Abstract
Previous studies have shown the predominance of mycolic acid-containing filamentous actinomycetes (mycolata) in foam layers in activated sludge systems. Gordona (formerly Nocardia) amarae often is considered the major representative of this group in activated sludge foam. In this study, small-subunit rRNA genes of four G. amarae strains were sequenced, and the resulting sequences were compared to the sequence of G. amarae type strain SE-6. Comparative sequence analysis showed that the five strains used represent two lines of evolutionary descent; group 1 consists of strains NM23 and ASAC1, and group 2 contains strains SE-6, SE-102, and ASF3. The following three oligonucleotide probes were designed: a species-specific probe for G. amarae, a probe specific for group 1, and a probe targeting group 2. The probes were characterized by dissociation temperature and specificity studies, and the species-specific probe was evaluated for use in fluorescent in situ hybridizations. By using the group-specific probes, it was possible to place additional G. amarae isolates in their respective groups. The probes were used along with previously designed probes in membrane hybridizations to determine the abundance of G. amarae, group 1, group 2, bacterial, mycolata, and Gordona rRNAs in samples obtained from foaming activated sludge systems in California, Illinois, and Wisconsin. The target groups were present in significantly greater concentrations in activated sludge foam than in mixed liquor and persisted in anaerobic digesters. Hybridization results indicated that the presence of certain G. amarae strains may be regional or treatment plant specific and that previously uncharacterized G. amarae strains may be present in some systems.
Activated sludge treatment plants in different parts of the world with widely varying wastewater characteristics and operating conditions have experienced foaming episodes in their aeration basins and anaerobic digesters (9, 12, 30, 38, 45). Measures that have been used to control foaming have not produced consistent results; consequently, the adoption of control mechanisms remains empirical (39). Foaming can lead to numerous problems, such as reduction in effluent quality, severe operational difficulties in anaerobic digesters, and clogging of gas collection systems (29). The recent isolation of a pathogenic Nocardia sp. from activated sludge foam (44) suggests that foaming also may represent a health concern because of the possible spread of pathogens in windblown scum (8).
Mycolic acid-containing actinomycetes or mycolata (14) often have been isolated from foams and are considered the main cause of foam stabilization in many activated sludge systems throughout the world. The phylogeny of the mycolata recently has been elucidated by using small-subunit (SSU) rRNA sequence analyses. Goodfellow and coworkers distinguished two suprageneric lineages and proposed that the mycolata should be grouped into the following two families: the family Corynebacteriaceae, which encompasses the genera Corynebacterium, Dietza, and Turicella; and the family Mycobacteriaceae, containing the genera Gordona, Mycobacterium, Nocardia, Rhodococcus, Tsukamurella, and Skermania (13, 14). Stackebrandt et al. (40) proposed a new suborder within the order Actinomycetales, the suborder Corynebacterineae, which would include the families Nocardiaceae, Gordoniaceae, Mycobacteriaceae, Dietziaceae, Tsukamurellaceae, and Corynebacteriaceae. Some members of these groups, including Gordona (Nocardia) amarae and “Microthrix parvicella,” an unclassified actinomycete (11), commonly have been cited as predominant organisms in activated sludge foams (9, 12, 25, 38). Stackebrandt et al. (40) also proposed that the genus Gordona should be renamed Gordonia based on correct etymology.
In most studies in which it was determined that G. amarae was a major microorganism in activated sludge foam, morphology- and physiology-based identification methods were used. These methods are inadequate for describing microbial diversity in complex environments, such as activated sludge (37, 47). Filament identification techniques (20, 23), which are widely used in activated sludge microbiology, are hampered by the morphological similarities of actinomycetes (7) and the transitions that these organisms undergo depending on their growth stages (hyphal to coccal stages and vice versa) (39). Biases presented by culture-based methods and the recent finding that G. amarae is one of the easiest actinomycetes to isolate and maintain in pure culture (24) make quantification of this organism with culture-based techniques unreliable. Therefore, previous reports of G. amarae abundance in foam samples may have been inadequate. These observations raise new questions concerning the significance of this species in foaming.
Oligonucleotide probes that target rRNA may provide more reliable characterizations of microbial community structure (4, 5, 42). We previously designed a probe for G. amarae based on a limited number of sequences, probe S-S-G.am-0192-a-A-18 (probe nomenclature is based on the nomenclature of the Oligonucleotide Probe Database [2]) (17). However, G. amarae quantification was impaired after it was determined that the probe targeted only a few of the known G. amarae strains. As a result, assessment of the significance of G. amarae in foaming was not possible.
In this study, we report interstrain heterogeneity in the SSU ribosomal DNA (rDNA) sequences of G. amarae, which explains the variable responses of various G. amarae strains with the previously designed species-specific probe (17). We describe the development and application of a probe for G. amarae that targets all available strains, as well as two probes that distinguish the interstrain sequence variability in this species.
MATERIALS AND METHODS
Organisms, culture techniques, and nucleic acid extractions.
The following seven strains of G. amarae were used in this study: SE-6 (= ATCC 27808) (type strain), SE-102 (= ATCC 27809), SE-149B (= ATCC 27810), NM23, ASAC1, RBI, and ASF3. Strains SE-6, SE-102, and SE-149B were isolated by plate dilution by Lechevalier and Lechevalier (25) from activated sludge foams in Andover, Fla. (Miami, Fla.), Bordentown Township, N.J., and Jamaica Bay, N.Y., respectively. Strain ASAC1 was obtained by micromanipulation from mixed liquor from the Sacramento, Calif., Regional Water Quality Plant by L. L. Blackall and H. Ho; strain RBI was isolated by plate dilution from mixed liquor from the Richmond, Calif., wastewater treatment plant by M. Richard (46); and strain ASF3 was isolated by micromanipulation from mixed liquor from the San Francisco, Calif., Southeast Water Pollution Control Plant. Strain NM23 was obtained from an activated sludge system in Queensland, Australia, by L. L. Blackall by micromanipulation (9). A variety of organisms not targeted by the probes designed in this study served as controls in specificity studies and fluorescent in situ hybridization (FISH) studies (see below).
The strains were grown in yeast extract-glucose broth (10 g of tryptone per liter, 7 g of NaCl per liter, 2.5 g of yeast extract per liter, 1 g of dextrose per liter; pH 7.0) for 3 to 5 days at 37°C with constant shaking. Strain ASF3 was grown on the mineral salts medium of Stanier et al. (43) after it was found that this organism did not grow well in yeast extract-glucose broth. The cells were harvested by centrifugation, and the cell pellets were immediately frozen on dry ice and kept at −80°C.
Genomic DNA was extracted by using a microwave protocol for gram-positive bacteria. This method involved washing of the cell pellets with TE buffer (10 mM Tris [pH 8.0], 10 mM EDTA), resuspension in TE buffer and 10% sodium dodecyl sulfate (SDS), and incubation at 65°C for 30 min to lyse the cells. The lysate was centrifuged, the tubes were heated in a microwave oven (1,000 W) twice for 1 min, and the resulting pellets were dissolved in TE buffer. DNA was extracted with chloroform-isoamyl alcohol-phenol (24:1:25, vol/vol/vol), precipitated with ethanol (overnight, −20°C), and centrifuged, and the resulting pellets were resuspended in water. DNA concentrations were determined spectrophotometrically by assuming that 1 mg of DNA per ml was equivalent to 20 optical density units at a wavelength of 260 nm. DNA quality was assessed by gel electropheresis on 1% agarose gels, and bands were viewed with a charge-coupled device camera (Bioimaging Technologies, Elburn, Ill.).
RNA was extracted by a low-pH hot-phenol method (41). The quality of the extracted RNA was evaluated by polyacrylamide gel electrophoresis (1), and the extracted RNA was quantified with the BIT Image Analysis software (Bioimaging Technologies).
Amplification and cloning of SSU rDNA.
The SSU rDNA was amplified by PCR with primers S-D-Bact-0011-a-S-17 and S-D-Bact-1492-a-A-21 by using the following protocol: one cycle consisting of 4 min at 94°C, 1 min at 50°C, and 1.5 min at 72°C; 28 cycles consisting of 1 min at 92°C, 1 min at 50°C, and 5 min at 72°C; and temporary storage at 4°C. The sizes and quantities of the PCR products were evaluated by agarose gel electrophoresis, and the PCR products were quantified with the BIT Image Analysis software.
Cloning and transformation were carried out by using instructions provided with an Invitrogen TA cloning kit, version B (Invitrogen Corporation, San Diego, Calif.). Transformants were screened further by preparing plasmids and performing restriction enzyme digestions (36).
Sequencing of the SSU rDNA.
Cell pellets were prepared from overnight growth of transformed Escherichia coli cells on Luria-Bertani broth, and sequencing was performed at the University of Illinois Biotechnology Center Sequencing Laboratory with four primers specific for the bacterial domain, S-*-Bact-0343-a-A-15 (TACGGGAGGCAGCAG), S-*-Bact-1100-a-S-16 (AGGGTTGCGCTCGTTG), S-*-Bact-0519-a-S-18 (GTATTACCGCGGCTGCTG), and S-*-Bact-0907-a-A-20 (AAACTCAAATGAATTGACGG) (19), as well as the M13(-20) forward and M13 reverse primers (Invitrogen Corporation). The sequences obtained for strains SE-102, ASAC1, ASF3, and NM23 were assembled and edited by using the Fragment Assembly program of the Genetics Computer Group software, version 9.0, Wisconsin Package, and the Sequencher program (Gene Codes Corp., Ann Arbor, Mich.). Phylogenetic analyses were done with programs available in the PHYLIP package (version 3.5) (21), such as the distance and maximum-likelihood methods.
Design and characterization of oligonucleotide probes.
Based on the SSU rDNA sequences obtained for strains NM23, ASAC1, ASF3, and SE-102 and the sequence of type strain SE-6 available from the Ribosomal Database Project (RDP) (26), oligonucleotide probes were designed to target the G. amarae strains. The newly designed oligonucleotide probes, their target groups, and their sequences are shown in Table 1 and Fig. 1. Additional probes used in this study also are listed in Table 1. The oligonucleotide probes were synthesized with a DNA synthesizer (Applied Biosystems, Foster City, Calif.) at the University of Illinois Biotechnology Center DNA Synthesis Laboratory and were purified with an oligonucleotide purification cartridge (Applied Biosystems).
TABLE 1.
Oligonucleotide probes used in membrane hybridizations
| Probea | Target group | Optimum Td (°C) | Reference |
|---|---|---|---|
| S-*-Univ-1390-a-A-18 | Almost all organisms | 44 | 50 |
| S-D-Bact-0338-a-A-18 | Domain Bacteria | 56 | 6 |
| S-*-Myb-0736-b-A-22 | Mycolata | 51 | 17 |
| S-G-Gor-0596-a-A-22 | Genus Gordona | 54 | 17 |
| S-S-G.am-0205-a-A-19 | Gordona amarae | 53 | This study |
| S-*-G.am1-0439-a-A-19 | G. amarae group 1 strains | 66 | This study |
| S-*-G.am2-0439-a-A-19 | G. amarae group 2 strains | 61 | This study |
Probe names are standardized according to the Oligonucleotide Probe Database as follows: S or L for large- or small-subunit rRNA as the target; D for domain, O for order, F for family, G for genus, S for species, Ss for subspecies, and a wild-card symbol (∗) for universal probes, probes designed to target unidentified environmental isolates, or probes that do not circumscribe a coherent taxonomic group; letters designating the target group of the oligonucleotide probe; nucleotide position (E. coli numbering) in the target where the 3′ end of the probe binds; letter designating the version of the probe (a for version 1, b for version 2, etc.); S or A for sense or antisense direction; and number indicating the length (in nucleotides) of the probe (2).
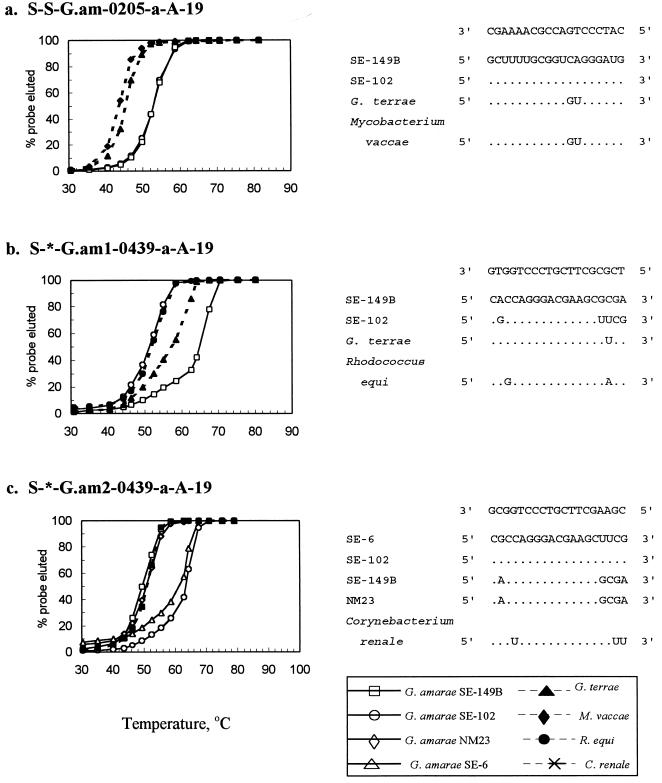
FIG. 1.
Td studies for probes S-S-G.am-0205-a-A-19, S-*-G.am1-0439-a-A-19, and S-*-G.am2-0439-a-A-19. Adjacent to the probe dissociation results are the SSU rRNA sequences of target and nontarget species and probe sequences. The top SSU rRNA sequence in each list of sequences is the target sequence. Dots in the sequences below this sequence indicate identical nucleotides, and replacement nucleotides are nucleotides that differ from the nucleotides in the target sequences.
To determine the optimal hybridization wash temperatures, dissociation temperature (Td) studies were performed (17, 50) by using RNAs from a variety of target and nontarget organisms (Fig. 1). Oligonucleotide probes were 5′ end labeled with [γ-32P]ATP (ICN Radiochemicals, Irvine, Calif.) (32) and were purified with a Nensorb 20 cartridge (DuPont Corp., Boston, Mass.) (42). Prehybridization, hybridization, washing, and scintillation counting were carried out as described by de los Reyes et al. (17). The temperature at which 50% of the hybridized probe washed off was determined to be the Td of the probe.
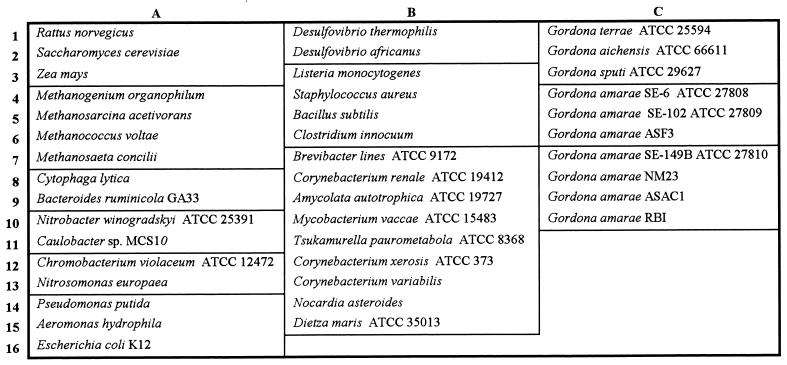
The specificities of the probes were assessed experimentally by hybridizing them with RNAs from 41 target and nontarget organisms (Fig. 2). The final wash of the hybridization membranes was performed at the previously determined Td, the membranes were exposed to PhosphorImager screens (Molecular Dynamics, Sunnyvale, Calif.), and hybridization signals were viewed and quantified with a PhosphorImager (Molecular Dynamics) (17).
FIG. 2.
Membrane template used for the probe specificity study (see Fig. 4).
Characterization of environmental samples.
The environmental samples used and the wastewater treatment plants from which they were obtained are listed in Table 2. All of the plants experienced foaming at the time of sampling or had a history of recurring foaming. The samples were stored in 50-ml conical centrifuge tubes (Corning Costar Corp., Cambridge, Mass.), immediately put on ice, and brought or mailed overnight to the laboratory. Aliquots were centrifuged, and cell pellets were frozen in ethanol with dry ice and stored at −80°C until they were used for RNA extraction. RNA was extracted by a low-pH hot-phenol extraction method (41).
TABLE 2.
Summary of samples used in membrane hybridizations
| Wastewater treatment plant | Type of activated sludge system, wastewater treated | Sample locations | Foaming frequency |
|---|---|---|---|
| San Francisco Southeast Water Pollution Control Plant, San Francisco, Calif. | High-purity oxygen, municipal wastewater | RAS, anaerobic digester sludge | Always |
| Richmond Wastewater Treatment Plant, Richmond, Calif. | Conventional, municipal wastewater | RAS, anaerobic digester sludge | Intermittent |
| City of Sacramento Regional Wastewater Treatment Plant, Sacramento, Calif. | High-purity oxygen, municipal wastewater | RAS, anaerobic digester sludge | Intermittent (summer) |
| Oakland East Bay Municipal Utilities District (EBMUD), Oakland, Calif. | High-purity oxygen, municipal wastewater | RAS, anaerobic digester sludge | Intermittent |
| Urbana-Champaign Sanitary District Northeast Treatment Plant, Urbana, Ill. | Contact stabilization municipal wastewater | Aeration tank foam, aeration tank mixed liquor, anaerobic digester foam, anaerobic digester sludge | Recurrent |
| Wisconsin | Completely mixed, milk-processing wastewater | Aeration tank foam, aeration tank mixed liquor | Frequent |
Quantification of extracted RNA and hybridizations with radioactive oligonucleotide probes were performed as previously described (17). All hybridization membranes were prepared in duplicate; for each set of membranes, one membrane was hybridized with a universal probe, and the other membrane was hybridized with a specific probe (Table 1). The hybridization responses of triplicate applications of RNAs obtained from samples and of a dilution series of RNA obtained from a pure culture (target organism) were used to determine the relative concentrations of target SSU rRNAs in the samples. Hybridization results were expressed in terms of the relative SSU rRNA concentrations of target groups, which were calculated by dividing the amount of rRNA (in nanograms) targeted by a specific probe by the amount of rRNA (in nanograms) bound to the universal probe.
FISH and microscopy.
A tetramethyl rhodamine (TRITC)-labeled version of the species-specific probe S-S-G.am-0205-a-A-19 was obtained from Genosys Biotechnologies, Inc. (The Woodlands, Tex.). Pure cultures and wastewater treatment plant samples were fixed in 4% paraformaldehyde for 1 min at room temperature (17). FISH were performed on SuperCured heavy Teflon-coated slides (Cel-Line Associates Inc., Newfield, N.J.) as previously described (17), with minor modifications. The hybridization buffer consisted of 30% formamide, 0.9 M NaCl, 0.1 M Tris (pH 7.2), and 0.1% SDS. Hybridizations were conducted in saturated humidity chambers at 46°C for 2 h. The slides were rinsed once and immersed in 50 ml of wash solution (80 mM NaCl, 0.05 M sodium phosphate buffer [pH 7.2], 0.1% SDS, 5 mM EDTA) at 48°C for 20 min. These hybridization and wash conditions were found to be optimal for in situ hybridization with this probe. In some cases, dual staining with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) was performed (17). The slides were then washed with ice-cold water, air dried, and mounted in Citifluor (UKC Chemical Laboratory, Canterbury, United Kingdom).
The slides were visualized with an epifluorescence microscope (Axioskop; Carl Zeiss, Oberkochen, Germany) fitted with filter sets for DAPI and TRITC (Chroma Technology Corp., Brattleboro, Vt.) (17). Images were captured with a charge-coupled device camera (Photometrics Ltd., Tucson, Ariz.) and IPLab Spectrum image analysis software (Signal Analytics, Vienna, Va.). Images were imported to Adobe Photoshop 3.0 (Adobe, Seattle, Wash.) for printing.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank database under accession no. AF020329 for ASAC1, accession no. AF020330 for SE-102, accession no. AF020331 for ASF3, and accession no. AF020332 for NM23.
RESULTS AND DISCUSSION
Sequence analysis.
Complete SSU rDNA sequences for strains NM23, ASAC1, ASF3, and SE-102 and partial sequences for RBI and SE-149B were obtained. Alignment with sequences deposited in the RDP and GenBank-EMBL databases showed that the sequences of all of the strains were most similar to the G. amarae SE-6 sequence (accession no. X80635). An analysis to determine the presence of chimeras produced negative results for all sequences.
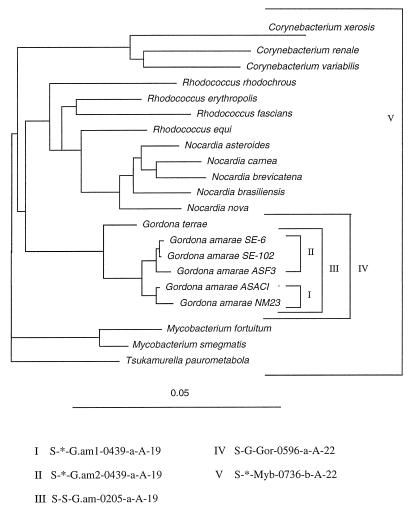
A comparison of the full-length sequences and similarity values obtained from the Pileup program of the Genetics Computer Group software indicated that strains ASAC1 and NM23 were more similar to each other than to the other strains, while strains ASF3, SE-102, and SE-6 clustered together (data not shown). This apparent grouping of the G. amarae strains into two clusters was supported by the results of phylogenetic analyses in which we used maximum-likelihood and distance-based neighbor-joining methods, which produced similar phylogenetic trees. Figure 3 shows the phylogenetic tree obtained with a neighbor-joining–unweighted pair group method with arithmetic averages (distances were calculated by using a Kimura two-parameter distance setting), which shows the distinct branching of the strains into two groups; strains NM23 and ASAC1 cluster together in one group (group 1), and strains SE-102, ASF3, and SE-6 cluster together in another group (group 2). The partial sequences of strains SE-149B and RBI indicated that these organisms belong to group 1, and this was confirmed by the results of probe specificity and Td studies (see below). The evolutionary relationships among mycolata reflected in the phylogenetic tree are consistent with the evolutionary relationships observed by Chun et al. (14).
FIG. 3.
Phylogenetic tree showing the positions of representative mycolata, inferred from a comparison of SSU rRNA sequences. The tree was constructed by using a neighbor-joining method. The bar represents 5 estimated changes per 100 nucleotides. The oligonucleotide probes are shown with their respective target groups. Probes I, II, and III were designed in this study; probes IV and V have been described previously (17).
Intraspecific variation in SSU rDNA sequences has been attributed to sequencing errors, misidentification of strains, and rRNA operon heterogeneity (16, 27, 28, 31, 35). The interstrain variability observed among the sequences obtained in this study (0.5 to 1%) is higher than the estimated 0.1% random sequencing error for sequences deposited in GenBank (15). Moreover, alignment of the sequences showed that the strains were properly classified as G. amarae. Interoperon differences cannot be ruled out as a potential source of the interstrain variability observed in this study. This can be tested if intrastrain variability is compared with interstrain variability (31). Even though we did not evaluate this possibility through additional sequence analyses, the results of probe specificity studies indicated that it is unlikely (see below).
Design of oligonucleotide probes.
The following three oligonucleotide probes were designed and characterized based on the results of the sequence analysis: S-S-G.am-0205-a-A-19, which is species specific for G. amarae; S-*-G.am1-0439-a-A-19, which targets group 1 G. amarae strains; and S-*-G.am2-0439-a-A-19, which is specific for group 2 strains (Fig. 1 and 3). The specificities of the probes were initially assessed by performing a Basic Local Alignment Search Tool (BLAST) in GenBank (3) and by using the CHECK_PROBE program provided by the RDP (26). The species-specific probe perfectly matched all G. amarae strains for which sequences were available and was an improvement over the previously designed G. amarae probe (17), which was found to have two mismatches with the group 1 strains. Probe S-S-G.am-0205-a-A-19 had at least two consecutive mismatches (at positions 216 and 217 [Escherichia coli numbering]) with the SSU rRNA sequences of nontarget species.
Probe S-*-G.am1-0439-a-A-19 perfectly matched all group 1 strains. A few closely related nontarget species (Gordona terrae, Gordona rubropertincta, and Gordona bronchialis) had one mismatch. Other nontarget organisms had at least two mismatches, while group 2 strains had five mismatches. Similarly, probe S-*-G.am2-0439-a-A-19 perfectly matched all group 2 strains, had at least three mismatches with nontarget organisms, and had five mismatches with group 1 strains.
Other factors considered during probe design were G+C content, probe length, and the presence of self-complementary regions. The G+C content and probe length were adjusted as much as possible to obtain similar theoretical Td values. This is important when multiple probes are used in simultaneous membrane or in situ hybridizations (34). No regions of internal complementarity were present in S-S-G.am-0205-a-A-19 and S-*-G.am1-0439-a-A-19, whereas probe S-*-G.am2-0439-a-A-19 contained a 4-base region of self-complementarity, but this internal complementarity region likely did not affect the hybridization response, as discussed previously (34).
Optimization of wash temperatures.
The Td values were experimentally determined for each probe and were used as posthybridization wash temperatures to ensure dissociation of duplexes with one or more mismatches (42). Figure 1 shows the Td curves obtained for the three probes and the sequence alignments for the probes and the target and nontarget organisms used in the Td studies.
Strains SE-149B (group 1) and SE-102 (group 2) had overlapping Td curves (Td, 53.5°C), indicating that they had identical responses with the species-specific probe S-S-G.am-0205-a-A-19. G. terrae and Mycobacterium vaccae, both of which had two mismatches with this probe, had Td values between 42 and 45°C.
The group 1 probe S-*-G.am1-0439-a-A-19 had a Td of 65°C for group 1 strain SE-149B and lower Td values for the following nontarget organisms: 57°C for G. terrae (one mismatch), 53°C for Rhodococcus equi (three mismatches), and 52.5°C for group 2 strain SE-102 (five mismatches). These results indicate that the group 1 probe can be used to distinguish a target strain from a nontarget strain with a single nucleotide mismatch when a posthybridization wash temperature of approximately 65°C is used.
The Td values for group 2 strains SE-6 and SE-102 were 61 and 63°C, respectively, for group 2 probe S-*-G.am2-0439-a-A-19. Corynebacterium renale (three mismatches) had a Td of 51°C, while the group 1 strains SE-149B and NM23 (five mismatches) exhibited Td values between 50 and 51°C.
Probe specificity studies.
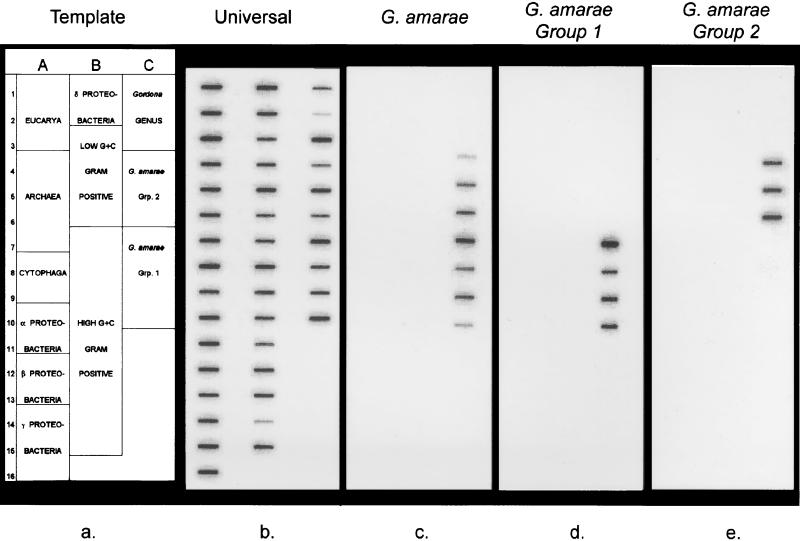
A diverse selection of RNAs obtained from target and nontarget organisms (Fig. 2) were hybridized with the oligonucleotide probes to assess their specificities. Figure 4 shows the hybridization membranes used for this specificity study. Membrane b was hybridized with a universal probe (S-*-Univ-1390-a-A-18 [50]), which targeted all RNA samples applied to the membrane. Variations in the signal intensities on this membrane were likely due to differences in the amounts of RNA applied. Membrane c shows the results of hybridization with the species-specific probe; this probe targeted all seven G. amarae strains, including strains SE-149B and RBI, for which only partial sequences were available. The variations in signal intensity for the G. amarae strains were generally consistent with the variations observed with the universal probe. The specificity results for the group 1 probe (membrane d) show that this probe hybridized with all group 1 strains, including strains SE-149B and RBI. We used a wash temperature of 66°C for this probe; this temperature was 1°C higher than the experimentally derived Td, which resulted in minimal nonspecific binding for closely related nontarget organisms. For example, the hybridization responses for G. terrae and M. vaccae RNAs comprised only 2.6 and 3.1%, respectively, of the signal for the target group 1 RNAs. Membrane e shows that the group 2 probe hybridized only with group 2 strains. The wash temperature that resulted in strong hybridization responses for the target strains and negligible signals for nontarget organisms was 61°C. Overall, the specificity studies showed that the three probes were specific to their target organisms, which was consistent with the results of database searches.
FIG. 4.
Results of the probe specificity study. Membrane hybridization results were analyzed with a PhosphorImager and were scanned and printed with Adobe Photoshop 3.0. (a) Template. Names of individual organisms are shown in Fig. 2. (b) Probe S-*-Univ-1390-a-A-18. (c) Probe S-S-G.am-0205-a-A-19. (d) Probe S-*-G.am1-0439-a-A-19. (e) Probe S-*-G.am2-0439-a-A-19.
Characterization of environmental samples.
The probes designed in this study, as well as probes for the genus Gordona, the mycolata, and the bacterial domain (Table 1), were used to determine whether G. amarae strains were involved in foaming problems experienced in six full-scale wastewater treatment plants in Illinois, California, and Wisconsin (Table 2). Hybridization results for samples from these treatment plants are summarized in Table 3. Samples were obtained from activated sludge foam and mixed liquor from the Illinois and Wisconsin treatment plants. Because three of the California plants have high-purity oxygen-activated sludge systems, samples were taken from the return activated sludge (RAS) lines. The hybridization results for the Illinois plant revealed similar rRNA levels for all target groups for RAS and mixed liquor (data not shown), suggesting that RAS data can be used to estimate target group abundance in mixed liquor. In addition to the activated sludge samples, anaerobic digester sludge samples were obtained from the Illinois and California plants, and an anaerobic digester foam sample was taken at the Illinois plant.
TABLE 3.
Quantitative hybridization results for full-scale wastewater treatment plants
| City or state | Sample | % of total SSU rRNA
|
|||||
|---|---|---|---|---|---|---|---|
| Domain Bacteria | Mycolata | Genus Gordona | G. amarae | G. amarae group 1 | G. amarae group 2 | ||
| Urbana, Ill. | Activated sludge foam | 67.0 | 22.9 | 26.9 | 17.4 | 9.6 | 0.4 |
| Activated sludge mixed liquor | 52.2 | 3.0 | 4.7 | 2.4 | 1.7 | <0.1 | |
| Anaerobic digester foam | 87.9 | 7.8 | 11.4 | 6.2 | 3.6 | <0.1 | |
| Anaerobic digester sludge | 70.0 | 14.4 | 21.3 | 14.8 | 7.0 | 0.4 | |
| San Francisco, Calif. | RAS | 54.3 | 6.3 | 5.9 | 4.3 | 1.1 | 2.3 |
| Anaerobic digester sludge | 68.2 | 8.8 | 8.6 | 5.5 | 1.4 | 1.7 | |
| Richmond, Calif. | RAS | 68.4 | 5.9 | 6.3 | 5.3 | 2.5 | 0.3 |
| Anaerobic digester sludge | 79.3 | 9.2 | 8.5 | 8.3 | 3.3 | 0.4 | |
| Sacramento, Calif. | RAS | 63.2 | 1.8 | 0.7 | 0.2 | 0.4 | <0.1 |
| Anaerobic digester sludge | 64.7 | 5.0 | 1.3 | 0.9 | 1.1 | <0.1 | |
| Oakland Calif. | RAS | 71.9 | 2.0 | 1.7 | 0.6 | 0.7 | 0.2 |
| Anaerobic digester sludge | 84.7 | 3.7 | 3.1 | 1.2 | 1.7 | 0.6 | |
| Wisconsin | Activated sludge foam | 63.9 | 5.0 | 7.1 | 3.4 | 1.2 | <0.1 |
| Activated sludge mixed liquor | 68.8 | 4.8 | 5.3 | 2.5 | 0.8 | <0.1 | |
| Avg SD | 1.9 | 0.8 | 0.2 | 0.1 | 0.2 | 0.1 | |
The hybridization results for the Urbana, Ill., plant indicated that activated sludge foam had significantly higher rRNA levels than mixed liquor for the bacterial domain, mycolata, the genus Gordona, G. amarae, and group 1 strains. This finding supports the results of previous studies in which researchers observed a greater abundance of filamentous actinomycetes in activated sludge foam than in mixed liquor (12, 49). Gordona strains appeared to be the only mycolata representatives present in this activated sludge system since the relative rRNA levels obtained with the Gordona-specific probe were slightly higher than the levels obtained with the myclolata probe in foam and mixed liquor (Table 3). Furthermore, a significant percentage of the Gordona strains were G. amarae strains (65% for activated sludge foam, 51% for mixed liquor), the majority of which were group 1 strains (55% for activated sludge foam, 71% for mixed liquor). Group 2 strains probably were not present in this system since the hybridization response was close to or below the detection limit (Table 3). Thus, it appears that G. amarae group 1 strains were involved in the foaming problem observed in the Urbana treatment plant. However, since group 1 strains contributed only approximately one-half of the G. amarae rRNA in foam, previously uncharacterized G. amarae strains likely were present in this system.
Results obtained with samples from a foaming anaerobic digester at the Urbana treatment plant indicate that the relative rRNA levels for mycolata, the genus Gordona, and G. amarae were higher in anaerobic digester sludge than in activated sludge mixed liquor. This implies that there was a transfer of mycolata from the activated sludge basin to the anaerobic digester through waste-activated sludge feeding. These findings also suggest that rRNAs from strictly aerobic filamentous actinomycetes persisted under anaerobic conditions. The survival of significant quantities of metabolically competent Gordona (Nocardia) filaments in anaerobic digesters has previously been observed by Hernandez and Jenkins (22). In continuous-flow digestion experiments, these authors observed only 37% filament reduction after a 14-day retention time and noted that 60% of the filaments retained their ability to respire, as judged by 2-(p-iodophenyl)-3-(p-nitrophenyl)-S-phenyltetrazolium chloride reduction, after 14 days of anaerobic digestion. A discrepancy between ribosome content and metabolic activity has also been observed for a methanogenic reactor which was amended with sulfate (33); the methanogenic activity ceased immediately after sulfate was added (as indicated by a cessation of methane production), but the methanogen rRNA levels decreased at a much lower rate. Thus, in accordance with previous observations obtained with oligonucleotide probes (33, 48), our results suggest that rRNA levels may be poor indicators of reduced metabolic activity in environmental samples. However, foaming is a physical phenomenon caused by the entrapment of gas within a film of liquid and stabilized by the presence of filamentous microorganisms with hydrophobic cell walls (23). Even when filamentous actinomycetes are metabolically inactive, their presence can contribute to foam formation and stabilization. Therefore, the fact that cellular rRNA is present for considerable time periods even if organisms are metabolically inactive might make it possible to develop an early-warning system for foaming based on hybridizations with rRNA-targeted probes.
In previous studies (22, 45) workers have observed selective accumulation of mycolata strains in anaerobic digester foam, and these findings contradict our findings that mycolata rRNA is more abundant in anaerobic digester sludge than in anaerobic digester foam (Table 3). In these studies, however, the researchers used conventional and immunofluorescent stains, which are not dependent on cellular metabolism for visualization. The rRNA content of mycolata cells in anaerobic digester foam may be low enough to account for this discrepancy. This is because the partitioning of digester foam from digesting sludge may lead to longer retention times for digester foam. Increased retention times in anaerobic digesters significantly increase the degradation of Gordona filaments and reduce their viability (22). It is also possible that biogas production in the Urbana anaerobic digester resulted in a level of selective accumulation of mycolata strains in anaerobic digester foam lower than the level of selective accumulation caused by aeration in activated sludge basins. In any case, since foaming in the Urbana anaerobic digester appeared to be linked to the presence of mycolata, control strategies should take into consideration the recycling of filamentous actinomycetes; the seeding of anaerobic digesters with activated sludge foam can cause foaming problems in anaerobic digesters, and recycling of anaerobic digester supernatant to activated sludge systems may result in reseeding of the aeration basin.
The mycolata, Gordona, and G. amarae rRNA levels in the RAS of the San Francisco and Richmond plants were significantly higher than those in the mixed liquor of the Urbana plant (Table 3), suggesting that filamentous actinomycetes should also be very abundant in the activated sludge foam of these California plants. Almost all of the mycolata rRNA was Gordona rRNA, and the majority of the Gordona rRNA was G. amarae rRNA in these plants. On the other hand, the rRNA levels in the Oakland and Sacramento plants were significantly lower than those in the Urbana treatment system, suggesting that filamentous microorganisms not targeted by our probes were responsible for foaming. Gordona spp. other than G. amarae and members of genera other than the genus Gordona constituted a significant fraction of the mycolata. This further supported the observation that G. amarae was not contributing to foaming problems in the Oakland and Sacramento plants.
The combined abundance of group 1 and group 2 rRNAs approximated the abundance of G. amarae rRNA in Sacramento and Oakland samples. For the San Francisco and Richmond samples, the group probes targeted 56 to 79 and 44 to 53%, respectively, of the G. amarae strains. These results indicate that novel strains of G. amarae that are not targeted by the group probes may have been present in the San Francisco and Richmond plants. Hybridization data obtained with the group probes also demonstrated the predominance of G. amarae strains in sites from which they were isolated originally. For example, group 2 strains were more abundant in San Francisco samples, from which strain ASF3 (group 2) was originally isolated, and the group 1 strain rRNA levels were higher in the Sacramento and Richmond treatment plants, the sites of isolation of strains ASAC1 and RBI (group 1), respectively. Since some of the strains were isolated by micromanipulation techniques, these results indicate that micromanipulation can be a reliable tool for isolating predominant strains from environmental samples, as suggested by Soddell and Seviour (39).
Hybridization results obtained with samples from a milk-processing wastewater treatment plant in Wisconsin, which was experiencing severe foaming at the time of sampling, showed that the mycolata, Gordona, and G. amarae rRNA levels were only slightly higher in activated sludge foam than in mixed liquor (Table 3), suggesting that mycolata did not selectively accumulate in foam. In addition, the levels of these organisms in foam were significantly lower than the levels in the foam of the Urbana plant. The genus Gordona was found to be the predominant genus within the mycolata, but G. amarae rRNA constituted less than one-half of the Gordona rRNA. Group 1 rRNA contributed approximately one-third of the G. amarae rRNA, while group 2 strains were absent.
Since mycolata apparently were not responsible for foaming in this milk-processing wastewater treatment plant, surfactants present in the wastewater or filamentous microorganisms not targeted by our probes may have caused the foaming problem. For example, it is possible that “Microthrix parvicella,” a common constituent of activated sludge foam (8, 12, 30), was involved in foaming in this plant. Using the recently published SSU rRNA sequence of “Microthrix parvicella” (10), we determined that none of our probes target this organism.
FISH.
The results of membrane hybridizations indicate the contributions of organisms involved in filamentous foaming to total activity, as reflected by rRNA levels. However, it is important to develop a method that relates the abundance of filamentous microorganisms (not only rRNA levels) to the severity of foaming. FISH has better potential to accomplish this goal. Therefore, we addressed problems with permeabilizing cell walls of gram-positive bacteria for probe accessibility in a previous study (17). We demonstrated that fixation with 4% paraformaldehyde for very short time periods (1 min) rendered most mycolata permeable for labeled probes (17). Schuppler et al. (37b) recently demonstrated that ethanol fixation combined with mutanolysin incubation after dehydration also resulted in excellent FISH results for mycolata. We also developed a method for quantifying members of the genus Gordona on a mass basis in foaming activated sludge systems and anaerobic digesters by using FISH (17, 18). Additional studies are necessary to relate this and other quantitative approaches to foaming potential.
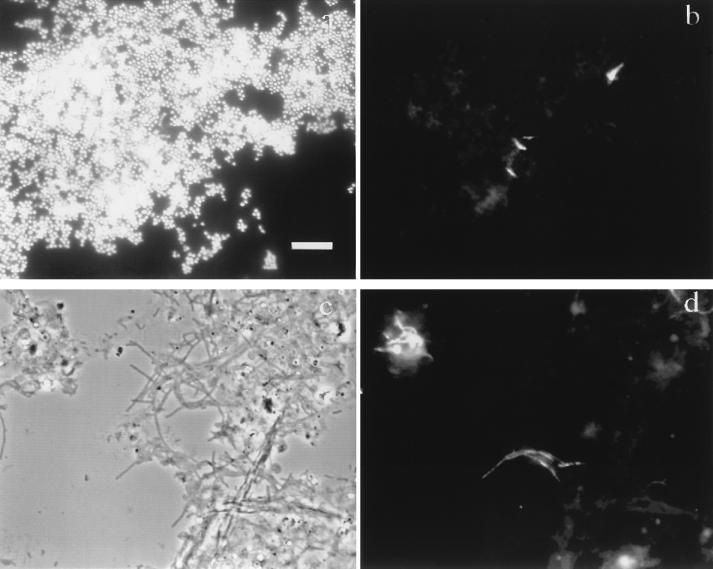
To demonstrate the potential of FISH for such studies, we used a TRITC-labeled version of the G. amarae-specific probe S-S-G.am-0205-a-A-19. The optimum formamide concentration in the hybridization buffer was determined to be 30%, and the optimum NaCl concentration in the corresponding wash solution was found to be 80 mM. Figures 5a and b show the specificity of the probe when it was hybridized with a mixture of cultures of R. rhodochrous and G. amarae SE-102. Figure 5a shows the mixed culture stained with DAPI, while Fig. 5b shows the same image field viewed with a TRITC filter set. A comparison of Fig. 5a and b indicates that G. amarae can be distinguished clearly from nontarget cells at the hybridization stringency used. Figures 5c and d show the application of FISH to a RAS sample from the San Francisco treatment plant. Figure 5c shows the phase-contrast image, while Fig. 5d shows the same image field when a TRITC filter set was used. These images demonstrate the ability of FISH to distinguish target cells inside and extending from activated sludge flocs without regard for morphology. The formation of G. amarae filamentous clusters was observed, which confirmed previous observations (17). In addition, we observed very low levels of nonspecific probe binding and activated sludge floc autofluorescence. This is particularly encouraging for future FISH studies with this probe.
FIG. 5.
Phase-contrast and epifluorescence images after FISH of mixed cultures and activated sludge. Bar = 10 μm. (a and b) Mixture of cultures of R. rhodochrous and G. amarae SE-102 hybridized with TRITC-labeled probe S-S-G.am-0205-a-A-19 and dual stained with DAPI. Panels a and b show the same image field, as viewed by epifluorescence microscopy, prepared with the DAPI and TRITC filter sets, respectively. (c and d) RAS sample from San Francisco Southeast Water Pollution Control Plant hybridized with probe S-S-G.am-0205-a-A-19 labeled with TRITC. Panels c and d show the same image field, as viewed by phase-contrast microscopy and epifluorescence microscopy, respectively.
Conclusions.
In this study, we developed oligonucleotide probes for G. amarae and two subgroups within this species. The probes allowed identification and quantification of G. amarae strains often implicated in foaming activated sludge and anaerobic digester systems by membrane hybridization and FISH. Use of a nested set of probes permitted a quantitative assessment of G. amarae strains in foaming samples, which is necessary when the importance of this species in foaming is evaluated. The quantities and types of G. amarae rRNA were found to vary in different treatment plants. Future studies on the analysis of foaming occurrences should benefit from the use of phylogenetic probes specific for foam-related microorganisms, such as the probes designed in this study. In particular, correlating FISH and membrane hybridization results to changes in foaming potential could provide threshold values which could be used to predict foaming incidents.
ACKNOWLEDGMENTS
We thank the following personnel of the wastewater treatment plants for providing samples: Tim Bachman, Urbana-Champaign Sanitary District; Paul Pitt, City and County of San Francisco; David Fretias, Oakland East Bay Municipal Utilities District; William Louis, City of Sacramento Regional Wastewater Treatment Plant; and the operators of the Richmond Wastewater Treatment Plant.
This research was supported by grant BES 9410476 from the U.S. National Science Foundation.
REFERENCES
- 1.Alm, E. W., and D. Stahl. Evaluation of key parameters for extraction of native rRNA from different environmental matrices. Submitted for publication.
- 2.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Amann R, Ludwig W, Schleifer K-H. Identification and in situ detection of individual bacterial cells. FEMS Microbiol Lett. 1992;100:45–50. [Google Scholar]
- 5.Amann R, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awong J, Bitton G, Koopman B. ATP, oxygen uptake rate and INT-dehydrogenase activity of actinomycete foams. Water Res. 1985;19:917–921. [Google Scholar]
- 8.Blackall L, Harbers A, Greenfield P, Hayward A. Actinomycete scum problems in Australian activated sludge plants. Water Sci Technol. 1988;20:23–29. [Google Scholar]
- 9.Blackall L, Harbers A, Greenfield P, Hayward A. Foaming in activated sludge plants: a survey in Queensland, Australia and an evaluation of some control strategies. Water Res. 1991;25:313–317. [Google Scholar]
- 10.Blackall L, Seviour E M, Cunningham M A, Seviour R J, Hugenholz P. “Microthrix parvicella” is a novel, deep branching member of the actinomycetes subphylum. Syst Appl Microbiol. 1994;17:513–518. [Google Scholar]
- 11.Blackall L, Seviour E, Bradford D, Stratton H, Cunningham M, Hugenholz P, Seviour R. Towards understanding the taxonomy of some of the filamentous bacteria causing bulking and foaming in activated sludge plants. Water Sci Technol. 1996;34:137–144. [Google Scholar]
- 12.Blackbeard J R, Ekama G A, Marais G v R. A survey of bulking and foaming in activated sludge plants in South Africa. Water Pollut Control. 1986;85:90. [Google Scholar]
- 13.Chun J, Blackall L, Kang S, Hah Y, Goodfellow M. A proposal to reclassify Nocardia pinensis Blackall et al. as Skermania piniformis gen. nov., comb. nov. Int J Syst Bacteriol. 1997;47:127–131. doi: 10.1099/00207713-47-1-127. [DOI] [PubMed] [Google Scholar]
- 14.Chun J, Kang S-O, Hah Y, Goodfellow M. Phylogeny of mycolic acid-containing actinomycetes. J Ind Microbiol. 1996;17:205–213. [Google Scholar]
- 15.Clark A G, Whittam T S. Sequencing errors and molecular evolutionary analysis. Mol Biol Evol. 1992;9:744–752. doi: 10.1093/oxfordjournals.molbev.a040756. [DOI] [PubMed] [Google Scholar]
- 16.Clayton R A, Sutton G, Hinkle P S J, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syt Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 17.de los Reyes F, Ritter W, Raskin L. Group-specific small-subunit rRNA hybridization probes to characterize filamentous foaming in activated sludge systems. Appl Environ Microbiol. 1997;63:1107–1117. doi: 10.1128/aem.63.3.1107-1117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de los Reyes, F. L., D. B. Oerther, M. F. de los Reyes, M. Hernandez, and L. Raskin. Characterization of filamentous foaming in activated sludge systems using oligonucleotide hybridization probes and antibody probes. Water Sci. Technol., in press.
- 19.Dorsch M, Stackebrandt E. Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J Microbiol Methods. 1992;16:271–279. [Google Scholar]
- 20.Eikelboom D, van Buijsen H. Microscopic sludge investigation manual. 2nd ed. Delft, The Netherlands: TNO Research Institute of Environmental Hygiene; 1983. [Google Scholar]
- 21.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez M, Jenkins D. The fate of Nocardia in anaerobic digestion. Water Environ Res. 1994;66:828–835. [Google Scholar]
- 23.Jenkins D, Richard M, Daigger G T. Manual on the causes and control of activated sludge bulking and foaming. Chelsea, Mich: Lewis Publishers, Inc.; 1993. [Google Scholar]
- 24.Kampfer P. Detection and cultivation of filamentous bacteria from activated sludge. FEMS Microbiol Ecol. 1997;23:169–181. [Google Scholar]
- 25.Lechevalier M P, Lechevalier H A. Nocardia amarae sp. nov., an actinomycete common in foaming activated sludge. Int J Syst Bacteriol. 1974;24:278–288. [Google Scholar]
- 26.Maidak B, Larsen N, McCaughey M J, Overbeek O, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nubel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersson B, Leitner T, Ronaghi M, Bolske G, Uhlen M, Johansson K. Phylogeny of the Mycoplasma mycoides cluster as determined by sequence analysis of the 16S rRNA genes from the two rRNA operons. J Bacteriol. 1996;178:4131–4142. doi: 10.1128/jb.178.14.4131-4142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitt P, Jenkins D. Causes and control of Nocardia in activated sludge. Res J Water Pollut Control Fed. 1990;62:143–150. [Google Scholar]
- 30.Pujol R, Duchene P, Schetrite S, Canler J P. Biological foams in activated sludge plants: characterization and situation. Water Res Rev. 1991;25:1399–1404. [Google Scholar]
- 31.Rainey F, Ward-Rainey N, Janssen P, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308 contains multiple 16S rRNA genes with heterogenous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 32.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raskin L, Rittmann B E, Stahl D A. Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62:3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raskin L, Capman W C, Sharp R, Poulsen L K, Stahl D A. Molecular ecology of gastrointestinal ecosystems. In: Mackie R I, White B A, Isaacson R E, editors. Gostrointestinal microbiology. 2. Gastrointestinal microbiology and host interactions. New York, N.Y: Chapman and Hall; 1997. pp. 243–298. [Google Scholar]
- 35.Ruimy R, Boiron P, Biovin V, Christen R. A phylogeny of the genus Nocardia deduced from the analysis of small-subunit ribosomal DNA sequences, including transfer of Nocardia amarae to the genus Gordona as Gordona amarae comb. nov. FEMS Microbiol Lett. 1994;123:261–268. doi: 10.1111/j.1574-6968.1994.tb07234.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schuppler M, Mertens F, Schon G, Gobel U B. Molecular characterization of nocardioform actinomycetes in activated sludge by 16S rRNA analysis. Microbiology. 1995;141:513–521. doi: 10.1099/13500872-141-2-513. [DOI] [PubMed] [Google Scholar]
- 37b.Schuppler M, Wagner M, Schon G, Gobel U B. In situ identification of nocardioform actinomycetes in activated sludge using fluorescent rRNA-targeted oligonucleotide probes. Microbiology. 1998;144:249–259. doi: 10.1099/00221287-144-1-249. [DOI] [PubMed] [Google Scholar]
- 38.Seviour E M, Williams C J, Seviour R J, Sodell J A, Lindrea K C. A survey of filamentous bacterial populations from foaming activated sludge plants in eastern states of Australia. Water Res. 1990;24:493. [Google Scholar]
- 39.Soddell J A, Seviour R J. A review: microbiology of foaming in activated sludge plants. J Appl Bacteriol. 1990;69:145–176. [Google Scholar]
- 40.Stackebrandt E, Rainey F, Ward-Rainey N. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- 41.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons, Ltd.; 1991. pp. 205–248. [Google Scholar]
- 43.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads. A taxonomic study. J Gen Microbiol. 1966;43:159–277. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 44.Stratton H, Seviour R, Soddell J, Blackall L, Muir D. The opportunistic pathogen Nocardia farcinica is a foam-producing bacterium in activated sludge plants. Lett Appl Microbiol. 1996;22:342–346. doi: 10.1111/j.1472-765x.1996.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 45.van Niekerk A, Kawahigashi J, Reichlin D, Malea A, Jenkins D. Foaming in anaerobic digesters—a survey and laboratory investigation. Res J Water Pollut Control Fed. 1987;59:249–253. [Google Scholar]
- 46.von Weber J. The nature and location of Nocardia amarae cell components that have the ability to stabilize foams in activated sludge wastewater treatment plants. Ph.D. thesis. Davis: University of California at Davis; 1992. [Google Scholar]
- 47.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner M, Rath G, Amann R, Koops H, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]
- 49.Wheeler D, Rule A. The role of Nocardia in the foaming of activated sludge: laboratory studies. Savannah, Ga: Georgia Water and Pollution Control Association; 1980. [Google Scholar]
- 50.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]