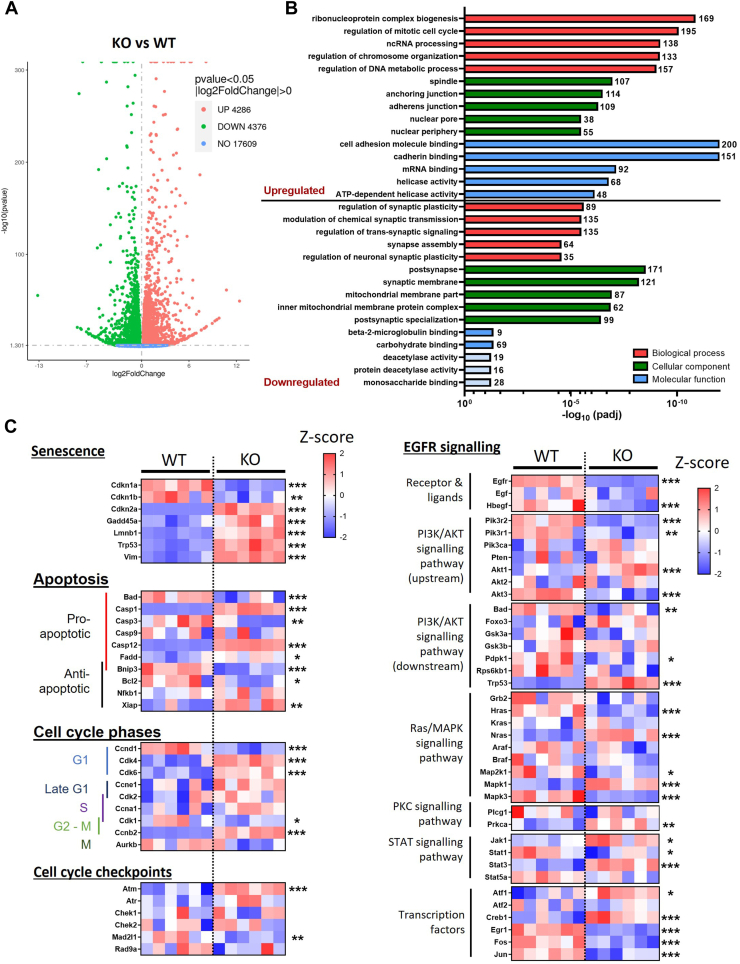
To examine in more detail the differences between the WT and PrP KO NSCs that result in the different growth and senescent phenotypes, we performed bulk RNA-seq. A substantial number of genes were found to be significantly changed in the PrP KO NSCs (Fig. 3A). When the gene ontology was examined, genes that were increased in the PrP KO NSCs were associated with the cell cycle, including regulation of chromosomes, DNA, and spindle, as well as cell adhesion (Fig. 3B). Those genes downregulated were associated with the ability to form synapses and membrane composition especially of the mitochondria. We further interrogated a selection of the genes associated with senescence, apoptosis, and the cell cycle. This confirmed significant changes in the PrP KO NSCs associated with senescence and variable changes in genes associated with apoptosis, which is consistent with it being inhibited in senescent cells ((24); Fig. 3C). A number of cell cycle genes were significantly increased in the PrP KO NSCs, indicating an attempt to drive the cell cycle but cyclin D1 (Ccnd1), which is essential for transition through the G1 phase of the cell cycle, was significantly reduced.
Figure 3.
Altered senescence and EGFR pathways in PrP KO cells as detected by bulk RNA sequencing.A, volcano plot showing changed transcripts between WT and PrP KO NSCs. B, gene ontology analyses showing the five most changed categories in biological process (red), cellular component (green), and molecular function (blue) for genes that are upregulated and downregulated in PrP KO NSCs. The number of genes changed in each category is indicated at the end of each bar. Pale blue coloring of the molecular function bars denotes that this category did not reach the threshold for statistical significance. C, heatmaps (Z-scores) of gene selections associated with senescence, apoptosis, cell cycle, and EGFR signaling broken down into pathways showing relative changes in PrP KO NSCs. ∗∗∗Padj < 0.001, ∗∗ Padj < 0.01, and ∗Padj < 0.05. EGFR, epidermal growth factor receptor; NSC, neural stem cell; PrP, prion protein.
EGFR activation influences several canonical pathways including PI3K, MAPK, PKC, and STAT signaling. When we examined a subset of genes from these pathways it was apparent that the upstream activators, EGFR and heparin-binding egf-like growth factor, are significantly downregulated and the downstream transcription factors were substantially changed (Fig. 3D). However, the intermediates in the pathways did not show a preference for influencing any individual signaling pathway with all pathways showing increases in some genes with decreases in others. Altogether the transcript changes support a senescent phenotype in the KO NSCs associated with changed EGFR signaling.
Manipulation of available EGF
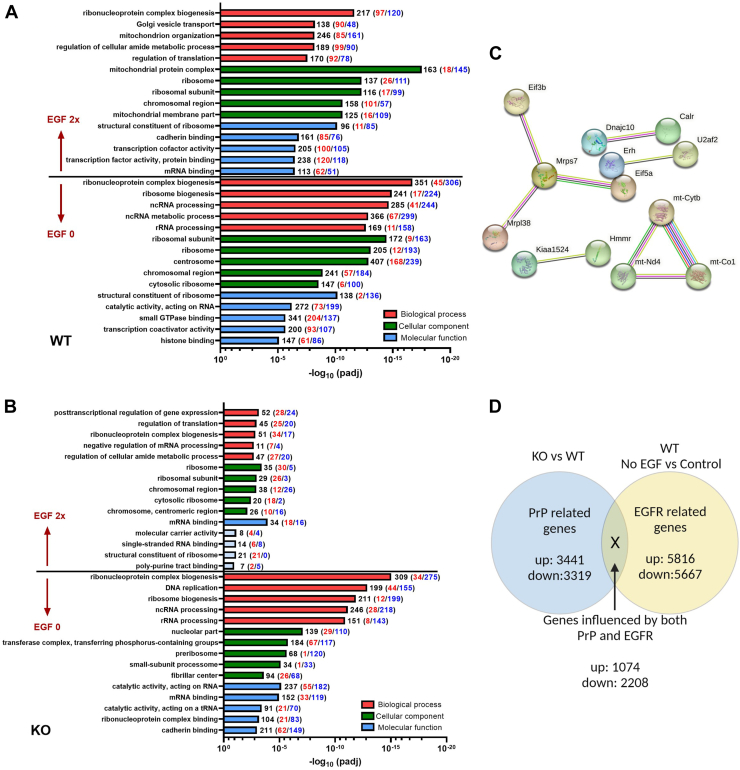
To examine the influence of EGFR signaling and look for further changes associated with PrP KO in the NSCs, we compared transcriptomes following either withdrawal of EGF (0 ng/ml EGF) or doubling the media concentration (40 ng/ml) of EGF with standard culture conditions (20 ng/ml EGF). NSCs were treated for 24 h (the treatment time after which the changes in senescence had been measured, Fig. 2C) and, therefore, these data represent the pathways affected by the senescence shifts as opposed to the acute gene expression changes that effect the senescence changes. Removal of EGF from the culture media resulted in overall more gene changes in both WT and PrP KO NSCs with similarities in the gene ontology including changes in ribosomes and nuclear gene families, all of which pointed to the expected decrease in cellular protein production and cell cycle that would be associated with a decrease in EGFR signaling (Fig. 4, A and B). Doubling the media concentration resulted in more significant gene ontology changes in WT NSCs than PrP KO, likely ascribable to the decreased expression of EGFR in the PrP KO NSCs and therefore a lesser response to stimulation (Fig. 4, A and B). Although there were fewer gene changes in the KO NSCs, many of these were associated with transcription and translation. This indicates that the EGF-stimulated KO NSCs have increased these activities allowing them to maintain their cell cycle without exiting to senescence, albeit while impacting few other pathways. This analysis also showed that with increased media EGF, the WT NSCs changed a large number of genes associated with mitochondrial structure and protein complexes, which were not observed in the PrP KO NSCs (Fig. 4, A and B). Over activation of EGFR is known to cause translocation of EGFR to the mitochondria (25), which would occur at a much lesser extent in the KO NCSs with low levels of EGFR. However, when STRING analysis (26, 27, 28) for protein–protein interactions was applied to the genes with significant changes (padj <0.05 from control EGF levels) in the PrP KO NSCs, the gene ontology also showed significant changes in mitochondrial protein interactions (Fig. 4C and Supporting information 4). This might be contributing to the decrease in cells progressing to senescence after 24 h of increased EGF.
Figure 4.
Genes ontology pathways altered by EGF.A and B, gene ontology analyses showing the five most changed categories in biological process (red), cellular component (green), and molecular function (blue) for genes altered by EGF manipulation compared with normal media control by category. The number of genes changed in each category is indicated at the end of the bar with increased genes indicated in red and decreased in blue. Pale blue coloring of the molecular function bars denotes that this category did not reach the threshold for statistical significance. C, STRING analysis of PrP KO NSCs treated with double EGF as compared with their normal EGF level controls (see also Supporting information 4). D, schematic for investigating genes that were influenced by both PrP KO and EGF withdrawal. EGF, epidermal growth factor; NSC, neural stem cell; PrP, prion protein.
Comparison of the overlap in changed genes between PrP KO and EGF withdrawal identifies common metabolic process transcription changes
To examine what common pathways may be changed in the PrP KO NSCs and in response to EGF withdrawal, simulating lowered EGFR signaling, we performed gene ontology analysis on only the genes that were commonly changed in the PrP KO NSCs and the WT no EGF conditions compared with the WT normal media EGF control (Fig. 4D). This analysis showed significant changes in the EGFR signaling pathways as expected but also substantial changes in cellular energy metabolism, especially involving the mitochondria (Table 1).
Table 2.
| Figure 4 | Eif3b | Eukaryotic translation initiation factor 3 subunit B |
| Mrps7 | 28S ribosomal protein S7, mitochondrial | |
| Mrpl38 | 39S ribosomal protein L38, mitochondrial | |
| Eif5a | Eukaryotic translation initiation factor 5A-1 | |
| Dnajc10 | Dnaj heat shock protein family member c10 | |
| Calr | Calreticulin | |
| Erh | Erh mrna splicing and mitosis factor | |
| U2af2 | U2 small nuclear ribonucleoprotein auxiliary factor 2 | |
| Kiaa1524 | Cell proliferation regulating inhibitor of protein phosphatase 2a | |
| Hmmr | Hyaluronan mediated motility receptor | |
| mt-Cytb | Cytochrome b | |
| mt-Nd4 | NADH-ubiquinone oxidoreductase chain 4 | |
| mt-Co1 | Component of the cytochrome c oxidase | |
| Figure 7 | BPG | 2,3-Bisphosphoglyceric acid |
| DHAP | Dihydroxyacetone phosphate | |
| F6P | Fructose 6-phosphate | |
| FBP | Fructose 1,6-bisphosphate | |
| G1P | Glucose 1-phosphate | |
| G6P | Glucose 6-phosphate | |
| GAP | Glyceraldehyde 3-phosphate | |
| GlcN6P | Glucosamine-6-phosphate | |
| GlCNAc6P | N-Acetyl-D-glucosamine 6-phosphate | |
| GSH | Glutathione | |
| GSSG | GSH/glutathione disulfide | |
| PEP | Phosphoenolpyruvic acid | |
| PG | Phosphoglycerate | |
| PRPP | Phosphoribosylpyrophosphate | |
| UDP-GlcNAc | Uridine diphosphate N-acetylglucosamine | |
| UDP-glucose | Uridine diphosphate glucose | |
| Figure 8 | Dlat | Dihydrolipoamide S-acetyltransferase |
| Dld | Dihydrolipoamide dehydrogenase | |
| LDHA | Lactate dehydrogenase A | |
| LDHB | Lactate dehydrogenase B | |
| PCx | Pyruvate carboxylase | |
| Pdha1 | PDHA1 pyruvate dehydrogenase E1 subunit alpha 1 | |
| Pdhab | PDHA1 pyruvate dehydrogenase E1 subunit beta | |
| Pdhx | PDHX pyruvate dehydrogenase complex component X |
Hsp40, heat shock protein family; U2AF, U2 small nuclear ribonucleoprotein auxiliary factor.
Table 1.
Top ten gene ontology classes for genes changed in both PrP KO NSCs and EGF-WT NSCs as compared with the WT normal EGF control cells
| Genes upregulated (p < 0.01) | Genes downregulated (p < 0.01) |
|---|---|
| Regulation of epidermal growth factor receptor signaling pathway (GO:0042058) | Mitochondrial electron transport, ubiquinol to cytochrome c (GO:0006122) |
| Epidermal growth factor receptor signaling pathway (GO:0007173) | Negative regulation of Notch signaling pathway (GO:0045746) |
| Autophagosome maturation (GO:0097352) | Apoptotic mitochondrial changes (GO:0008637) |
| ERBB signaling pathway (GO:0038127) | Glycosphingolipid biosynthetic process (GO:0006688) |
| Regulation of ERBB signaling pathway (GO:1901184) | Mitochondrial ATP synthesis coupled electron transport (GO:0042775) |
| Lysosomal transport (GO:0007041) | ATP synthesis coupled electron transport (GO:0042773) |
| Protein targeting to vacuole (GO:0006623) | Protein localization to endoplasmic reticulum (GO:0070972) |
| Fatty acid beta-oxidation (GO:0006635) | Aerobic electron transport chain (GO:0019646) |
| Regulation of cell shape (GO:0008360) | Ribonucleoside diphosphate metabolic process (GO:0009185) |
| Negative regulation of protein-containing complex assembly (GO:0031333) | Purine ribonucleoside diphosphate metabolic process (GO:0009179) |
Italics denote categories associated with EGFR signaling, and bold indicates categories associated with energy production.
Both PrP KO and EGF withdrawal are associated with mitochondrial transcriptome changes
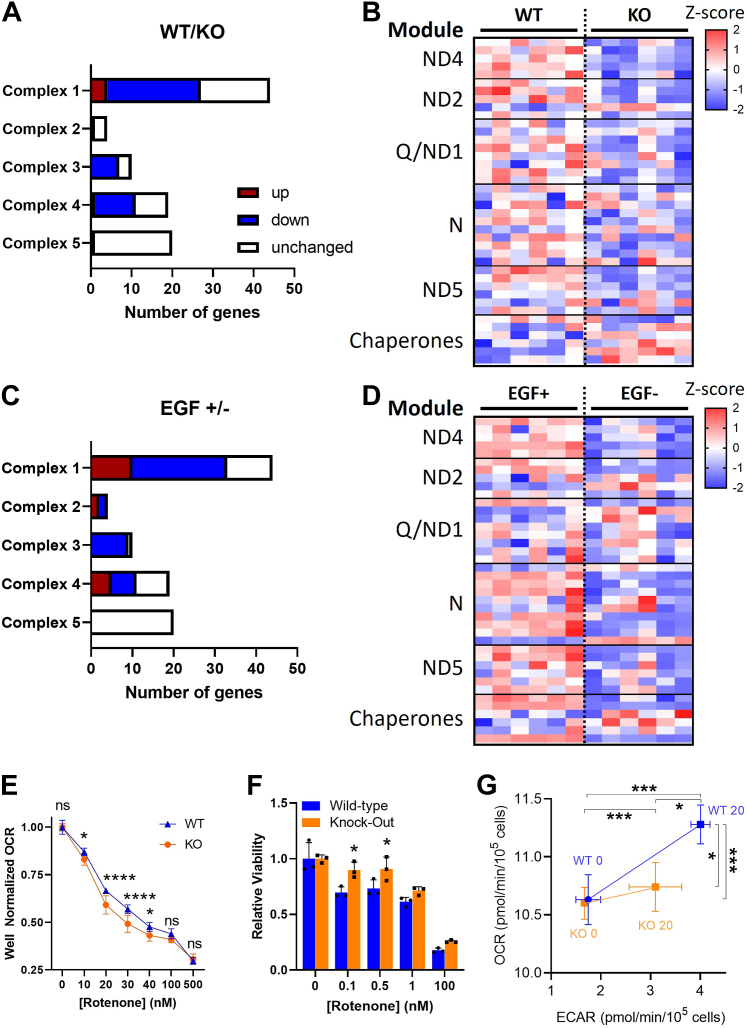
Since the above transcriptomic analyses had identified changes in mitochondrial gene expression, both basally and in response to EGF manipulation, between WT and PrP KO NSCs, we analyzed the genes involved in mitochondrial electron transport more closely. In the PrP KO NSCs, mitochondrial transcripts were sizably reduced in complexes 1, 3, and 4, with complex 1 showing the most changed transcripts (this complex is encoded by the largest number of genes) (Fig. 5A). Complex 5, ATP synthase, was least affected by transcriptomic changes. Complex 1 assembly is via a series of module constructions that are chaperoned into the final complete complex (29). When transcripts from each of the complex 1 modules were compared, it was evident that components of the complex are downregulated in the KO NSCs at all points of assembly (Fig. 5B). When a similar analysis was performed comparing the WT NSCs with normal EGF supplementation to those where EGF was withdrawn from the culture, similar changes in the mitochondrial transcripts were observed with a large number of downregulated genes (Fig. 5C), although a few more were increased than observed in the PrP KO NSCs. Further analysis of the complex 1 modules likewise indicated that the depletion of EGF was also downregulating the whole complex without specifically affecting any modules (Fig. 5D).
Figure 5.
Mitochondrial complex 1 is transcriptionally and functionally downregulated in PrP KO NSCs.A, mitochondrial gene changes in KO NSCs compared with WT. Statistically increased genes (p < 0.05) are shown in red with decreased genes in blue. B, heatmap displaying Z-scores of complex 1 gene expression in the PrP KO compared with WT NSCs reflecting their assembly modules and select chaperones. C, mitochondrial gene changes in WT NSCs deprived of EGF compared with those in normal levels of EGF. Statistically increased genes (p < 0.05) are shown in red with decreased genes in blue. D, heatmap displaying Z-scores of complex 1 gene expression in WT NSCs deprived of EGF compared with those in normal levels of EGF reflecting their assembly modules and select chaperones. E, Seahorse analysis of oxygen consumption rate (OCR) with increasing concentrations of the complex 1 inhibitor, rotenone, of KO and WT NSCs. F, Prestoblue analysis of cell viability with increasing concentrations of rotenone of PrP KO and WT NSCs. G, energy maps of WT and KO NSCs before and after injection of 1× EGF (20 ng) into the assay media. For B and D, see Supporting informations 3 and 4 for full gene and statistical analyses. For E–G, two-way ANOVA analysis ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, and ∗p < 0.05. Graphs in E and F are displayed relative to the average of the untreated (0 μM/nM rotenone) media conditions for WT or KO NSCs (n = 9 in E and 3 in F). EGF, epidermal growth factor; NSC, neural stem cell; PrP, prion protein.
Complex 1 function is impaired in PrP KO NSCs
With the downregulation of complex 1 in the PrP KO NSCs, we would expect mitochondrial respiration to be sensitive to disruptions in complex 1, with greater relative drops in oxygen consumption since there is likely less complex to saturate with an inhibitor. Conversely, as the cells are likely to be compensating for the reduced mitochondrial activity, inhibition of complex 1 should be less toxic. As expected, when selectively inhibiting complex 1 with an acute treatment of increasing rotenone concentrations, we observed a more rapid reduction in oxygen consumption rate (OCR) in the PrP KO NSCs (Fig. 5E). Likewise, a slower decline in cell viability was observed after 24 h of treatment at lower concentrations of rotenone (selected to avoid the complete death of the cultures observed when using the concentrations with acute activity) (Fig. 5, F). To consider how this impairment of function influences the response to EGF, we equilibrated WT and KO NSCs into zero EGF media overnight then measured their OCR and extracellular acidification rate (indicative of glycolytic response) before and immediately after the addition of normal cultures levels of EGF (20 ng; Fig. 5G). This showed the KO cells were less able to respond metabolically to the addition of EGF than the WT cells, with their mitochondrial response negligible. Together this suggests that complex 1 is transcriptionally decreased by knockout of PrP and this translates into the expected reduction in complex function.
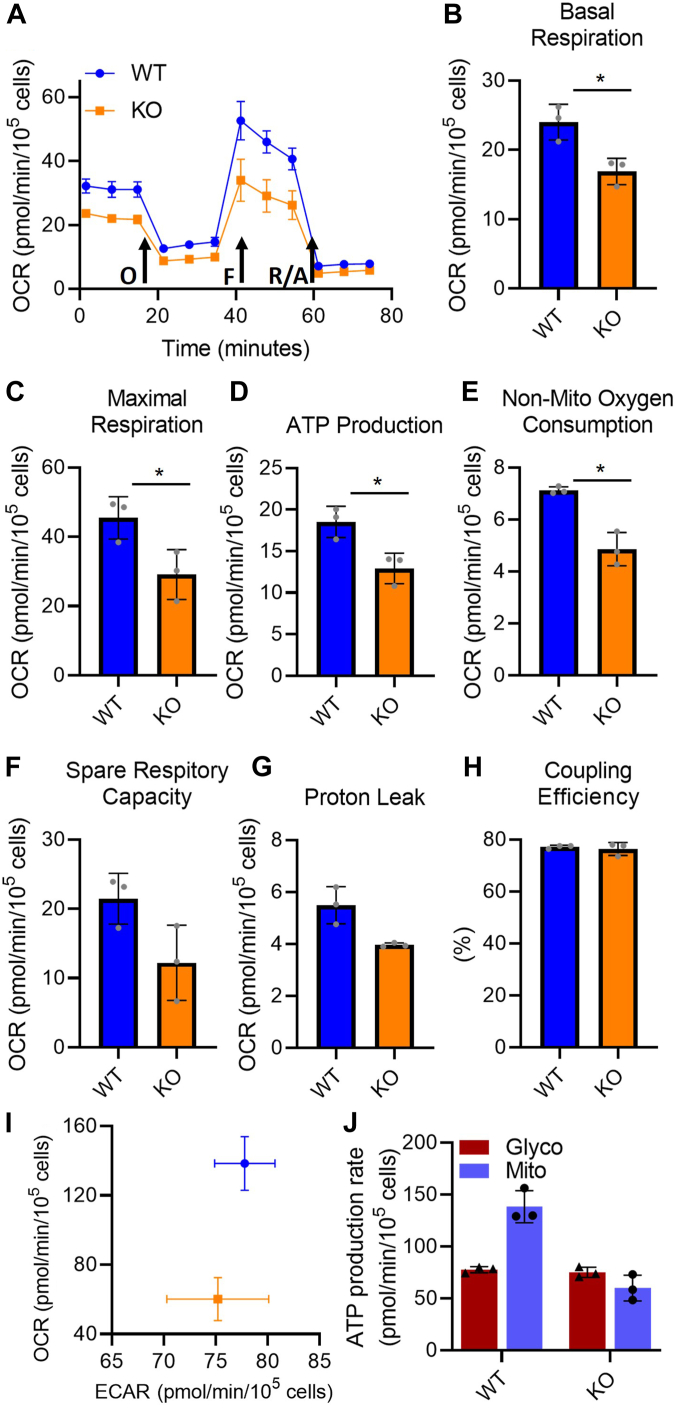
Mitochondrial function is reduced in PrP KO NSCs
The reduced mitochondrial transcripts in PrP KO NSCs are not limited to complex 1 and indicated a probable overall decrease in mitochondrial electron transport chain (ETC) function. To determine the extent of the reduced ETC transcript influence on overall mitochondrial function, we performed a mitochondrial stress test using a Seahorse analyser. The stress test uses sequential addition of oligomycin to block ATPase activity, followed by FCCP to simulate maximal activity, and finally rotenone with antimycin A to completely block respiration. The profile of the stress test is shown in Figure 6A, and the parameters calculated from the profile data shown in Figure 6, B–H. All parameters, with the exception of percentage coupling efficiency (Fig. 6H), are decreased showing an overall decrease in mitochondrial function within PrP KO NSC mitochondria both basally and upon stimulation. This appears to be a direct reflection of the reduction of component intermediates shifting the overall KO mitochondrial activity lower. The change in mitochondrial output is also reflected in the basal cellular energy map (Fig. 6I), comparing cellular ATP produced by mitochondria with that generated from glycolysis and in the ATP production rates from glycolysis and mitochondria (Fig. 6J). This confirms that the PrP KO NSCs are producing substantially less ATP from mitochondrial respiration than WT NSCs.
Figure 6.
Overall mitochondrial function and energy production are decreased in PrP KO NSCs.A, example traces showing oxygen consumption rate (OCR) following stimulation or inhibition of the electron transport chain in WT and PrP KO NSCs. Arrows indicate injections of oligomycin (O) to inhibit ATP production, FCCP (F) to stimulate maximum respiration and rotenone/antimycin A (R/A) to completely shut down the electron transport chain. Traces show the mean and SD of three replicate wells. Two-way ANOVA analysis ∗∗∗∗p < 0.0001, ∗∗p < 0.01, n = 3. B–H, graphical representation of the indicated parameters for mitochondrial function calculated from the OCR traces (n = 3). Results were compared by two-tailed unpaired student’s t test with Welch’s correction, ∗p < 0.05. I, energy map of mitochondrial versus glycolytic ATP production and (J) quantitation of ATP production rate from glycolysis and OXPHOS (n = 3). NSC, neural stem cell; PrP, prion protein.
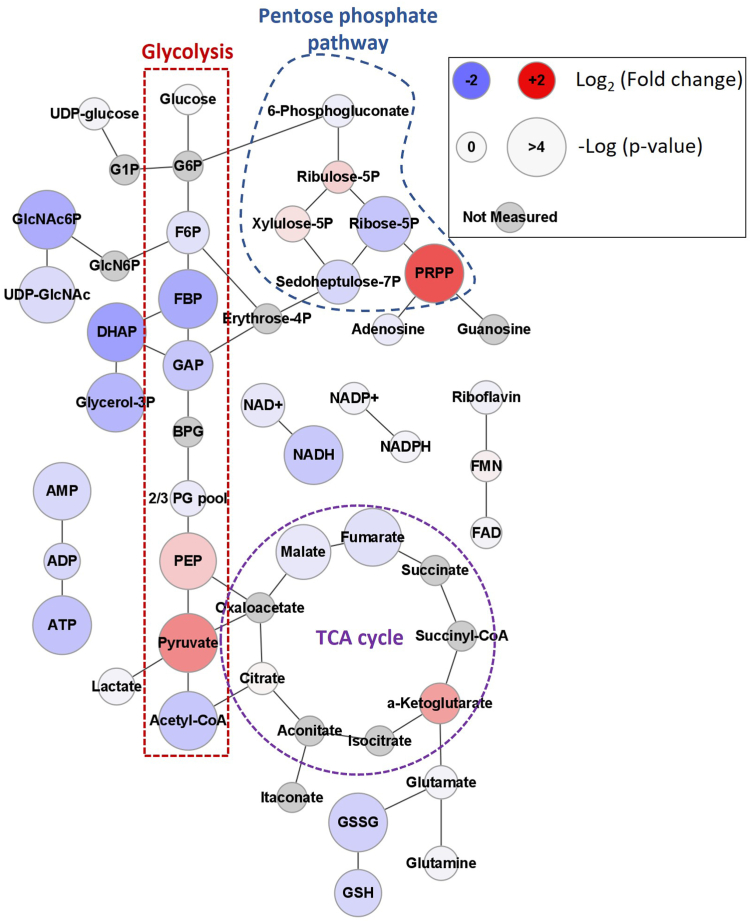
PrP KO NSCs accumulate pyruvate
Electron flow into the ETC is regulated by the balance of central carbon metabolism processes including glycolysis, the tricarboxylic acid cycle (TCA), and fatty acid oxidation. Glycolysis was identified as changed in the analysis of EGFR-related genes in PrP KO NSCs and EGF withdrawal (Table 1). We therefore performed metabolomics analysis on the PrP KO and WT NSCs to examine changes in cellular metabolites. Significant changes in glycolysis, the pentose phosphate pathway, and the TCA cycle were observed. Accumulations of pyruvate and phosphoribosyl pyrophosphate (PRPP) are apparent with reductions in a number of TCA cycle intermediates including acetyl-CoA, malate, and fumarate. Also evident was a decrease in purine containing compounds including AMP, ATP, NAD+, and NADH (Fig. 7 and Supporting information 5).
Figure 7.
PrP KO alters NSC cellular metabolic profile. Web diagram of affected metabolite pools in energy producing metabolism associated with the loss of PrP in NSCs. Coloring of each node indicates the base 2 logarithm of the fold change of the PrP KO versus the WT while size reflects the negative logarithm of the p-value from an unpaired t test. Positions of glycolysis, the pentose phosphate pathway, and tricarboxylic acid (TCA) cycle are indicated. n = 6. Explanation of abbreviations can be found in Table 2. NSC, neural stem cell; PrP, prion protein.
This overall central metabolic pattern indicated a potential dysfunction at the terminal end of glycolysis resulting in pyruvate accumulation. Based on the depletion of upstream glycolytic intermediates glycolysis appeared to be turning over in PrP KO NSCs but pyruvate failed to be converted to downstream metabolites. Separate but potentially related to this glycolytic pattern, an imbalance in the levels of depleted Ribose-5P compared to accumulated PRPP was evident that suggested that the pentose phosphate pathway was turning over but that subsequent conversion of PRPP to nucleic acids was impeded. The buildup of pyruvate and the reduction of both acetyl-CoA and the TCA intermediates directly upstream of acetyl-CoA integration suggest that the TCA cycle is starved for carbon input in PrP KO NSCs. This starvation of the TCA cycle likely explains the trend toward a decreased NADH/NAD+ signal ratio in the PrP KO NSC’s (0.3 ± 0.06) compared to the WT (0.4 ± 0.08). The broader reduction in NAD(H) regardless of redox state may be coupled with the broader observed decrease in purine containing nucleotides. This secondary purine centred effect was likely derived from the observed accumulation of PRPP, which may indicate a failure to covert PRPP to downstream nucleotides.
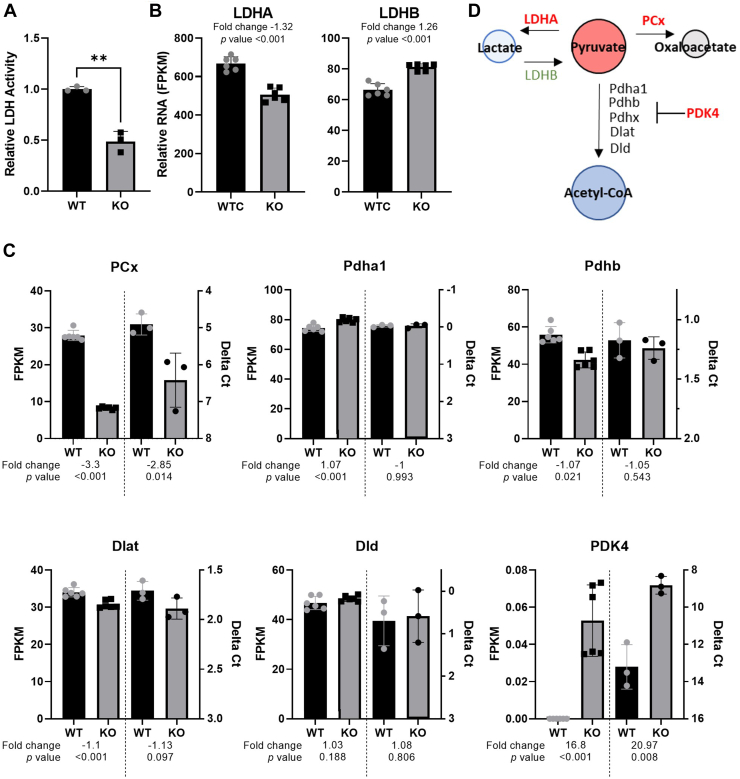
Pyruvate forms a hub in central metabolism that links together glycolysis with the TCA-cycle via multiple routes as well as fatty acid metabolism. To determine which pyruvate-associated pathway was responsible for the observed pyruvate accumulation we examined the enzymes responsible for the conversion of pyruvate to lactate, acetyl-CoA, or oxaloacetate. Measurement of cellular lactate dehydrogenase (LDH) activity (Fig. 8A) and examination of genes associated with catabolism of pyruvate (Fig. 8, B and C and Supporting information 6) indicated reduced activity or enzyme levels of all pathways that utilize pyruvate as a substrate (summarized in Fig. 8D). Additionally, the levels of LDHB, which has been shown to facilitate lactate conversion to pyruvate, were increased in PrP KO NSCs. This combination of changes in metabolic transcripts, central carbon metabolite pools, and pyruvate associated enzymes suggest a failure of the PrP NSCs to maintain effective coupling between glycolysis and the TCA cycle and failure to maintain purine biosynthesis.
Figure 8.
Pyruvate metabolism is reduced in PrP KO NSCs.A, LDH activity assay of WT and KO NSCs with (B) LDHA and LDHB gene expression. C, changes in the pyruvate dehydrogenase complex genes and pyruvate carboxylase transcripts associated with PrP KO in NSCs with the RNA-seq FPKM shown on the left y-axis and the qRT-PCR Delta Ct shown on the right y-axis for comparison. RNA samples were harvested for RNAseq and qRT-PCR independently. D, schematic showing the enzymes that catalyze pyruvate with those shown in red reduced and those shown in green increased in PrP KO NSCs. Graph in panel A is displayed relative to the average LDH activity of WT (n = 3). Results in A were compared by two-tailed unpaired student’s t test with Welch’s correction, ∗∗p < 0.01. For B and C, fold changes and p-values appropriate to the assay (see methods) are shown with each plot. Explanation of abbreviations can be found in Table 2. FPKM, Fragments per kilobase million; LDH, lactate dehydrogenase; NSC, neural stem cell; PrP, prion protein; qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
Discussion
PrP has been repeatedly linked with cell proliferation (15, 16, 17, 22, 30). PrP expression in NSCs promotes cell proliferation while PrP KO has the opposing effect, suppressing the cell cycle (15, 31). Cellular proliferation is not only influenced by the speed of cell cycling but also by the number of cells that irreversibly exit the cell cycle and become senescent. Our results herein support that PrP KO increases senescence in NSCs and that this is linked with reduced EGFR signaling and compromised mitochondrial function. The latter resulted in reduced cellular energy and intermediates for biomass production. Further changes within glycolysis, blocking the catabolism of pyruvate, additionally contribute to a lack of intermediates for the TCA cycle, respiration, and therefore ATP production. Altogether, we observed that PrP KO NSCs exhibit cellular deficiencies that reduce growth and promote senescence.
The link between PrP and cell proliferation has been further strengthened with recent findings that PrP can promote metastases or drug resistance in certain cancers (32, 33). Liang et al. (33) found that PrP promoted cell cycle through facilitating the G1/S phase transition by upregulation of Ccnd1 in human gastric cancer cell lines. Herein, Ccnd1 was significantly reduced in the RNA-seq analysis, suggesting this could be contributing to the permanent cell cycle exit. Depletion of Ccnd1 has also been found in PrPnull-1C11 cells where cell proliferation was significantly decreased (34). The authors linked the reduced proliferation with changes in the Sonic Hedgehog pathway, which is also critical for stem cell renewal and exhibits cross-talk with the EGFR pathway (35, 36).
The EGFR pathway has been a long-time target for anticancer drugs as overexpression or activation of the EGFR pathway has been linked with multiple types of cancers. Expression of EGFR is modulated by PrP (15, 17, 22, 30), with decreased PrP expression causing a decrease in EGFR expression (22). The data presented herein supports the decreased expression of EGFR in PrP KO cells as well as observations from others who have shown that blocking or knocking down EGFR downregulated Ccnd1 and arrested cell growth by inducing senescence (21) in human bronchial or mammary epithelial cells. Furthermore, both EGFR (37) and PrP expression (38) influence LDHA expression in cystic fibrosis lung epithelial cells and primary neurons or human embryonic kidney cells, respectively. LDHA is responsible for converting pyruvate to lactate. Under normal conditions, cell proliferation is aided by the production of NAD+ when pyruvate is converted to lactate by LDHA (39). In addition to lactate, pyruvate can be carboxylated to oxaloacetate by pyruvate carboxylase or decarboxylated to acetyl-CoA by the pyruvate dehydrogenase complex, which both feed the TCA and promote mitochondrial respiration. Abnormally low (39) or high (40) levels of pyruvate, caused by dysregulation in any of the above pathways, could trigger fibroblast senescence in a concentration dependent manner. Knockdown of EGFR for example decreases LDHA expression and activity (37), which could in turn cause the cell to accumulate pyruvate and trigger senescence. Additionally, knockdown of PrP can increase expression of pyruvate dehydrogenase kinase 4 which in turn blocks the transport of pyruvate into the mitochondria and its conversion to acetyl-CoA, impeding the cells’ ability to process glucose (41). Transport of pyruvate into the mitochondria is a critical step in the activation and shift from quiescence to proliferation of NSCs (42) Similarly, suppression of LDHA slows tumor progression by disrupting cell respiration, as evidenced by a decreased OCR (43), increasing ROS levels, reducing ATP production (44), and inhibiting cell proliferation thought Ccnd1 downregulation (45). These changes are all reproduced by the PrP KO phenotype in the NSCs.
Mitochondrial morphology is disrupted following prolonged LDHA inhibition (43) and can be modulated by EGFR (25). Similar morphological changes, including mitochondrial shape and decreased cristae density, have been observed in PrP KO mice (46). Since mitochondria are also a major site of biomass production, reduced ATP production from the ETC, coupled with reduced capacity to produce biomass limits growth. Additionally, the observed blockage at PRPP to synthesis of adenosine and guanosine may contribute both to the lower cellular energy production and to a deficiency of nucleotides for synthesis of DNA and RNA, thereby blocking the cell cycle due to lack of resources. Decreases in NADH and malate, substrates for complex 1 activity, also likely influences the decrease in complex 1 subunits and activity.
In contrast to our results, one study showed that PrP KO mice have increased superoxide formation, which they attribute to increased complex 1 activity (47). However, we found that many essential complex 1 genes were downregulated in our adult NSC cultures when PrP was knocked out. The variation in findings could be due to the cell types analyzed. NSCs are only a very small population in the adult brain, and they maintain low ROS levels to preserve their stemness. An additional study reported similar findings of increased superoxide radicals in cortical neurons, however their findings support impaired mitochondrial respiration and an inhibition in the ETC (48). Furthermore, decreased LDHA, as seen in the PrP KO-NSCs, has been shown to cause increased rates of electron leakage from complex 1, resulting in increased levels of cellular ROS in promyeloblasts (43). If this leakage from complex 1 was producing high enough levels of ROS this might have caused complex 1 (and other complexes) to be downregulated to preserve cell viability. In non–small cell lung cancer cells, ROS levels have also been shown to be increased with impaired EGFR signaling (49) and in turn trigger additional degradation of EGFR, resulting in mitochondrial dysfunction and pushing the cell toward apoptosis (50, 51). In the context of our observations, apoptosis is an unlikely outcome as it requires more energy than is available so instead the cells commit to senescence.
In the context of the whole mouse PrP KO model, our findings could explain some of the observed phenotypes. As previously mentioned, KO mice, while mostly normal, do show impaired exercise capacity (7). Although the authors did not find evidence of mitochondrial dysfunction in this study, they demonstrated a lack of endurance in the PrP KO mice, which they hypothesize could be due to abnormal accumulation or expenditure of unidentified metabolites. Furthermore, PrP KO mice have impaired glucose tolerance resulting in delayed recovery from hyperglycaemia (52). This phenotype could be explained by a blockage in glycolysis, corresponding with buildup of pyruvate, which could feasibly lead to delayed catabolism of glucose. PrP KO mice have also been demonstrated to have defects in olfaction that were directly linked with altered electrophysiology of the olfactory bulb (53). In mice, NSCs produce new progenitors in the subventricular zone that migrate down the rostral migratory stream to differentiate and integrate into the olfactory bulb (54, 55). In PrP KO mice, increased senescence with reduced proliferative capacity of NSCs likely results in fewer progenitors making this migration in turn causing altered neuronal activity. Therefore, the defects observed in the PrP KO NSCs may underly many of the, albeit mild, manifestations of PrP loss in mice.
Together, these findings suggest that absence of PrP results in reduced proliferation and increased senescence in NSCs through a reduction in EGFR signaling. The decreased proliferation and increased senescence were associated with impaired mitochondrial function, which was at least partially due to downregulation of components of the ETC. A blockage at the end of glycolysis causing accumulation of pyruvate along with an inhibition in nucleic acid production from PRPP may also be contributing to an overall reduction in cellular intermediates needed for growth. The lack of ATP resulting from the mitochondrial impairment would then drive the cells toward senescence. These data therefore confirm the critical role of PrP in NSC signaling and proliferation and also demonstrate how its loss or dysfunction through mutation or disease could result in detrimental senescence.
Experimental procedures
Animal ethics statement
Use of mice for the scientific research included in this manuscript was approved by the Institutional Animal Care and Use Committee of the Rocky Mountain Laboratories protocol 2016-058. Mouse handling and housing was conducted adhering to the institution’s guidelines for animal use and followed the guidelines and basic principles in the United States Public Health Service Policy on Humane Care and Use of Laboratory Animals and the Guide for the Care and Use of Laboratory Animals. Work was carried out by certified staff in an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility.
Mouse NSC culture
The establishment of the NSC cultures has been described previously (56, 57). Briefly, the subventricular zone was removed from three C57BL/10SnJ WT, PrP-KO, and Tg44 GPI anchorless mice (58) and separated into a cell suspension using the Stem Cell Technologies NeuroCult Enzymatic Dissociation Kit for Adult CNS Tissue (Mouse and Rat) as per the manufacturer’s instructions. NSC suspensions were cultured in low adhesion plasticware in complete proliferation media (NeuroCult Proliferation Kit [Stem Cell Technologies] supplemented with 20 ng/ml EGF, 10 ng/ml FGF, and 2 μg/ml heparin). Media was half exchanged every 3 to 4 days or when needed and neurospheres were mechanically dissociated and passaged when they reached approximately 150 μm in size. Cells are periodically tested to ensure they are free from mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza).
Ki67 analysis
Ki67 analysis of proliferating cells was done using the Muse Ki67 assay kit (Luminex) as per the manufacturer’s instructions and results collected using a Muse Cell AnalyZer (Luminex). Mechanically dissociated cells were passed through a cell strainer and immediately assayed at 1 × 105 cells/sample. Unlabeled cells were used to gate for background/Ki67-negative cells.
Senescence assays
Senescence was determined using a β-galactosidase staining kit (Cell Signaling Technologies, #9860), following the manufacturer's instructions. Briefly, mechanically dissociated cells were passed through a cell strainer and then plated into 24-well dishes precoated with growth factor reduced Matrigel (Roche) at 2.2 × 105 cells/well and allowed to adhere for 24 h. Treatments and media manipulations were performed during this 24-h period. β-galactosidase staining was then performed, and the wells were imaged on an EVOS-FL-auto light microscope (Invitrogen). Multiple fields were captured at 4× magnification and stitched together using the EVOS software (Invitrogen). Images were quantified using ImageJ (NIH; https://imagej.nih.gov/ij/download.html) as the number of stained cells out of the total number of cells per well. Raw data are shown in Supporting information 3.
RNA extraction
Mechanically dissociated cells were passed through a cell strainer and then plated into 6-well dishes precoated with growth factor reduced Matrigel (Roche) at 3 × 106 cells per well in either completed proliferation media, proliferation media without EGF, or proliferation media with 2× EGF for 24 h. Cells were lysed and RNA was isolated using the Zymo Research Quick-RNA Miniprep Kit. RNA quality was checked by 2100 Bioanalyser (Agilent) and samples were frozen at −80 °C in DNase/RNase free water containing RNase inhibitors (Ambion/Thermo Fisher Scientific) prior to submission to Novogene for next-generation sequencing (NGS) or use in quantitative RT-PCR.
Next-generation sequencing
Following submission to Novogene, samples entered the Novogene RNA-seq workflow and analysis pipeline, which involves sample quality control, library construction and quality control, sequencing using the Illumina NovaSeq 6000 platform (utilizing paired-end 150 bp sequencing strategy), data quality control, and bioinformatic analysis. Raw data were transformed to sequence reads using CASAVA base recognition, single base error was <1%, and ATCG content was equal. Data was filtered to remove reads with adapter contamination, reads with uncertain nucleotide (N > 10%), and reads where low-quality nucleotides constitute more than 50% of the read. Alignments were done with HISAT2 v2.0.5 (http://daehwankimlab.github.io/hisat2/download/) and >90% of reads mapped to exons. Quality control information is presented in Supporting information 7. Differential analysis was performed using DESeq2 v1.20.0 (https://genepattern.github.io/DESeq2/v1/index.html) and enrichment analysis using clusterProfiler v3.8.1 (https://bioconductor.org/packages/3.17/bioc/html/clusterProfiler.html). Further gene ontology analysis was performed using PANTHER GO-slim analysis (pantherdb.org (59)). Briefly, common gene lists with p-values >0.01 from NGS were compared between either KO versus WT NSCs and WT NSCs without EGF versus WT control NSCs. This identified common upregulated or downregulated genes that were influenced by both PrP and EGFR (Fig. 4D). These gene lists were submitted to the PANTHER GO-slim algorithm for biological processes against the Mus musculus database. The top ten results from each list are displayed in Table 1. STRING analysis was performed by submitting the relevant gene lists to a multiple protein search. The minimum required interaction score was set to high confidence (0.7).
The expanded methods provided by Novogene can be found in Supporting information 8 and the fragments per kilobase million values and Deg lists used for the analyses herein are found as Supporting informations 9 and 10, respectively. The complete dataset can be accessed in the NCBI Sequence Read Archive repository.
Quantitative RT-PCR and analysis
Four hundred nanograms of RNA from each sample was reverse transcribed to synthesize (complementary deoxyribonucleic acid) (cDNA) using the RT2 First Stand Kit per manufacturer’s instructions (Qiagen). Each cDNA reaction was mixed with 2× RT2 SYBR Green qPCR Mastermix purchased from Qiagen with RNase-free water to a final volume of 1.3 ml. Ten microliters of the mixture was then added to each well of a 384-well format plate of the Mouse Glucose Metabolism array (PAMM-006ZE, Qiagen).
The assay was carried out on an Applied Biosystems ViiA 7 Real-Time PCR System with a 384-well block using the following conditions: one cycle at 10 min, 95 °C; 40 cycles at 15 s, 95 °C then 1 min, 60 °C with fluorescence data collection. Melting curves were generated at the end of the completed run to determine the quality of the reaction products. Raw threshold cycle (CT) data was collected with a CT of 35 as the cut-off. CT data was analyzed using the web-based RT2 Profiler PCR Array Data Analysis from Qiagen. All CT values were normalized to the arithmetic mean of the CT values for the housekeeping genes Actb, B2m, Gapdh, Gusb, and Hsp90ab1. Changes in transcription were calculated by the software using the ΔΔCT based method (60). Statistical analysis was performed using the two-tailed unpaired Student's t test of the replicate 2−ΔΔCT values for each gene in the control group versus experimental groups. A mean of ≥ or ≤ 2-fold change and p-value of ≤0.05 considered significant. For quantitative reverse-transcription polymerase chain reaction data, the p-values were not adjusted for multiple comparisons, since we were interested in only controlling for the individual error rate, where an adjustment for multiple tests is deemed unnecessary.
Seahorse analyses
Mitochondrial function was assessed using the SeaHorse XFp analyzer (Agilent) with the Mito Stress Assay and ATP rate kits as described in the manufacturer’s protocol. Seahorse Dulbecco's modified Eagle's media (Agilent) was made up to the following specifications throughout; 25 mM glucose, 1 mM pyruvate, and 2 mM glutamine (Agilent). FCCP calibrations were performed on control cells and the optimal concentration was 0.5 μM. Cell density was titrated to an optimal 1.5 × 104 cells per well. Cells were mechanically dissociated, passed through a cell strainer, and plated to the desired density in the Seahorse plate coated with growth factor reduced Matrigel (Roche). Complex 1 function was assayed by monitoring OCR during exposed to increasing concentrations of rotenone through subsequent injections. Results were analyzed using Agilent’s Wave software (https://www.agilent.com/en/products/cell-analysis/software-download-for-wave-desktop) normalized to cell number.
Prestoblue viability assay
Cell metabolism was measured by PrestoBlue Cell Viability Reagent (Invitrogen Cat. A13262) as described previously (61). NSCs were incubated in PrestoBlue solution (1:10 dilution in culture media) for 15 min in a CO2 incubator. The fluorescence emission of of the PrestoBlue solution was measured at 590 nm (excitation 560 nm) in a ClarioStar plate reader (BMG).
Metabolomic analyses
NSC samples were collected for metabolomics analysis from cell culture via immersion in ice cold methanol followed by scraping. Following sample collection metabolites were extracted as described previously (62). Aqueous metabolites were analyzed using a combination of a previously established ion pairing method operating exclusively in negative ionization mode and a pentafluorophenylpropyl (F5) column method operating exclusively in positive ionization mode. Aqueous fraction samples from the metabolite extraction were separated using a Sciex ExionLC AC system and measured using a Sciex 5500 QTRAP mass spectrometer. Quality control samples were injected regularly to monitor for signal stability. For negative mode analysis, peaks were resolved on a Waters Atlantis T3 column (100 Å, 3 μm, 3 mm × 100 mm) using a binary gradient from 5 mM tributylamine, 5 mM acetic acid in 2% isopropanol, 5% methanol, 93% water (v/v) to 100% isopropanol over 15 min. For positive mode analysis, a Phenomenex Kinetex F5 column (100 Å, 2.6 μm, 2.1 mm × 100 mm) column was used with a binary gradient from 100% water with 0.1% formic acid to 95% acetonitrile with 0.1% formic acid over 5 min at a flow rate of 0.3 ml/min. Each metabolite was identified using at least two distinct multiple reaction monitoring (MRM) signals and a defined retention time. All signals were integrated using MultiQuant Software 3.0.3 (https://sciex.com/br/products/software/multiquant-software). Signals with greater than 50% missing values were discarded and remaining missing values were replaced with the lowest registered signal value. All signals with a QC coefficient of variance greater than 30% were discarded. Metabolites with multiple MRMs were quantified with the higher signal to noise MRM. Filtered datasets were total sum normalized prior to analysis and stitched together using a common signal for glutamate. Single and multivariate analysis was performed in MarkerView Software 1.3.1 (https://sciex.com/products/software/markerview-software). All univariate comparisons were subjected to a Benjamini–Hochberg cut-off for false discovery as indicated. Raw data can be found in Supporting information 5.
LDH activity assay
LDH activity was measured using the Lactate Dehydrogenase Activity Assay Kit (MAK066 Sigma). Mechanically dissociated cells were passed through a cell strainer. One million cells per samples were lysed in assay buffer and then centrifuged at 10,000g for 15 min at 4 °C. Protein concentrations of the supernatants were normalized using the Rapid Gold BCA Protein Assay Kit (Pierce) prior to assaying for LDH activity. The LDH activity was measured following manufacturer’s instructions using a ClarioStar plate reader (BMG).
Western blotting
Lysates were denatured by boiling for 5 min in 1 × sample buffer containing ∼6 (v/v) beta-mercaptoethanol. Denatured lysates were resolved in Bolt 4 to 12% Bis-Tris gels (Invitrogen) and transferred to a polyvinylidene difluoride membrane (Millipore). Membranes were probed with anti-EGFR (Cell Signaling #2232) at a 1:1000 dilution, anti-PrP (6D11; Biolegend #808001) at a 1:5000 dilution, anti-p21 (AbCam; ab188224) at a 1:1000 dilution, or anti-β-actin (AbCam; ab8227) at a 1:1000 dilution. The protein bands were visualized using ECL Select (Amersham and imaged by iBright imaging system (Invitrogen). Total protein was visualized by Coomassie staining. Quantifications were performed using densitometry measurements generated in Fiji ImageJ 1.52n (https://imagej.net/software/fiji/downloads) and are shown relative to total protein levels for normalization.
Data and statistical analyses
All graphs shown were generated using GraphPad Prism (https://graphpad-prism.software.informer.com/5.0/) and represent the mean and SD of the biological repeats unless otherwise stated. The “n” numbers displayed in the figures come from averages of individual reaction wells seeded with cells plated at the same time. All experiments were repeated at least three times with similar results. Data was tested for normal distribution where appropriate using GraphPad Prism’s normality test. Heat maps show Z-scores.
Data availability
Data can be found in this manuscript and in the National Library of Medicine, National Center for Bioinformatic Information, Sequence Read Archive, under BioProject ID: PRJNA961204. https://www.ncbi.nlm.nih.gov/sra/PRJNA961204.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors would like to thank Sue Priola and Jason Hollister for critical review of the manuscript, Brent Race for collection of mouse brains for NSC generation, and Forrest Jessop for critical discussions.
Author contributions
B. R. G. and C. L. H. conceptualization; B. R. G. and C. L. H. methodology; B. R. G., B. S., E. B., S. T. F., J. A. C., and A. R. W. investigation; B. R. G., B. S., E. B., J. A. C., and C. L. H. formal analysis; C. M. B. and C. L. H. supervision; B. R. G. and C. L. H. writing–original draft; B. R. G., B. S., E. B., S. T. F., J. A. C., A. R. W., C. M. B., and C. L. H. writing–review and editing.
Funding and additional information
This research was supported by the Intramural Research Program of the NIH (NIAID). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Elizabeth J. Coulson
Supporting information
Secreted PrP from GPI anchorless PrP NSCs shows less senescence than KO NSCs but more than WT NSCs.A, example images of beta-galactosidase staining in WT, KO, and GPI-anchorless PrP NSCs. The scale bar indicates 1000 μm. B, quantification of the number of cells stained positive for beta-galactosidase relative to the WT control. NSC, neural stem cell; PrP, prion protein.
PrP expression.A, Western blot analysis and quantitation (B) of prion protein expression. B, RNA expression levels of PrP. B (n = 4) and C (n = 6) were analyzed by two-way ANOVA ∗∗∗∗p < 0.0001, ∗p < 0.05. Graphs are displayed relative the average of the WT (B and C). PrP, prion protein.
Senescence staining raw data.A, quantification of beta-galactosidase staining by cells counts (shown normalized to the respective control in the main text). B, alternative analysis approach of quantifying the area of beta-galactosidase staining shows very similar results to cell counts.
Expanded STRING analysis. Analysis of the protein–protein interactions associated with the decreased senescence phenotype of the PrP KO NSCs treated with 2× (40 ng) EGF and contrasted with the increased senescence phenotype of the WT NSCs following EGF withdrawal. Only 41 genes were significantly changed (padj < 0.05) in the PrP KO NSC, increased EGF analysis so only these were included. For consistency, only the most significantly changed 41 genes were used for the WT NSC, no EGF analysis, despite a much larger number being significantly changed overall. The minimum required interaction score was set to high confidence (0.7). Where more than five categories were significant, only the top five based on network strength are shown. EGF, epidermal growth factor; NSC, neural stem cell; PrP, prion protein.
Raw data for the metabolomic analyses.
qPCR raw data file.
RNA-seq quality control data.
Expanded bulk RNA-seq methods.
FPKM data.
DEG lists.
References
- 1.Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 2.Caughey B., Kraus A. Transmissibility versus pathogenicity of self-propagating protein aggregates. Viruses. 2019;11:1044. doi: 10.3390/v11111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steele A.D., Lindquist S., Aguzzi A. The prion protein knockout mouse: a phenotype under challenge. Prion. 2007;1:83–93. doi: 10.4161/pri.1.2.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H.-P., DeArmond S.J., et al. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 5.Tobler I., Gaus S.E., Deboer T., Achermann P., Fischer M., Rülicke T., et al. Altered circadian activity rhythms and sleep in mice devoid of prion protein. Nature. 1996;380:639–642. doi: 10.1038/380639a0. [DOI] [PubMed] [Google Scholar]
- 6.Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., et al. Axonal prion protein is required for peripheral myelin maintenance. Nat. Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- 7.Nico P.B., Lobao-Soares B., Landemberger M.C., Marques W., Jr., Tasca C.I., de Mello C.F., et al. Impaired exercise capacity, but unaltered mitochondrial respiration in skeletal or cardiac muscle of mice lacking cellular prion protein. Neurosci. Lett. 2005;388:21–26. doi: 10.1016/j.neulet.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 8.Carulla P., Llorens F., Matamoros-Angles A., Aguilar-Calvo P., Espinosa J.C., Gavin R., et al. Involvement of PrP(C) in kainate-induced excitotoxicity in several mouse strains. Sci. Rep. 2015;5 doi: 10.1038/srep11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spudich A., Frigg R., Kilic E., Kilic U., Oesch B., Raeber A., et al. Aggravation of ischemic brain injury by prion protein deficiency: role of ERK-1/-2 and STAT-1. Neurobiol. Dis. 2005;20:442–449. doi: 10.1016/j.nbd.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Weise J., Sandau R., Schwarting S., Crome O., Wrede A., Schulz-Schaeffer W., et al. Deletion of cellular prion protein results in reduced Akt activation, enhanced postischemic caspase-3 activation, and exacerbation of ischemic brain injury. Stroke. 2006;37:1296–1300. doi: 10.1161/01.STR.0000217262.03192.d4. [DOI] [PubMed] [Google Scholar]
- 11.Haigh C.L., Brown D.R. Prion protein reduces both oxidative and non-oxidative copper toxicity. J. Neurochem. 2006;98:677–689. doi: 10.1111/j.1471-4159.2006.03906.x. [DOI] [PubMed] [Google Scholar]
- 12.Bertuchi F.R., Bourgeon D.M., Landemberger M.C., Martins V.R., Cerchiaro G. PrPC displays an essential protective role from oxidative stress in an astrocyte cell line derived from PrPC knockout mice. Biochem. Biophys. Res. Commun. 2012;418:27–32. doi: 10.1016/j.bbrc.2011.12.098. [DOI] [PubMed] [Google Scholar]
- 13.Klamt F., Dal-Pizzol F., Conte da Frota M.L., Jr., Walz R., Andrades M.E., da Silva E.G., et al. Imbalance of antioxidant defense in mice lacking cellular prion protein. Free Radic. Biol. Med. 2001;30:1137–1144. doi: 10.1016/s0891-5849(01)00512-3. [DOI] [PubMed] [Google Scholar]
- 14.Wong B.S., Liu T., Li R., Pan T., Petersen R.B., Smith M.A., et al. Increased levels of oxidative stress markers detected in the brains of mice devoid of prion protein. J. Neurochem. 2001;76:565–572. doi: 10.1046/j.1471-4159.2001.00028.x. [DOI] [PubMed] [Google Scholar]
- 15.Steele A.D., Emsley J.G., Ozdinler P.H., Lindquist S., Macklis J.D. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3416–3421. doi: 10.1073/pnas.0511290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins S.J., Tumpach C., Li Q.X., Lewis V., Ryan T.M., Roberts B., et al. The prion protein regulates beta-amyloid-mediated self-renewal of neural stem cells in vitro. Stem Cell Res. Ther. 2015;6:60. doi: 10.1186/s13287-015-0067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y.J., Baskakov I.V. Treatment with normal prion protein delays differentiation and helps to maintain high proliferation activity in human embryonic stem cells. J. Neurochem. 2010;114:362–373. doi: 10.1111/j.1471-4159.2010.06601.x. [DOI] [PubMed] [Google Scholar]
- 18.Collins S.J., Tumpach C., Groveman B.R., Drew S.C., Haigh C.L. Prion protein cleavage fragments regulate adult neural stem cell quiescence through redox modulation of mitochondrial fission and SOD2 expression. Cell Mol. Life Sci. 2018;75:3231–3249. doi: 10.1007/s00018-018-2790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Y.S., Kim S.M., Lee J.H., Jung S.K., Noh H., Lee S.H. Melatonin protects chronic kidney disease mesenchymal stem cells against senescence via PrP(C) -dependent enhancement of the mitochondrial function. J. Pineal Res. 2019;66 doi: 10.1111/jpi.12535. [DOI] [PubMed] [Google Scholar]
- 20.Boilan E., Winant V., Dumortier E., ElMoualij B., Quatresooz P., Osiewacz H.D., et al. Role of Prion protein in premature senescence of human fibroblasts. Mech. Ageing Dev. 2018;170:106–113. doi: 10.1016/j.mad.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Alexander P.B., Yuan L., Yang P., Sun T., Chen R., Xiang H., et al. EGF promotes mammalian cell growth by suppressing cellular senescence. Cell Res. 2015;25:135–138. doi: 10.1038/cr.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Lannerée S., Halliez S., Hirsch T.Z., Hernandez-Rapp J., Passet B., Tomkiewicz C., et al. The cellular prion protein controls Notch signaling in neural stem/progenitor cells. Stem Cells. 2017;35:754–765. doi: 10.1002/stem.2501. [DOI] [PubMed] [Google Scholar]
- 23.Martellucci S., Manganelli V., Santacroce C., Santilli F., Piccoli L., Sorice M., et al. Role of Prion protein-EGFR multimolecular complex during neuronal differentiation of human dental pulp-derived stem cells. Prion. 2018;12:117–126. doi: 10.1080/19336896.2018.1463797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang E. Senescent human fibroblasts resist programmed cell death, and failure to suppress bcl2 is involved. Cancer Res. 1995;55:2284–2292. [PubMed] [Google Scholar]
- 25.Che T.F., Lin C.W., Wu Y.Y., Chen Y.J., Han C.L., Chang Y.L., et al. Mitochondrial translocation of EGFR regulates mitochondria dynamics and promotes metastasis in NSCLC. Oncotarget. 2015;6:37349–37366. doi: 10.18632/oncotarget.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Signes A., Fernandez-Vizarra E. Assembly of mammalian oxidative phosphorylation complexes I-V and supercomplexes. Essays Biochem. 2018;62:255–270. doi: 10.1042/EBC20170098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y.J., Baskakov I.V. The cellular form of the prion protein is involved in controlling cell cycle dynamics, self-renewal, and the fate of human embryonic stem cell differentiation. J. Neurochem. 2013;124:310–322. doi: 10.1111/j.1471-4159.2012.07913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos T.G., Silva I.R., Costa-Silva B., Lepique A.P., Martins V.R., Lopes M.H. Enhanced neural progenitor/stem cells self-renewal via the interaction of stress-inducible protein 1 with the prion protein. Stem Cells. 2011;29:1126–1136. doi: 10.1002/stem.664. [DOI] [PubMed] [Google Scholar]
- 32.Atkinson C.J., Kawamata F., Liu C., Ham S., Győrffy B., Munn A.L., et al. EGFR and Prion protein promote signaling via FOXO3a-KLF5 resulting in clinical resistance to platinum agents in colorectal cancer. Mol. Oncol. 2019;13:725–737. doi: 10.1002/1878-0261.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang J., Pan Y., Zhang D., Guo C., Shi Y., Wang J., et al. Cellular prion protein promotes proliferation and G1/S transition of human gastric cancer cells SGC7901 and AGS. FASEB J. 2007;21:2247–2256. doi: 10.1096/fj.06-7799com. [DOI] [PubMed] [Google Scholar]
- 34.Halliez S., Martin-Lannerée S., Passet B., Hernandez-Rapp J., Castille J., Urien C., et al. Prion protein localizes at the ciliary base during neural and cardiovascular development and its depletion affects α-tubulin post-translational modifications. Sci. Rep. 2015;5 doi: 10.1038/srep17146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangelberger D., Kern D., Loipetzberger A., Eberl M., Aberger F. Cooperative Hedgehog-EGFR signaling. Front Biosci. (Landmark Ed. 2012;17:90–99. doi: 10.2741/3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinchisi G., Parada M., Lois P., Oyanadel C., Shaughnessy R., Gonzalez A., et al. Sonic Hedgehog modulates EGFR dependent proliferation of neural stem cells during late mouse embryogenesis through EGFR transactivation. Front. Cell Neurosci. 2013;7:166. doi: 10.3389/fncel.2013.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massip-Copiz M.M., Valdivieso A.G., Clauzure M., Mori C., Asensio C.J.A., Aguilar M.A., et al. Epidermal growth factor receptor activity upregulates lactate dehydrogenase A expression, lactate dehydrogenase activity, and lactate secretion in cultured IB3-1 cystic fibrosis lung epithelial cells. Biochem. Cell Biol. 2021;99:476–487. doi: 10.1139/bcb-2020-0522. [DOI] [PubMed] [Google Scholar]
- 38.Ramljak S., Schmitz M., Zafar S., Wrede A., Schenkel S., Asif A.R., et al. Cellular prion protein directly interacts with and enhances lactate dehydrogenase expression under hypoxic conditions. Exp. Neurol. 2015;271:155–167. doi: 10.1016/j.expneurol.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 39.Xu D., Finkel T. A role for mitochondria as potential regulators of cellular life span. Biochem. Biophys. Res. Commun. 2002;294:245–248. doi: 10.1016/S0006-291X(02)00464-3. [DOI] [PubMed] [Google Scholar]
- 40.Kim J.Y., Lee S.H., Bae I.H., Shin D.W., Min D., Ham M., et al. Pyruvate protects against cellular senescence through the control of mitochondrial and lysosomal function in dermal fibroblasts. J. Invest. Dermatol. 2018;138:2522–2530. doi: 10.1016/j.jid.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 41.Arnould H., Baudouin V., Baudry A., Ribeiro L.W., Ardila-Osorio H., Pietri M., et al. Loss of prion protein control of glucose metabolism promotes neurodegeneration in model of prion diseases. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrelli F., Scandella V., Montessuit S., Zamboni N., Martinou J.C., Knobloch M. Mitochondrial pyruvate metabolism regulates the activation of quiescent adult neural stem cells. Sci. Adv. 2023;9 doi: 10.1126/sciadv.add5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y.J., Fan X.Y., Wang A.D., Xia Y.Z., Fu W.R., Liu J.Y., et al. LDHA suppression altering metabolism inhibits tumor progress by an organic arsenical. Int. J. Mol. Sci. 2019;20:6239. doi: 10.3390/ijms20246239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le A., Cooper C.R., Gouw A.M., Dinavahi R., Maitra A., Deck L.M., et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di H., Zhang X., Guo Y., Shi Y., Fang C., Yuan Y., et al. Silencing LDHA inhibits proliferation, induces apoptosis and increases chemosensitivity to temozolomide in glioma cells. Oncol. Lett. 2018;15:5131–5136. doi: 10.3892/ol.2018.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miele G., Jeffrey M., Turnbull D., Manson J., Clinton M. Ablation of cellular prion protein expression affects mitochondrial numbers and morphology. Biochem. Biophys. Res. Commun. 2002;291:372–377. doi: 10.1006/bbrc.2002.6460. [DOI] [PubMed] [Google Scholar]
- 47.Paterson A.W.J., Curtis J.C., MacLeod N.K. Complex I specific increase in superoxide formation and respiration rate by PrP-null mouse brain mitochondria. J. Neurochem. 2008;105:177–191. doi: 10.1111/j.1471-4159.2007.05123.x. [DOI] [PubMed] [Google Scholar]
- 48.De Mario A., Peggion C., Massimino M.L., Norante R.P., Zulian A., Bertoli A., et al. The link of the prion protein with Ca(2+) metabolism and ROS production, and the possible implication in abeta toxicity. Int. J. Mol. Sci. 2019;20:4640. doi: 10.3390/ijms20184640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y., Han C.Y., Duan F.G., Fan X.X., Yao X.J., Parks R.J., et al. p53 sensitizes chemoresistant non-small cell lung cancer via elevation of reactive oxygen species and suppression of EGFR/PI3K/AKT signaling. Cancer Cell Int. 2019;19:188. doi: 10.1186/s12935-019-0910-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X., Fan X.X., Jiang Z.B., Loo W.T., Yao X.J., Leung E.L., et al. Shikonin inhibits gefitinib-resistant non-small cell lung cancer by inhibiting TrxR and activating the EGFR proteasomal degradation pathway. Pharmacol. Res. 2017;115:45–55. doi: 10.1016/j.phrs.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 51.Weng M.S., Chang J.H., Hung W.Y., Yang Y.C., Chien M.H. The interplay of reactive oxygen species and the epidermal growth factor receptor in tumor progression and drug resistance. J. Exp. Clin. Cancer Res. 2018;37:61. doi: 10.1186/s13046-018-0728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strom A., Wang G.S., Scott F.W. Impaired glucose tolerance in mice lacking cellular prion protein. Pancreas. 2011;40:229–232. doi: 10.1097/mpa.0b013e3181f7e547. [DOI] [PubMed] [Google Scholar]
- 53.Le Pichon C.E., Valley M.T., Polymenidou M., Chesler A.T., Sagdullaev B.T., Aguzzi A., et al. Olfactory behavior and physiology are disrupted in prion protein knockout mice. Nat. Neurosci. 2009;12:60–69. doi: 10.1038/nn.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akter M., Kaneko N., Sawamoto K. Neurogenesis and neuronal migration in the postnatal ventricular-subventricular zone: similarities and dissimilarities between rodents and primates. Neurosci. Res. 2021;167:64–69. doi: 10.1016/j.neures.2020.06.001. [DOI] [PubMed] [Google Scholar]
- 55.Curtis M.A., Monzo H.J., Faull R.L. The rostral migratory stream and olfactory system: smell, disease and slippery cells. Prog. Brain Res. 2009;175:33–42. doi: 10.1016/S0079-6123(09)17503-9. [DOI] [PubMed] [Google Scholar]
- 56.Carroll J.A., Foliaki S.T., Haigh C.L. A 3D cell culture approach for studying neuroinflammation. J. Neurosci. Methods. 2021;358 doi: 10.1016/j.jneumeth.2021.109201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carroll J.A., Groveman B.R., Williams K., Moore R., Race B., Haigh C.L. Prion protein N1 cleavage peptides stimulate microglial interaction with surrounding cells. Sci. Rep. 2020;10:6654. doi: 10.1038/s41598-020-63472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Race B., Baune C., Williams K., Striebel J.F., Hughson A.G., Chesebro B. Second passage experiments of chronic wasting disease in transgenic mice overexpressing human prion protein. Vet. Res. 2022;53:111. doi: 10.1186/s13567-022-01130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas P.D., Ebert D., Muruganujan A., Mushayahama T., Albou L.P., Mi H. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 2022;31:8–22. doi: 10.1002/pro.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Groveman B.R., Foliaki S.T., Orru C.D., Zanusso G., Carroll J.A., Race B., et al. Sporadic Creutzfeldt-Jakob disease prion infection of human cerebral organoids. Acta Neuropathol. Commun. 2019;7:90. doi: 10.1186/s40478-019-0742-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foliaki S.T., Smith A., Schwarz B., Bohrnsen E., Bosio C.M., Williams K., et al. Altered energy metabolism in Fatal Familial Insomnia cerebral organoids is associated with astrogliosis and neuronal dysfunction. PLoS Genet. 2023;19 doi: 10.1371/journal.pgen.1010565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Secreted PrP from GPI anchorless PrP NSCs shows less senescence than KO NSCs but more than WT NSCs.A, example images of beta-galactosidase staining in WT, KO, and GPI-anchorless PrP NSCs. The scale bar indicates 1000 μm. B, quantification of the number of cells stained positive for beta-galactosidase relative to the WT control. NSC, neural stem cell; PrP, prion protein.
PrP expression.A, Western blot analysis and quantitation (B) of prion protein expression. B, RNA expression levels of PrP. B (n = 4) and C (n = 6) were analyzed by two-way ANOVA ∗∗∗∗p < 0.0001, ∗p < 0.05. Graphs are displayed relative the average of the WT (B and C). PrP, prion protein.
Senescence staining raw data.A, quantification of beta-galactosidase staining by cells counts (shown normalized to the respective control in the main text). B, alternative analysis approach of quantifying the area of beta-galactosidase staining shows very similar results to cell counts.
Expanded STRING analysis. Analysis of the protein–protein interactions associated with the decreased senescence phenotype of the PrP KO NSCs treated with 2× (40 ng) EGF and contrasted with the increased senescence phenotype of the WT NSCs following EGF withdrawal. Only 41 genes were significantly changed (padj < 0.05) in the PrP KO NSC, increased EGF analysis so only these were included. For consistency, only the most significantly changed 41 genes were used for the WT NSC, no EGF analysis, despite a much larger number being significantly changed overall. The minimum required interaction score was set to high confidence (0.7). Where more than five categories were significant, only the top five based on network strength are shown. EGF, epidermal growth factor; NSC, neural stem cell; PrP, prion protein.
Raw data for the metabolomic analyses.
qPCR raw data file.
RNA-seq quality control data.
Expanded bulk RNA-seq methods.
FPKM data.
DEG lists.
Data Availability Statement
Data can be found in this manuscript and in the National Library of Medicine, National Center for Bioinformatic Information, Sequence Read Archive, under BioProject ID: PRJNA961204. https://www.ncbi.nlm.nih.gov/sra/PRJNA961204.